Abstract
In eukaryotic cells, the cell cycle checkpoint proteins Rad9, Rad1, and Hus1 form the 9-1-1 complex which is structurally similar to the proliferating cell nuclear antigen (PCNA) sliding clamp. hMSH2/hMSH6 (hMutSα) and hMSH2/hMSH3 (hMutSβ) are the mismatch recognition factors of the mismatch repair pathway. hMutSα has been shown to physically and functionally interact with PCNA. Moreover, DNA methylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) treatment induces the G2/M cell cycle arrest that is dependent on the presence of hMutSα and hMutLα. In this study, we show that each subunit of the human 9-1-1 complex physically interacts with hMSH2, hMSH3, and hMSH6. The 9-1-1 complex from both humans and Schizosaccharomyces pombe can stimulate hMutSα binding with G/T-containing DNA. Rad9, Rad1, and Hus1 individual subunits can also stimulate the DNA binding activity of hMutSα. Human Rad9 and hMSH6 colocalize to nuclear foci of HeLa cells after exposure to MNNG. However, Rad9 does not form foci in MSH6 defective cells following MNNG treatment. In Rad9 knockdown untreated cells, the majority of the MSH6 is in cytoplasm. Following MNNG treatment, Rad9 knockdown cells has abnormal nuclear morphology and MSH6 is distributed around nuclear envelop. Our findings suggest that the 9-1-1 complex is a component of the mismatch repair involved in MNNG-induced damage response.
Keywords: DNA repair, Mismatch repair, MutS homologs, Cell cycle checkpoint, Rad9/Rad1/Hus1, Protein-protein interaction
1. Introduction
All organisms replicate their genome with high fidelity and repair their damaged genomic DNA caused by endogenous and environmental agents. Eukaryotes have evolved complex regulatory mechanisms that coordinate DNA replication and DNA repair with cell cycle regulation [1,2]. Upon replication block and DNA damage, the signal is recognized by cell cycle checkpoint sensors and then transferred to transducers and effectors. This leads to cell cycle arrest and permits time for DNA repair or, when repair cannot be completed, triggers cell apoptosis [3,4]. Two protein kinases, ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related protein) are the central components of the DNA damage response pathway [4]. Upon activation, these protein kinases phosphorylate downstream substrates to enforce DNA repair, transcriptional activation, cell cycle arrest and/or apoptosis [1,5]. Rad9, Rad1, and Hus1 checkpoint sensors form a heterotrimeric complex (the 9-1-1 complex) and are required to activate ATR or ATM and to coordinate DNA damage response [6–8]. Recently, the structure of the 9-1-1 complex has been determined [9–11] and shown to exhibit structural similarity with the sliding clamp proliferating nuclear antigen (PCNA) [12–14]. While PCNA is loaded onto DNA by the clamp loader replication factor C (RFC1-5) complex, the 9-1-1 complex is loaded onto DNA by Rad17-RFC2-5 [15–17]. It has been shown that the 9-1-1 complex interacts with and stimulates the enzymes involved in base excision repair (BER) including DNA glycosylases [18–21], AP endonuclease (APE1) [22], DNA polymerase β[23], flap endonuclease (FEN1) [24,25], replication protein A (RPA) [26], and DNA ligase 1 [27,28]. These findings indicate a new role for the 9-1-1 complex: it not only serves as a damage sensor to activate checkpoint control, but it also serves as a BER component.
Mismatch repair (MMR) enhances replication fidelity by correcting replicative errors that escape the proofreading of DNA polymerases (reviewed by[29–33]). Germline mutations in human mismatch repair genes can lead to genetic mutations and microsatellite instability in hereditary nonpolyposis colon cancer (HNPCC) and also other forms of cancer [34–36]. The reconstitution of human MMR has been established with a DNA substrate containing a single G/T mismatch (or an insertion/deletion loop), a strand break and purified proteins: MutSα (or MutSβ), MutLα, replication protein A (RPA), high-mobility group box 1 (HMGB1), exonuclease 1 (EXO1), PCNA, RFC, DNA polymerase δ and DNA ligase I [37,38]. MutSα (MSH2/MSH6) and MutSβ (MSH2/MSH3) are mismatch recognition factors [29]. MutSα recognizes base/base mismatches and short insertion/deletion loops, while MutSβ recognizes larger insertion/deletion loops. Human MutLα (MLH1/PMS2) possesses ATPase and endonuclease activity, which introduces single-strand breaks 5’ and 3’ to the mismatch, and thus generates new entry points for the exonuclease EXOI to degrade the strand containing the mismatch [39].
MMR stimulates checkpoint and cell death responses to DNA damage suggested by the resistance of MMR-defective tumor cells to several chemotherapeutic agents [40–43]. MMR-dependent cytotoxic response may result from futile repair or a direct induction of checkpoint response. Treatment of cells with SN1-type alkylating reagents, such as N-methyl-N’-nitro-N-nitrosoguanidine (MNNG), produces the cytotoxic O6-methyl-guanine (O6-meG) [44]. The O6-meG lesion can be repaired in a saturated manner by the suicide enzyme methylguanine methyltransferase (MGMT) [44]. However, MGMT is inactivated in most solid tumor cells, and the persistence of O6-meG causes cytotoxicity [45]. Thymine is incorporated with O6-meG on the template during DNA replication. O6-meG/T recognized by MutSα [46] induces ATR foci formation and cell cycle arrest until the second G2 phase after exposure [42,47–49]. MMR enzymes are proposed to act as molecular sensors/adaptors for cell cycle checkpoint proteins [42,43,50–52]. Human MSH2 interacts with the ATR in response to alkylating agents [53]. Brown et al. [54] have shown that MSH2 interacts with Chk2 checkpoint effecter and that MLH1 associates with ATM. In addition, MSH2, MSH6, MLH1 have been shown to be associated with a large complex such as BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, ATM, RAD50, and RFC [55]. Recently, Yoshioka et al. [56] have shown that ATR, but not RPA, is preferentially recruited to O6-meG/T mismatches in a MutSα- and MutLα-dependent manner. Their results provide direct evidence that MutSα and MutLα act as adaptors for checkpoint sensors.
The facts that MutSα physically and functionally interacts with PCNA [57–59], and that PCNA and the 9-1-1 complex share structural similarity, raise the possibility that hMutSα may interact with the 9-1-1 complex. He et al, [60] have reported that Rad9 plays an important role in mismatch repair through physical interaction with MLH1. In this study, we show that hMutSα and hMutSβ physically interact with every subunit of the 9-1-1 complex. The 9-1-1 complex and each subunit are able to stimulate hMutSα binding with the G/T mismatch substrate. Moreover, following treatment of HeLa cells with MNNG, hRad9 forms foci colocalized with hMSH6. The ability of hRad9 and hMSH6 foci formation depends on the presence of each other. Our results suggest that a mutual relationship between MutSα and Rad9-Rad1-Hus1 in response to cytotoxic effect of methylating agents.
2. Materials and methods
2.1. Cell culture and treatments
Human HeLaS3, HCT15, and LoVo cell lines were purchased from American Type Cell Culture (Manassas, VA, USA). The HeLa and LoVo cells were maintained in Ham’s F12K medium (Invitrogen) supplemented with 2 mM L-glutamine 1.5 g/L sodium bicarbonate, 1% penicillin-streptomycin, and 10% fetal bovine serum at 37°C and 5% CO2. HCT15 cells were grown in RPMI medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2. For immunofluorescence studies, 1×105 cells were transferred to each chamber of Lab-Tek chamber slides (Nalge Nunc International, Rochester, NY) and grown overnight. Then the cells were incubated with 25 µM O6-Benzylguanine (O6-BG) (Sigma-Aldrich, St. Louse, MO), a competitive inhibitor of MGMT, for 2 h. The cells were than treated with 10 µM MNNG (VWR, West Chester, PA) and 25 µM O6-BG or DMSO in serum free medium for 1 h and recovered in complete medium for 24 h.
