Abstract
Previous studies in our laboratory proved that certain functional groups are able to mimic the pyrophosphate moiety and act as leaving groups in the enzymatic polymerization of deoxyribonucleic acids by HIV-1 reverse transcriptase. When the potential leaving group possesses two carboxylic acid moieties linked to the nucleoside via a phosphoramidate bond, it is efficiently recognized by this error-prone enzyme, resulting in nucleotide incorporation into DNA. Here, we present a new efficient alternative leaving group, iminodiacetic acid, which displays enhanced kinetics and an enhanced elongation capacity compared to previous results obtained with amino acid deoxyadenosine phosphoramidates. Iminodiacetic acid phosphoramidate of deoxyadenosine monophosphate (IDA-dAMP) is processed by HIV-1 RT as a substrate for single nucleotide incorporation and displays a typical Michaelis–Menten kinetic profile. This novel substrate also proved to be successful in primer strand elongation of a seven-base template overhang. Modelling of this new substrate in the active site of the enzyme revealed that the interactions formed between the triphosphate moiety, magnesium ions and enzyme's residues could be different from those of the natural triphosphate substrate and is likely to involve additional amino acid residues. Preliminary testing for a potential metabolic accessibility lets us to envision its possible use in an orthogonal system for nucleic acid synthesis that would not influence or be influenced by genetic information from the outside.
INTRODUCTION
The aim of our project is to develop and introduce additional types of nucleic acids in living cells. As a long term objective, these nucleic acids could be used to avert genetic pollution by preventing dissemination of artificial hereditary messages in natural ecosystems, including the human body (1). In formal terms, this project amounts to systematical searching of the chemical space in order to identify an assortment of activated monomers carrying a set of complementary bases. Another aspect of this project is to discover nucleic acid enzymes that would use activated monomers as substrates in polymerization, ligation or recombination. The result of this interplay would be templated reproduction of polymers which would convey additional genetic information but not interfere with RNA and DNA metabolism and function.
Base and backbone variants of DNA and RNA are the topic of intense research in several laboratories, auguring well for the feasibility of xeno-nucleic acid (XNA) reproduction in vivo (2–4). Even though it will not be part of the propagated polymers per se, the leaving group to be used for activating XNA precursors is in our opinion crucial. Indeed, the chemical structure of the leaving group can be elaborated leading to segregation of the polymerization of XNA polymers from that of indigenous DNA and RNA monomers and establishing an informational enclave. Simultaneously, it could disentangle the XNA polymerization from the phosphoanhydride economy of the cell, hence establishing an energetic enclave as well. The dual role of nucleoside triphosphates, as both energetic currency and precursors of the informational polymers RNA and DNA, drastically limits the range of metabolic and genetic reprogramming that can be accomplished in living cells. The judicious implementation of novel leaving groups in metabolism could overcome the need for physical compartmentalization of unnatural information transfers and enable the launching and sustaining of autonomous hereditary procedures in vivo.
We previously showed that the common metabolite aspartate coupled to the phosphate group of deoxynucleotides by a P–N phosphoramidate bond provides unnatural adducts acting as competent substrates in the HIV-1 RT directed template-dependent DNA polymerization (5–7). Through broadening our search of the chemical space we addressed the synthesis and enzymatic assay of the deoxyadenylate adduct of iminodiacetate. Although this isomer is not commonly found in living cells, its biosynthesis and recycling from glycine and glyoxylate could be readily implemented in metabolism. Formation of the prototype metabolite triphospho-imino-diacetate (TPI) and its reaction with nucleosides can also be experimentally studied to investigate novel energetic and informational metabolism and its potential implementation in bacterial cells.
The enzymatic synthesis of nucleic acids makes use of nucleoside triphosphates as substrates and this process is driven forward by the release and hydrolysis of pyrophosphate. The selection of new leaving groups for the enzymatic synthesis of nucleic acids should be accompanied by a selection of appropriate polymerases (6), which can use the modified nucleotides as building blocks for gene synthesis independent of the cellular gene-synthesis machinery. Since HIV-1 RT shows broad substrate specificity, this enzyme was chosen as a primary polymerase for selection of potential leaving groups to activate nucleoside triphosphate mimics.