Transfection of small interfering RNA (siRNA) was carried out using Ribojuice siRNA transfection reagent (Novagen) according to the manufacturer's instructions. Hela cells (1×105) were transfected with 50 pmol of a Silencer-validated Rad9 siRNA (ID 5114, Ambion) or a scrambled control siRNA (Cat. No 1022564, Qiagen) in a total volume of 1 mL in twelve-well tissue culture plates. Untransfected cells were included to monitor the cytotoxicity of the transfection reagents. Cells were harvested after 72 hr. Whole cell extract was quantized by Bradford and Rad9 levels were analyzed by western blotting using the anti-Rad9 antibody (Stratagene). The cells with scrambled RNA and siRNA for Rad9 were subjected to treatment by O6-BG and MNNG as described above.
2.2. Proteins and antibodies
Human MutSα, MutSβ, Rad9, Rad1, Hus1, and the 9-1-1 complex were expressed in baculovirus/insect Sf9 system and purified as described [20,61]. S. pombe 9-1-1 complex was purified from E. coli as described [19]. Anti-hRad9 is from Imgenex (San Diego, CA). Anti-hMSH2, anti-hMSH3, anti-hMSH6 used in Western blotting are from BD Biosciences (San Diego, CA). anti-hMSH6 used in immunofluorencence staining (sc1243), anti-hHus1, and anti-GST are from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 594 goat anti-rabbit and Alexa Fluro 488 goat anti-mouse IgG antibodies are from Invitrogen (Carlsbad, CA).
2.3. HeLa whole cell extracts
HeLaS3 cells (3×107) were resuspended in 0.5 ml of buffer containing 50 mM potassium phosphate, pH 7.4, 50 mM KCl, 1 mM dithiothreitol, 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol. An equal volume of 0.1 mm glass beads was added to the cell suspension. The cells were disrupted by vigorous vortexing for 10 s at 4°C and cooled on ice for 20 s. This cycle was repeated 10 times. The mixture was then centrifuged at 12,000 × g for 15 min and the supernatant was saved. The protein concentration was determined by Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, CA).
2.4. GST pull-down assays
Glutathione-S-transferase (GST) fusion proteins of hHus1, hRad1, and hRad9 were immobilized on glutathione-sepharose 4B as described [19]. Purified hMutSα (400 ng) or HeLa cell extracts (750 µg) were incubated separately with 300 ng immobilized GST-hHusI, GST-hRad1, and GST-hRad9 in 100 µl reactions for 3.5 h at 4°C [19]. After centrifugation at 1000×g, the supernatants were saved. The pellets were washed five times in 800 µl buffer G (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 2 mM EDTA) with 0.2% Nonidet P-40. The pellets and supernatants (10 µl) were fractionated on an 8% SDS-polyacrylamide gel and Western blot analysis was performed with anti-hMSH2, anti-hMSH3 and anti-hMSH6 antibodies.
2.5. Far-Western analysis
Ten pmol of recombinant hMSH2/hMSH6 (hMutSα) and hMSH2/hMSH3 (hMutSβ) were separated on 8% SDS- polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked with 5% non-fat milk in phosphate-buffered saline for 1 h and then incubated with E. coli extracts containing GST-hHus1, GST-hRad1, GST-hRad9, or GST alone [19] at 4 °C overnight. After extensive washing with blocking solution (5% non-fat dry milk and Tween-20 in PBS), the membrane was incubated with anti-GST and subjected to Western blot analysis.
2.6. Gel shift assay
The DNA substrates were 87-mer homoduplex (G:C) or heteroduplex containing a G/T mismatch, (G denotes the guanine in G:C or G/T): 5’-CCA GAT GAC GTT GTG ACT ACC TGT AGC TAC TGC GTG CGA TTG GAT TAG CAG AGG CAT GCA ATG TCC TAA GAC TAG CCA ATA ATC CAG-3’ and its complementary strand (81-mer). The annealed duplex with a 6-nucleotide overhang at the 5’ end of the G-strand were labeled at the 3’ end with [α-32P]dATP as described [62]. The assays were performed with 5 or 10 nM purified hMutSα and various amounts of purified h9-1-1 complex, Sp9-1-1 complex, hHus1, hRad1, or hRad9 as denoted in the figure legend in 20 µl volume containing 5 fmol DNA substrate, 100 mM NaCl, 25 mM HEPES-NaOH (pH8.0), 1 mM MgCl2, 0.1 mM ADP, 1 mM dethiothreitol, 0.1 mM EDTA, 10% glycerol, 0.075 mg/ml bovin serum albumin (BSA) and 150 ng of 200 base pair homoduplex competitor [63]. The reactions were incubated at 37°C for 20 min and separated on a 5% polyacrylamide gel containing 2.5% glycerol. The electrophoresis was carried out at 4°C with 34 mA current in 50 mM Tris-borate buffer. The gels were dried and then quantified using ImageQuant (GE Health, Waukesha, WI).
2.7. Immunofluorencence studies
HeLaS3 cells grown on Lab-Tek chamber slides were treated with alkylating agent MNNG in a similar manner as described above. The cells were fixed with 4% formaldehyde in PBS for 10 min followed by permeabilization in 0.5% Triton X-100 in PBS for 10 min. The slides were blocked with 5 µg/ml of BSA in PBS for 16 h at 4°C and incubated with anti-hMSH6 (at 1:250 dilution) and anti-hRad9 (at 1:400 dilution) in blocking buffer for 2 h at 25°C. After three PBS washes, the slides were incubated with Alexa Fluor 594 goat anti-rabbit and Alexa Fluro 488 goat anti-mouse IgG antibodies (Invitrogen) at a 1:250 dilution in PBS for 1 h at 25°C. The cells were then washed three times in PBS. Nuclear DNA was counterstained with 4’,6’-diamidineno-a-phenylindole (DAPI) (Vector Laboratories, Inc., Burlingame, CA). Images were captured with a Zeiss LSM510 META laser scanning confocal microscope.
3. Results
3.1. Physical Interaction between human MutS homologs and the 9-1-1 complex
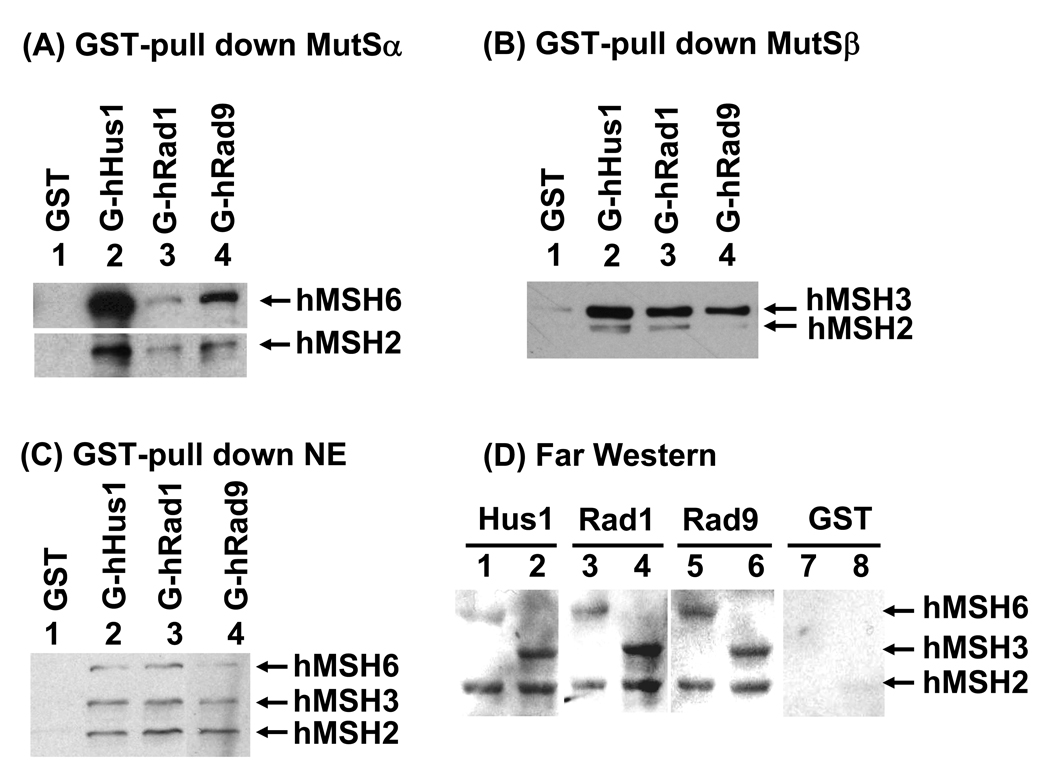
It has been shown that PCNA interacts and colocalizes with MutSα at replication foci [57–59]. Because the structure of the 9-1-1 complex [9–11] is similar to PCNA sliding clamp [64,65], we tested whether there is a similar interaction between hMutSα and h9-1-1 complex. Equal amounts of immobilized GST-hRad9, GST-hRad1, and GST-hHus1 were incubated with purified hMutSα, hMutSβ, or HeLa cell extracts. As shown in Fig. 1A, each subunit of the human 9-1-1 complex physically interacted with hMSH2/hMSH6. A similar GST pull-down assay with purified hMutSβ was also performed. hMSH2/hMSH3 complex interacted with hHus1, hRad1, and hRad9 (Fig. 1B). The different signal strength of hMSH2, MSH3, and hMSH6 in Fig. 1A and 1B is due to different affinities with their antibodies or the reactivity of Western blotting. We then performed the GST pull-down experiments with cell extracts. As shown in Fig. 1C, GST-tagged hRad9, hRad1, and hHus1 could pull down hMSH2, hMSH6, and hMSH3 from HeLa cell extracts. hMutSα and hMutSβ in HeLa cell extracts bind hHus1, hRad1 and hRad9 at similar levels (Fig. 1C). Therefore, each subunit of the h9-1-1 complex can interact with both hMSH2/hMSH6 and hMSH2/hMSH3 complexes.
Fig. 1.
Physical interactions between human 9-1-1 subunits and hMutSα and hMutSβ. (A) Pull-down of purified hMutSα by immobilized GST-hHus1, GST-hRad1 and GST-hRad9. hMutSα (450 ng) purified from baculovirus expression system was incubated with 300 ng of GST-hHus1, GST-hRad1, GST-hRad9 or GST alone immobilized on glutathione Sepharose 4B. The pellets were fractionated by 8% SDS-polyacrylamide gel followed by Western blot analysis with antibodies against hMSH2 and hMSH6. The exposure time for the blot with hMSH6 antibody was different from the one with hMSH2 antibody. (B) Pull-down of purified hMutSβ by immobilized GST-hHus1, GST-hRad1 and GST-hRad9. The procedures are similar as described in (A) except that hMutSβ (400 ng) was used and Western blot analysis was performed with antibodies against hMSH2 and hMSH3. (C) Pull-down of hMutSα and hMutSβ from HeLa cell extracts by immobilized GST-hHus1, GST-hRad1 and GST-hRad9. Similar experiments were performed as those in (A) but with 750 µg HeLa cell extracts instead of purified hMutSα and Western blot analysis was performed with antibodies against hMSH2, hMSH3 and hMSH6. (D) Far-Western analysis. Purified hMutSα (hMSH2/hMSH6) (odd lanes) and hMutSβ (hMSH2/hMSH3) (even lanes) (10 pmol each) were separated by 8% SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane. The membrane was incubated with E. coli extracts containing hHus1, hRad1, hRad9, or GST as indicated. Subsequent Western blot analysis was performed with antibodies against GST.
Next, we used Far-Western analysis to determine which subunit(s) of hMSH2/hMSH6 and hMSH2/hMSH3 interacts with the 9-1-1 complex. The individual subunits of hMutSα and hMutSβ were separated on SDS-polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was then incubated with E. coli extracts containing GST-hHus1, GST-hRad1, GST-hRad9, or GST alone [19]. Western blotting was carried out to detect GST-fusions or GST to the membrane through interaction with each subunit. The results showed that each subunit of the 9-1-1 complex interacted with hMSH2, hMSH3, and hMSH6 (Fig. 1D, lanes 1–6). Extracts containing GST alone did not bind hMSH2, hMSH3, and hMSH6 on membrane (Fig. 1D, lanes 7 and 8). Coomassie blue staining of the membrane indicated that similar amounts of hMSH2 and hMSH6 in hMutSα, and similar amounts of hMSH2 and hMSH3 in hMutSβ were immobilized onto the membrane (data not shown). This result is in contrast to the finding that PCNA interacts with hMSH2/hMSH6 and hMSH2/hMSH3 via the hMSH6 and hMSH3 subunits, respectively [57,59].