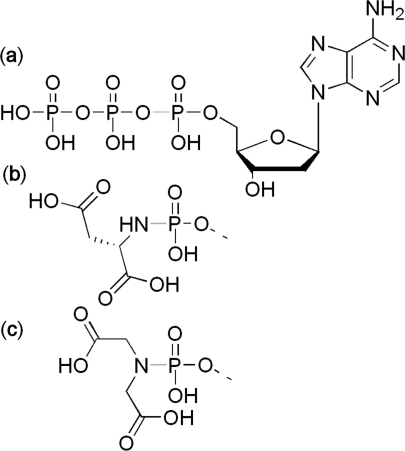
Despite the fact that such polymerases as HIV-1 reverse transcriptase (6), Taq DNA polymerase and Vent (exo-) DNA polymerase (6) are able to process selected substrates like amino acid dAMP phosphoramidates, HIV-1 RT is being the preferred enzyme. In our study the pyrophosphate leaving group (linked to a deoxynucleotide in Figure 1a) is replaced by a completely different molecule with potential chelating properties. It has been indicated that the L-aspartic acid derivatives of dATP (Figure 1b) (6), dGTP, dCTP and dTTP (7) demonstrated conserved single nucleotide incorporation and limited elongation properties. Incorporation of L-His-dAMP by HIV-1 RT however proceeded with lower efficiency. However in contrast to its congener L-Asp-dAMP, primer chain elongation was slightly improved when L-His-dAMP was used (6). It is important to note that regardless of a phosphoramidate mimic, DNA synthesis dramatically slowed down after incorporation of 2 nt. The reason for this stalling is not clear. The stalling issue might be a major hurdle for the use of such modified nucleotides for enzyme-catalyzed DNA synthesis in vivo.
Figure 1.
Chemical structure of (a) deoxyadenosine triphosphate (b) l-Asp-dAMP and (c) IDA-dAMP. The bond to be cleaved during enzymatic polymerisation is coloured in grey.
A model postulated by Steitz explains the way by which the incoming deoxynucleotide triphosphate (dNTP) is bound in the active site of the polymerase for incorporation in the growing DNA chain (8). This model takes into account the importance of the chelation of two divalent magnesium ions required in the polymerization mechanism and bringing the free 3′ OH group of the primer for in-line attack of the alpha phosphate group of dNTP. In line with these observations, Garforth et al. (9) suggest that enzyme processing could occur when only two potential chelating moieties are present.
In the case of phosphoramidate substrates, a necessity of the leaving group to carry two potential chelating moieties was confirmed recently as well (10). To broaden the chemical space and obtain further insight in the structure–activity relationship, we altered the substitution pattern of the nitrogen atom of the phosphoramidate moiety, while (i) taking care of potential chelating properties and (ii) staying focused on potential metabolic accessibility of the new leaving group. Enhanced coordination of the magnesium ion by the leaving group moiety of the modified nucleotide might influence incorporation kinetics and efficiency of the chain elongation reaction. In addition, iminodiacetic acid (Figure 1c) can be considered as potentially metabolically accessible and potentially prone to catabolism into non-toxic cellular constituents (i.e. glycine and a two-carbon atom fragment). As a consequence, this degradation reaction may have an effect on the equilibrium of the nucleotide incorporation reaction.
The iminodiacetate dAMP phosphoramidate (IDA-dAMP) (Figure 1c) possesses no chiral centre (ensuring less stereochemical constraint than for the chiral L-aspartic acid) and has chelating properties similar to EDTA. It has been shown recently that protonation of the pyrophosphate leaving group is important and facilitated by a neighbouring general acidic residue in the active site, at least for a polymerase (11). The presence of a nitrogen atom in iminodiacetate could be beneficial in this case.
MATERIALS AND METHODS
Analyses
Nuclear magnetic resonance (NMR) spectral analyses were carried out on a Brucker AvanceTM II 300 MHz or 500 MHz with PAXTI probe. The Bruker TopspinTM 2.1 software was used to process spectra. Chemical shifts are expressed in parts per million (ppm) by frequency. 1H and 13C NMR chemical shifts are referenced to an internal TMS signal (δ = 0.00 ppm), 31P NMR chemical shifts are referenced to an external 85% H3PO4 standard (δ = 0.00 ppm). Standard mass spectra were measured with a Finnigan LCQ DuO (Thermo Fischer Scientific) using the ionization by electron impact technique (ESI); data were acquired with the LAC/E32 system (Waters). Exact mass spectra were obtained with a Q-Tof 2TM (Micromass Ltd) coupled to a CapLCTM system (Waters). Chemicals of analytical or synthetic grade were obtained from commercial sources and were used as received [deoxyadenosine monophosphate: Sigma–Aldrich; dicyclohexylcarbodiimide (DCC), dimethyl iminodiacetic acid hydrochloride: Fluka; tert-butanol and triethylamine: Acros]. Technical solvents were obtained from Brenntag (Deerlijk, Belgium). Acetonitrile HPLC Grade was purchased from Fischer Scientific. Flash silica chromatography was performed on Davisil® silica gel 60, 0.040–0.063 mm (Grace Davison). Thin-layer chromatography was performed on Alugram® silica gel UV254 mesh 60, 0.20 mm (Macherey-Nagel).