3.2. The 9-1-1 complex and each subunit stimulate hMutSα binding towards G/T mismatch
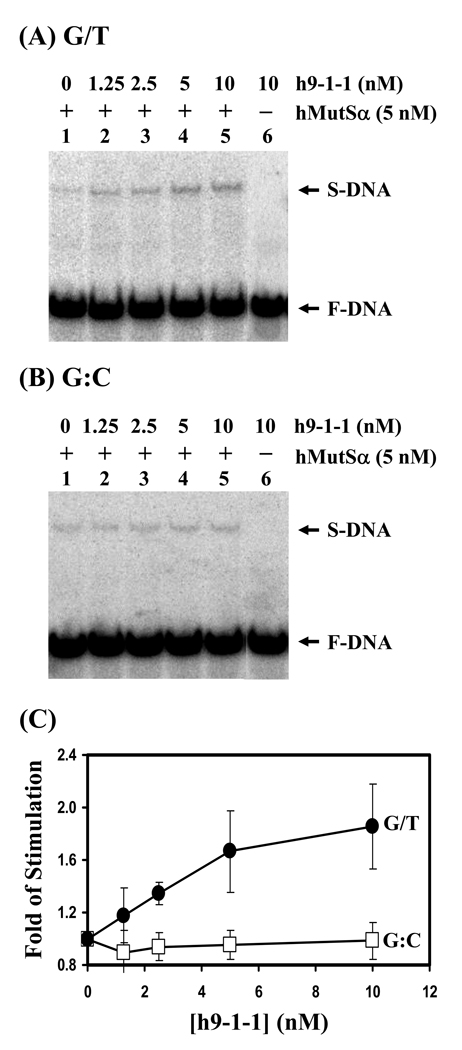
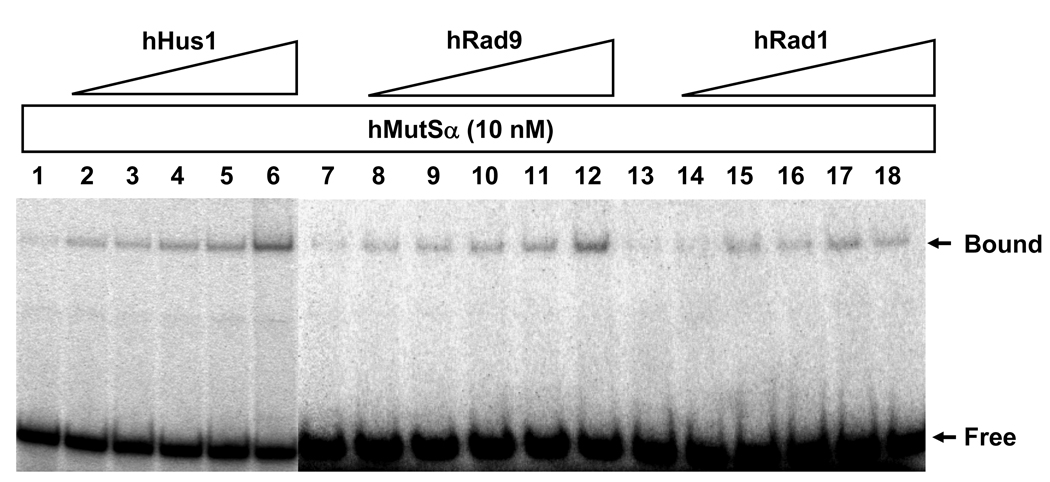
In order to check whether there is functional relevance to the interaction between h9-1-1 and hMutSα, we analyzed the effect of the h9-1-1 complex on hMutSα binding activity with an oligonucleotide duplex containing either a G/T mismatch or a normal G:C base pair. The h9-1-1 complex did not form a complex with the oligonucleotide duplexes (Fig. 2A and B, lanes 6). With increasing amounts of h9-1-1 complex (from 1.25 nM to 10 nM), the formation of hMutSα specific complex with G/T but not with G:C substrate was increased correspondingly. A two-fold excess of h9-1-1 complex (10 nM) could stimulate the hMutSα (5 nM) binding affinity to G/T mismatch by approximately twofold (Fig. 2C). Because MutSα could bind to each subunit of the 9-1-1 complex, we tested whether they could stimulate MutSα binding to mismatches. As shown in Fig. 3, hHus1, hRad1, and hRad9 at higher than 2-fold molar excess could also stimulate the G/T binding activity of hMSH2/hMSH6. hHus1, hRad1, and hRad9 at 6-fold molar excess could stimulate the binding ability of hMutSα to G/T mismatches by 3.8, 2.7, and 1.8-fold, respectively.
Fig. 2.
Human 9-1-1 complex stimulates hMutSα DNA binding activity. (A) hMutSα binding with G/T-containing DNA was stimulated by h9-1-1 complex. Lane 1, DNA substrates were incubated with 5 nM purified hMutSα. Lanes 2-5, reactions are similar to lane 1 but with increasing amounts of h9-1-1 complex (1.25, 2.5, 5, and 10 nM, respectively). Lane 6, DNA substrates were incubated with 10 nM h9-1-1 complex only. The arrows mark the free DNA substrate (F-DNA) and hMutSα-DNA complex (S-DNA). (B) hMutSα binding with homoduplex was not affected by h9-1-1 complex. The binding reactions were similar with those in (A) but with homoduplex (G:C) substrates. (C) Quantitative analyses of fold stimulation of h9-1-1 complex on hMutSα binding activity with G/T-containing DNA (closed circles) and homoduplex (opened squares) from three experiments. The fold of stimulation was calculated by dividing the percentage of DNA-protein complex in the presence of the 9-1-1 complex by the percentage of bound DNA in the absence of the 9-1-1 complex. The error bars reported are the standard deviations of the averages.
Fig. 3.
hMutSα binding activity with G/T-DNA can be stimulated by hHus1, hRad9, and hRad1. Reactions are similar to Fig. 2 but with 10 nM purified hMutSα and increasing amounts of hHus1, Rad9, and Rad1 (15.6, 31.2, 62.5, 125, and 250 nM, respectively). At 6-fold molar excess over hMutSα, hHus1, hRad1, and hRad9 could stimulate the binding ability of hMutSα to G/T mismatches by 3.8, 2.7, and 1.8-fold, respectively.
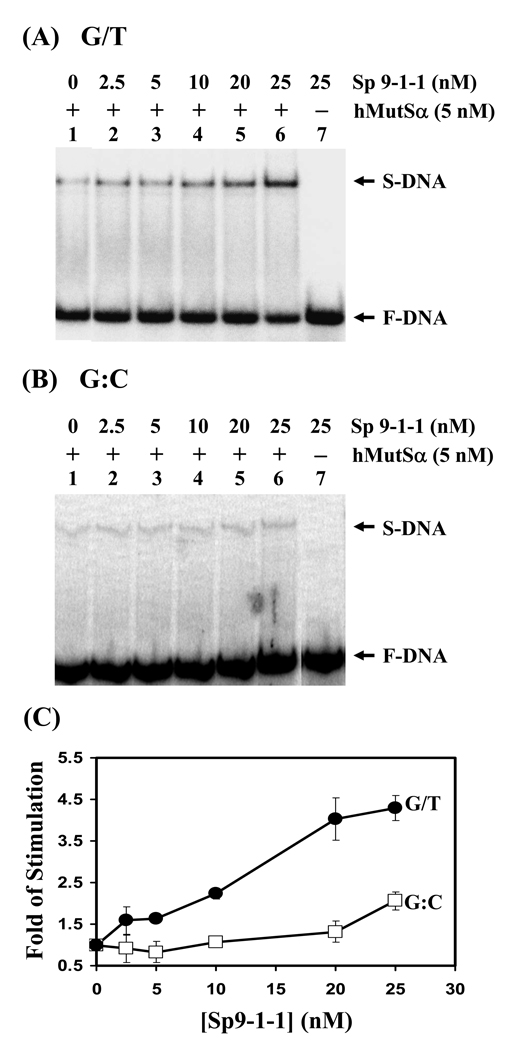
Both the MutSα and the 9-1-1 complex are conserved across species. Therefore, we tested whether the 9-1-1 complex from Schizosaccharomyces pombe (Sp9-1-1) could stimulate the hMutSα binding ability. As shown in Fig. 4A, the Sp9-1-1 could also stimulate the binding activity of hMutSα to G/T mismatches. Interestingly, the 9-1-1 complex of human and S. pombe stimulated hMutSα to a similar extent. The hMutSα binding towards homoduplex could be slightly stimulated by Sp9-1-1 at higher concentrations of Sp9-1-1 (Fig. 4B). Sp9-1-1 complex at 5-fold molar excess could stimulate the binding ability of hMutSα to G/T mismatches by 4.5-fold, but only by two-fold to homoduplex (Fig. 4C). In conclusion, the G/T binding activity of hMSH2/hMSH6 can be stimulated by Hus1, Rad1, Rad9, and the 9-1-1 complex.
Fig. 4.
S. pombe 9-1-1 complex can stimulate hMutSα binding activity. (A) hMutSα binding activity with G/T-containing DNA was stimulated by Sp9-1-1 complex. Reactions are similar to Fig. 2. Lane 1, DNA substrates were incubated with 5 nM purified hMutSα. Lanes 2-6, reactions are similar to lane 1 but with increasing amounts of Sp9-1-1 complex (2.5, 5, 10, 20, and 25 nM, respectively). Lane 7, DNA substrates were incubated with 25 nM Sp9-1-1 complex only. (B) hMutSα binding activity with homoduplex was stimulated by Sp9-1-1 complex to a less extent. The experiments were similar with those in (A) but with homoduplex (G:C) substrates. (C) Quantitative analyses of fold stimulation of Sp9-1-1 complex on hMutSα binding activity with G/T-containing DNA (closed circles) and homoduplex (opened squares) from three experiments. The fold of stimulation was calculated by dividing the percentage of DNA-protein complex in the presence of the 9-1-1 complex by the percentage of bound DNA in the absence of the 9-1-1 complex. The error bars reported are the standard deviations of the averages.
3.3. Human Rad9 and hMSH6 colocalize to nuclear foci following MNNG treatment
Cellular response to MNNG depends on cell types and treatment doses. Cells or mice with defective MGMT are more sensitive to MNNG [66,67]. MNNG (0.2 µM) treatment can induce G2/M cell cycle arrest in 293T Lα+ cells whose MGMT gene is transcriptionally silenced by cytosine methylation [68] in a MutSα-dependent manner [42]. However, higher doses of MNNG (>15 µM) activate DNA damage signaling cascades in a MMR-independent manner in 293T Lα+ cells [69]. We did not observe a cell density decrease and cell enlargement of HeLaS3 (MGMT proficient) cells with 0.2 µM MNNG treatment in the 2–48 hr range. When the MNNG concentrations were higher than 10 µM, the cells treated with MNNG grow more slowly than the untreated cells (data not shown). The difference in MNNG sensitivities of HeLaS3 cells and 293T Lα+ cells may be due to their MGMT status because MGMT removes the O6-meG lesion [44].
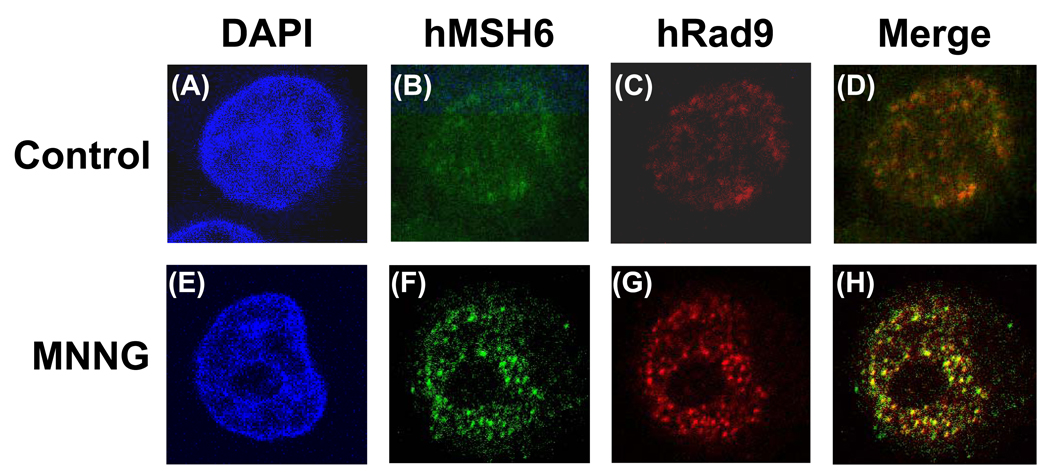
The MMR-dependent response to MNNG involves ATR, Chk1 and SMC1 (structural maintenance of chromosome 1) [42,53,56]. The 9-1-1 complex facilitates Chk1 activation by ATR (reviewed in [2]). Thus, we suspect that interaction between h9-1-1 complex and hMSH2/hMSH6 may contribute to the MNNG-induced DNA damage response. The HeLa cells were treated with different doses of MNNG for 1 h and recovered in complete medium for 24 h. The cells were stained with anti-hRad9 and anti-hMSH6 antibody and examined by confocal microscope. In the untreated cells, the hMSH6 and hRad9 displayed dispersed staining patterns all over the nuclei (Fig. 5, upper panels). With 0.2 µM MNNG treatment, we did not observe any foci formation of hMSH6 and hRad9 (data not shown). In contrast, hMSH6 and hRad9 formed foci in the nuclei when cells were treated with 10 µM MNNG and 25 µM O6-BG (a competitive inhibitor of MGMT) (Fig. 5, lower panels) or with 20 µM MNNG (data not shown). The majority of the foci of hMSH6 colocalized with those of hRad9 (Fig. 5H, yellow spots).
Fig. 5.
Colocalization of hMSH6 and hRad9 in MNNG-induced nuclear foci as determined by immunofluorescence. HeLaS3 cells were untreated (upper panel) or treated with 10 µM MNNG in the presence of O6-BG (see Material and Methods) (lower panel). The cells were immunostained with antibody against hMSH6 (green, B and F) and anti-hRad9 antibody (Red, C and G). A and E, DAPI-stained nuclei. D is the merge image of B and C. H is the merge image of F and G. Colocalization of hMSH6 and hRad9 foci is visualized as yellow spots.
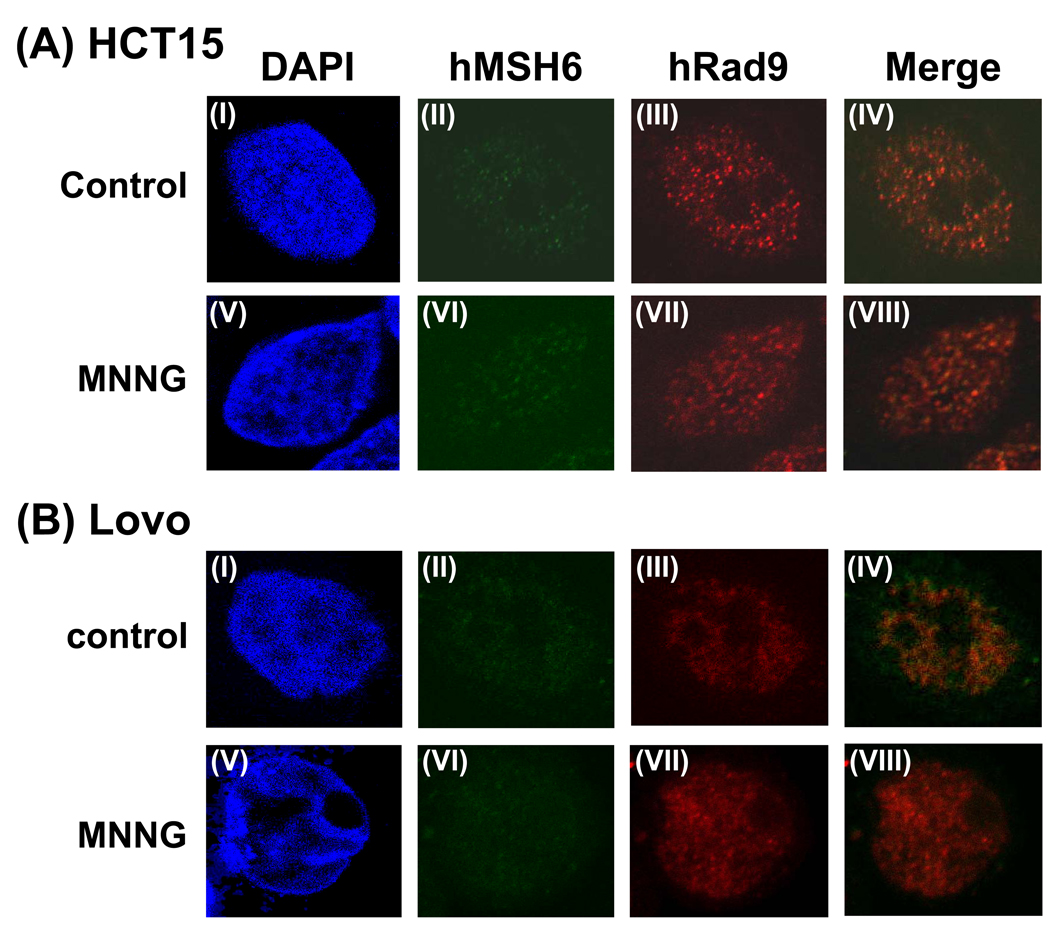
To test whether the foci formation of hRad9 is dependent on mismatch repair, we used two MMR defective cell lines. The human colon tumor line HCT15 which carries a frameshift mutation in the human MSH6 gene [70]. The antibody against the N-terminal domain of hMSH6 detected hMSH6 in the nucleus but the truncated hMSH6 did not form foci in the MNNG-treated cells (Fig. 6A, II and VI). Moreover, MSH6 defect prevented hRad9 foci formation in MNNG-treated cells (Fig. 6A, VII). Similarly, there is no hRad9 foci formation in MNNG-treated LoVo cell whose MSH2 gene contains a large deletion and MSH6 protein is unstable (Fig. 6B, VII). These data indicate that the translocation of the 9-1-1 complex to methylated DNA sites is dependent on hMutSα.
Fig. 6.
No hRad9 foci formation in MMR defective HCT15 and LoVo cells following MNNG treatment. (A) HCT15 cells which contain MSH6 frame-shift mutation were untreated (upper panel) or treated with 10 µM MNNG in the presence of O6-BG (lower panel). Immunofluorence staining was performed similarly to Fig. 5. (B) LoVo cells which contain a large deletion in MSH2 gene and unstable MSH6 were untreated (upper panel) or treated with 10 µM MNNG in the presence of O6-BG (lower panel). Immunofluorence staining was performed similarly to Fig. 5. IV is the merge image of II and III. VIII is the merge image of VI and VII.
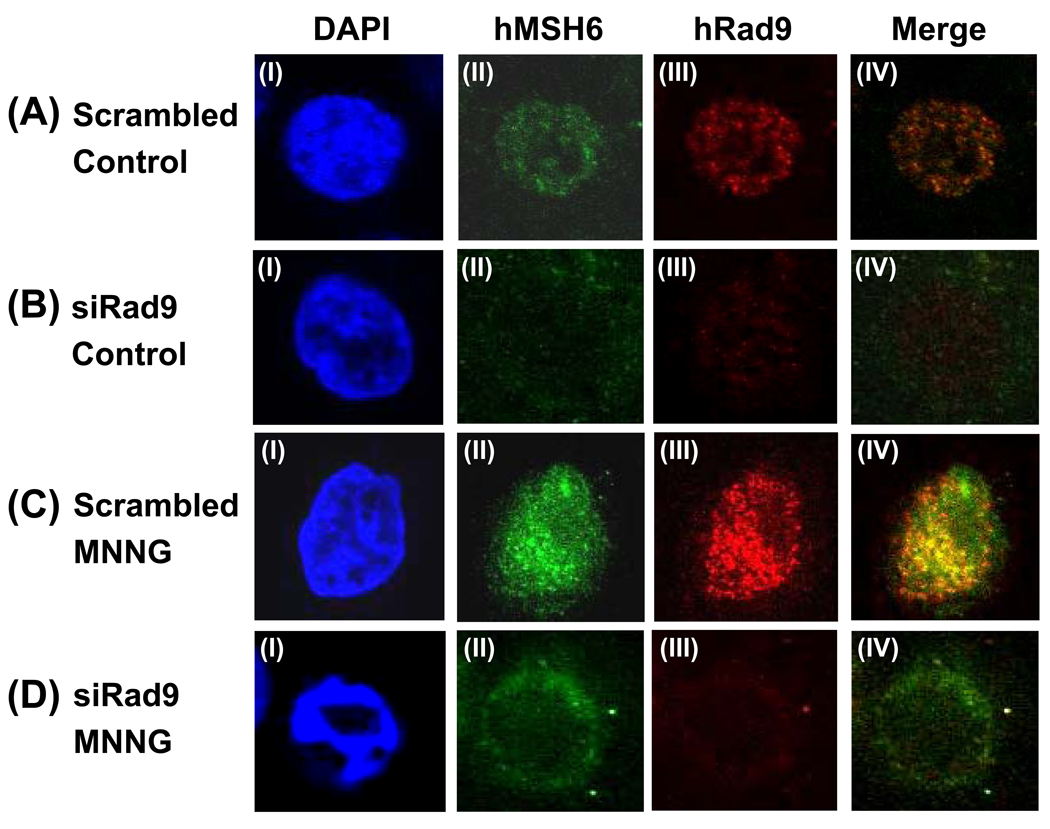
To test whether the foci formation of hMSH6 is dependent on the 9-1-1 complex, we used Rad9 specific siRNA to knockdown hRad9 expression. The hRad9 protein expression of HeLa cells was reduced by 70%, 72 hr after Rad9 siRNA treatment as compared to negative control siRNA (Fig. S1). With scrambled siRNA, the foci formation of both hMSH6 and hRad9 is similar to untrasfected HeLa cells (compare Fig. 7A and 7C to Fig. 5). However, when hRad9 was knocked down (Fig. 7B, III), the majority of the hMSH6 was observed in cytoplasm (Fig. 7B, II) in untreated cells. Following MNNG treatment, Rad9 knockdown cells has abnormal nuclear morphology (Fig. 7D, I) and hMSH6 was distributed around the outside of the nuclear envelop. Although some hMSH6 could be found inside the nucleus, there was no foci formation.
Fig. 7.
Altered hMSH6 distribution in hRad9 knockdown cells. (A) Untreated HeLa cells transfected with scrambled siRNA. (B) Untreated HeLa cells transfected with hRad9 specific siRNA. (C) HeLa cells transfected with scrambled siRNA were treated with 10 µM MNNG in the presence of O6-BG. (D) HeLa cells transfected with hRad9 specific siRNA were treated with 10 µM MNNG in the presence of O6-BG. Immunofluorence staining was performed similarly to Fig. 5. IV is the merge image of II and III. The expression of hRad9 is much reduced in (B, III) and (D, III). The majority of the hMSH6 was observed in cytoplasm (B, II) in untreated cells and was distributed around the outside of the nuclear envelop in MNNG treated cells (D, II). DAPI staining indicated that Rad9 knockdown cells has abnormal nuclear morphology following MNNG treatment (D, I).
4. Discussion
In eukaryotic cells, the mismatch repair proteins appear to be involved in at least two different processes, one to repair replication-associated errors in a strand-specific manner and the other to signal for DNA damage response following exposure to methylating agents. MMR stimulates checkpoint and cell death responses to DNA damage [40–43]. The 9-1-1 checkpoint clamp is proposed to function as damage sensor and in DNA repair [2,71]. The complex has been shown to play roles in homologous recombination repair [72], nucleotide excision repair [73], and base excision repair [18–28]. In the present study, we have shown that the 9-1-1 complex also has a role in MMR. Both hMutSα and hMutSβ protein complexes interact with the 9-1-1 checkpoint sensor. The DNA binding activity of hMutSα can be stimulated by the 9-1-1 complex and its individual subunits. Human Rad9 and hMSH6 colocalize to nuclear foci of HeLa cells after exposure to MNNG and the foci formation of hRad9 and hMSH6 in MNNG-treated cell is dependent on each other. Thus, the 9-1-1 complex is a component of the mismatch repair involved in MNNG-induced damage response.
Our data support that MMR proteins are molecular sensors in response to DNA damage [42,43,50–52] through recruiting the 9-1-1 complex. It has been shown that MSH2 protein interacts with the ATR kinase to form a signaling module and regulate the phosphorylation of Chk1 and SMC1 (structural maintenance of chromosome 1) [53]. MSH2 and MSH6 were found to be phosphorylated by ATR or ATM after irradiation (IR) [74]. Our results support the in vitro finding of Yoshioka et al [56] that MutSα may act as an adaptor to recruit the 9-1-1 complex to sites of cytotoxic O6-meG/T adducts. Moreover, He et al. [60] have reported that Rad9 defective cells have reduced MMR activity. Consistent with this finding, we have shown that MSH6 protein distributes differently in Rad9 knockdown cells. Thus, the signal flow between the 9-1-1 complex and MMR proteins is bidirectional. Similar model has been proposed that DNA glycosylases are adaptors to recruit the 9-1-1 complex to the sites of damage [18–21]. One of the DNA glycosylases, MutY homolog (MYH) DNA glycosylase, shares similar properties as hMutSα: they are involved in the initial steps of their respective postreplication mismatch repair pathways [32,75]; they cooperate to reduce 8oxoG level [76,77]; they are partners of PCNA [57–59,78] and the 9-1-1 complex ([19] and Fig. 1); and they interact to each other [79].
He et al. [60] have reported that Rad9 plays an important role in mismatch repair through physical interaction with MLH1. Because Rad9 also interacts with MSH2/MSH3 and MSH2/MSH6, their observed effect of Rad9 on MMR may not solely through interaction with MLH1. It is possible that the 9-1-1 complex is involved in several steps in MMR, a situation similar to BER [71]. hRad9 has been reported to have 3’ to 5’ exonuclease activity [80]. Whether this exonuclease activity is important for MME is not known. It is interesting to note that MLH1 only interacts with Rad9, but not with Rad1 and Hus1, while MSH2/MSH6 and MSH2/MSH3 interact with all three subunits of the 9-1-1 complex. Additionally, we also show that when hRad9 was knocked down, the majority of the hMSH6 was observed in cytoplasm and hMSH6 could not form foci inside nucleus following MNNG treatment. This may also contribute to the MMR deficiency in Rad9 defective cells. In this context, our results indicate that the 9-1-1 complex plays an important role in MMR.
Our results show that each subunit of the human 9-1-1 complex physically interacts with hMSH2, hMSH3, and hMSH6. Thus, the 9-1-1 subunits may bind to the common domains shared by hMSH2, hMSH3, and hMSH6. The hHus1 interaction motifs have been mapped to residues 295–350 on hMYH [19], residues 290–350 on hNEIL1 [20], and residues 67-110 on hTDG [20,21]. Structure-based sequence alignment of hMSH2, hMSH3 and hMSH6 shows many highly conserved regions among these three subunits [81–83] but shares limited similarity to residues 295–350 of hMYH. A common binding motif to bind the 9-1-1 complex awaits further analyses. It has been shown that certain MSH2 or MSH6 missense mutations can cause a deficiency in mismatch repair, whereas retaining the signaling functions that confer sensitivity to chemotherapeutic agents [84,85]. It will be interesting to test whether these mutants can still interact with the 9-1-1 complex.
Our finding that the G/T mismatch binding of hMutSα can be enhanced by the 9-1-1 complex and individual subunits suggests that the stimulatory effect of the 9-1-1 complex does not require the formation of the sliding clamp. This is similar to the observation with MYH, NEIL1, and TDG which can be stimulated by Hus1 [19–21]. The stimulation effect of the 9-1-1 complex on Ligase I also does not require the loading of the 9-1-1 clamp onto DNA [28]. Structures of hMutSα in complex with DNA substrates containing a G/T mispair, an unpaired nucleotide, and an O6-meG/T pair have been determined to be similar [83]. It is suggested that the control of cellular responses involves events downstream of the initial recognition step. It is possible that the 9-1-1 complex may replace PCNA to direct damage response signaling.
Although the 9-1-1 and PCNA share similar structure [12–14], they appear to have different roles in MMR. The important roles of PCNA in MMR include its involvement prior to DNA repair synthesis [86], coupling MMR to DNA replication [57], and increase the mismatch-binding specificity of MSH2-MSH6 [58]. PCNA also participates in strand excision and DNA synthesis (reviewed by[29–33]). After alkylation exposure, MutSα, MutLα, and PCNA bind to DNA to form an initial complex damaged [87–89]. These observations imply that PCNA has a role in O6-meG/T recognition and/or the induction of damage response. Interestingly, Mastrocola and Heinen [88] have shown that HeLa cells treated with 10 µM MNNG in the presence of O6-BG arrest at G2 after only one round of replication and contain persistent accumulation of hMSH2/hMLH1 complex, but not PCNA, on chromatin. We found that treating HeLaS3 cells with 20 µM MNNG or 10 µM MNNG in the presence of O6-BG resulted in colocalizing hMSH6 and hRad9 to nuclear foci. Our result suggests that the 9-1-1 complex may replace PCNA to participate the MMR-dependent signaling cascades upon detection of O6-meG adducts.
Supplementary Material
Acknowledgments
The authors thank Dr. Teresa M. Wilson for kindly providing hMutSα and hMutSβ. We appreciate the critical reading from Ms. M. Rebecca Saunders. This work was supported by National Institute of Health (NIH) Grants CA78391.
Abbreviations
- The 9-1-1 complex
Rad9/Rad1/Hus1 heterotrimer complex
- APE1
AP endonuclease
- ATM
ataxia telangiectasia mutated
- ATR
ATM- and Rad3-related protein
- BASC
BRCA1-associated genome surveillance complex
- BER
base excision repair
- BSA
bovine serine albumin
- DAPI
4’,6’-diamidineno-a-phenylindole
- EXO1
exonuclease 1
- FEN1
flap endonuclease 1
- GST
glutathione S-transferase
- h
human
- HMGB1
high-mobility group box 1
- MLH1
MutL homolog 1
- MMR
mismatch repair
- MNNG
N-methyl-N'-nitro-N-nitrosoguanidine
- MGMT
methylguanine methyltransferase
- MSH
MutS homolog
- MutLα
MLH1/PMS2
- MutSα
MSH2/MSH6
- MutSβ
MSH2/MSH3
- MYH
MutY homolog
- O6-meG
O6-methyl-guanine
- PBS
phosphate-buffered saline
- PCNA
proliferating cell nuclear antigen
- PMSF
post-meiotic segregation factor
- RFC
replication factor C
- RPA
replication protein A
- Sp
Schizosaccharomyces pombe
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 6.Al Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the Rad9-Rad1-Hus1 DNA Damage checkpoint complex- Implications for clamp loading and regulation. Mol. Cell. 2009:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 10.Sohn SY, Cho Y. Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp. J. Mol. Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J. Biol. Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 13.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-Rad1-Hus1 and Rad17-RFC. Genes to Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 14.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucl. Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS. Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 19.Shi G, Chang D-Y, Cheng CC, Guan X, Venclovas C, Lu A-L. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu A-L. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucl. Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan X, Madabushi A, Chang D-Y, Fitzgerald ME, Shi G, Drohat AC, Lu A-L. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucl. Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G, Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucl. Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toueille M, El Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucl. Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich-Heineken E, Toueille M, Tannler B, Burki C, Ferrari E, Hottiger MO, Hubscher U. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova E, Toueille M, Markkanen E, Hubscher U. The human checkpoint sensor and alternative DNA clamp Rad9-Rad1-Hus1 modulates the activity of DNA ligase I, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. Mechanism of stimulation of human DNA ligase I by the Rad9-Rad1-Hus1 checkpoint complex. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 29.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 30.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 31.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 32.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 34.Kolodner RD, Alani E. Mismatch repair and cancer susceptibility. Curr. Opin. Biotech. 1994;5:585–594. doi: 10.1016/0958-1669(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 35.Modrich P, Lahue RS. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 36.Lu A-L. Biochemistry of mammalian DNA mismatch repair. In: Hoelm H, Nicolaides NC, editors. DNA Repair in Higher Eukaryotes. Totowa, N J: Humana Press; 1998. pp. 95–118. [Google Scholar]
- 37.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5'-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 40.Branch P, Aquilina G, Bignami M, Karran P. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993;362:652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- 41.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Gu L, Wang H, Geacintov NE, Li GM. Mismatch repair processing of carcinogen-DNA adducts triggers apoptosis. Mol. Cell. Biol. 1999;19:8292–8301. doi: 10.1128/mcb.19.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sedgwick B, Lindahl T. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene. 2002;21:8886–8894. doi: 10.1038/sj.onc.1205998. [DOI] [PubMed] [Google Scholar]
- 45.Aquilina G, Zijno A, Moscufo N, Dogliotti E, Bignami M. Tolerance to methylnitrosourea-induced DNA damage is associated with 6-thioguanine resistance in CHO cells. Carcinogenesis. 1989;10:1219–1223. doi: 10.1093/carcin/10.7.1219. [DOI] [PubMed] [Google Scholar]
- 46.Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, Modrich P. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aquilina G, Crescenzi M, Bignami M. Mismatch repair, G(2)/M cell cycle arrest and lethality after DNA damage. Carcinogenesis. 1999;20:2317–2326. doi: 10.1093/carcin/20.12.2317. [DOI] [PubMed] [Google Scholar]
- 48.Cejka P, Stojic L, Mojas N, Russell AM, Heinimann K, Cannavo E, Di Pietro M, Marra G, Jiricny J. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawn MT, Umar A, Carethers JM, Marra G, Kunkel TA, Boland CR, Koi M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 50.Duckett DR, Bronstein SM, Taya Y, Modrich P. hMutSα and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc. Natl. Acad. Sci. USA. 1999;96:12384–12388. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl. Acad. Sci. USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr., Levrero M, Wang JY. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat. Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel T. Functional interaction of PCNA with MSH2/MSH6 and MSH2/MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 58.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 59.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He W, Zhao Y, Zhang C, An L, Hu Z, Liu Y, Han L, Bi L, Xie Z, Xue P, Yang F, Hang H. Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucl. Acids Res. 2008;36:6406–6417. doi: 10.1093/nar/gkn686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem. 1999;274:21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 62.Lu A-L. Repair of A/G and A/8-oxoG mismatches by MutY adenine DNA glycosylase. In: Vaughan P, editor. DNA Repair Protocols, prokaryotic systems. Totowa, New Jersey: Humana Press; 2000. pp. 3–16. [Google Scholar]
- 63.McDowell HD, Carney JP, Wilson TM. Inhibition of the 5' to 3' exonuclease activity of hEXO1 by 8-oxoguanine. Environ. Mol. Mutagen. 2008;49:388–398. doi: 10.1002/em.20398. [DOI] [PubMed] [Google Scholar]
- 64.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 66.Sanada M, Takagi Y, Ito R, Sekiguchi M. Killing and mutagenic actions of dacarbazine, a chemotherapeutic alkylating agent, on human and mouse cells: effects of Mgmt and Mlh1 mutations. DNA Repair (Amst) 2004;3:413–420. doi: 10.1016/j.dnarep.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Tsuzuki T, Sakumi K, Shiraishi A, Kawate H, Igarashi H, Iwakuma T, Tominaga Y, Zhang S, Shimizu S, Ishikawa TeA. Targeted disruption of the DNA repair methyltransferase gene renders mice hypersensitive to alkylating agent. Carcinogenesis. 1996;17:1215–1220. doi: 10.1093/carcin/17.6.1215. [DOI] [PubMed] [Google Scholar]
- 68.Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 69.Stojic L, Cejka P, Jiricny J. High doses of SN1 type methylating agents activate DNA damage signaling cascades that are largely independent of mismatch repair. Cell Cycle. 2005;4:473–477. doi: 10.4161/cc.4.3.1528. [DOI] [PubMed] [Google Scholar]
- 70.Boyer JC, Umar A, Risinger JI, Lipford R, Kane M, Barett JC, Kolodner RD, Kunkel TA. Microsatellite instability, mismatch repair deficiency and genetic defects in human cancer cell lines. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 71.Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 72.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, Roti Roti JL, Lieberman HB, Pandita TK. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 75.Lu A-L, Bai H, Shi G, Chang D-Y. MutY and MutY homologs (MYH) in genome maintenance. Front Biosci. 2006;11:3062–3080. doi: 10.2741/2033. [DOI] [PubMed] [Google Scholar]
- 76.Lu A-L, Li X, Gu Y, Wright PM, Chang D-Y. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophy. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 77.Russo MT, De Luca G, Casorelli I, Degan P, Molatore S, Barone F, Mazzei F, Pannellini T, Musiani P, Bignami M. Role of MUTYH and MSH2 in the control of oxidative DNA damage, genetic instability, and tumorigenesis. Cancer Res. 2009;69:4372–4379. doi: 10.1158/0008-5472.CAN-08-3292. [DOI] [PubMed] [Google Scholar]
- 78.Parker A, Gu Y, Mahoney W, Lee S-H, Singh KK, Lu A-L. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 79.Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog (hMYH), a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins hMSH2/hMSH6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 80.Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3' to 5' exonuclease. J. Biol. Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 81.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G.T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 82.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 83.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 84.Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, Umar A, Kucherlapati R, Lipkin M, Kunkel TA, Edelmann W. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 85.Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, Kucherlapati R, Kolodner RD, Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 86.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 87.Hidaka M, Takagi Y, Takano TY, Sekiguchi M. PCNA-MutSalpha-mediated binding of MutLalpha to replicative DNA with mismatched bases to induce apoptosis in human cells. Nucl. Acids Res. 2005;33:5703–5712. doi: 10.1093/nar/gki878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mastrocola AS, Heinen CD. Nuclear reorganization of DNA mismatch repair proteins in response to DNA damage. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schroering AG, Williams KJ. Rapid induction of chromatin-associated DNA mismatch repair proteins after MNNG treatment. DNA Repair (Amst) 2008;7:951–969. doi: 10.1016/j.dnarep.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.