Synthesis of 2′-deoxyadenosine-5′-(dimethyl iminodiacetate) phosphoramidate (intermediate for IDA-dAMP)
In a two neck flask, 2′-deoxyadenosine-5′-monophosphoric acid hydrate (100 mg, 0.286 mmol) and dimethyl iminodiacetate hydrochloride (283 mg, 1.430 mmol, 5 equiv.) were suspended in a mixture of 1,4-dioxane (9 ml) and N,N-dimethylformamide (DMF, 1 ml). A few drops of triethylamine were added to the solution to facilitate dissolution. Then, a solution of DCC (414 mg, 2.004 mmol, 7 equiv.) in 1,4-dioxane (1 ml) was added and the reaction mixture was heated for 3 h, while stirring under nitrogen atmosphere. The progress of the reaction was monitored by TLC (CHCl3:CH3OH:H2O 5:3:0.5). Upon completion, the reaction mixture was cooled down and the solvent was removed by rotary evaporation. The residue was resuspended in water (15 ml), extracted with diethyl ether (3 × 10 ml), and the aqueous phase was lyophilised. The resulting solid was subjected to column chromatography on silica gel using the following solvent gradient: CHCl3:CH3OH:H2O (5: 1: 0; 5: 2: 0.25; 5: 3: 0.25; 5: 3: 0.5). The product was obtained as a white solid (58 mg, 43%).
1H NMR (500 MHz, D2O): δ 8.46 (s, 1H, H8), 8.24 (s, 1H, H2), 6.50 (apparent t, 1H, JH1′–H2′ = 6.5 Hz, H1′), 4.72 (m, 1H, H3′), 4.22 (m, 1H, H4′), 4.00 (m, 2H, H5′), 3.78 (dd, 2H, J = 9.5 Hz, CH2COOCH3), 3.69 (dd, 2H, J = 11 Hz, CH2COOCH3), 3.56 (s, 6H, CH3), 2.89 (m, 1H, H2′a), 2.61 (m, 1H, H2′b) ppm.
13C NMR (125 MHz, D2O): δ 174.27, 156.07, 153.21, 149.36, 140.38, 119.14, 86.63, 84.12, 72.13, 64.67, 52.62, 49.28, 39.25 ppm.
31P NMR (121 MHz, D2O): δ 6.67 ppm.
HRMS: calculated for C16H22N6O9P 473.1186, found: 473.1180.
Synthesis of 2′-deoxyadenosine-5′-iminodiacetate phosphoramidate
A solution of 2′-deoxyadenosine-5′-dimethyl iminodiacetate phosphoramidate (50 mg, 0.105 mmol) in 0.4 M sodium hydroxide solution in MeOH:H2O (4:1) (2 ml), was stirred at room temperature under nitrogen for 2 h. The progress of the reaction was monitored by TLC (iPrOH/NH3/H2O 7:1:2). Upon completion, the reaction mixture was neutralized by addition of 1M triethylammonium bicarbonate. The solvent was removed under reduced pressure. The residue was purified by column chromatography and eluted with the following solvent gradient: CHCl3:CH3OH:H2O 5: 1: 0, 5: 2: 0.25, 5: 3: 0.5 and 5: 4: 1. The product was isolated and concentrated, affording a white solid (31 mg, 65% yield).
1H NMR (500 MHz, D2O): δ 8.46 (s, 1H, H8), 8.25 (s, 1H, H2), 6.50 (apparent t, 1H, 3JH1′–H2′ = 6.48 Hz, H1′), 4.70 (m, 1H, H3'), 4.26 (m, 1H, H4′), 4.02 (m, 2H, H5′), 3.64 (m, 4H, CH2COOH), 2.83 (m, 1H, H2′a), 2.59 (m, 1H, H2′b) ppm.
13C NMR (125 MHz, D2O): δ 171.96, 156.26, 153.35, 149.49, 140.68, 119.41, 86.62, 84.42, 71.87, 64.67, 49.67, 39.62 ppm.
31P NMR (121 MHz, D2O): δ 8.34 ppm.
HRMS (ESI): calculated for C14H19N6O9P 446.0951, found: 445.0864 (negative mode).
Metabolic availability
Trisodium trimetaphosphate (153 mg, 0.5 mmol) and iminodiacetic acid disodium salt (979 mg, 5 mmol, 10 equiv.) were dissolved in double-distilled water (6 ml) and the resulting clear mixture was let at r.t. for 1 day and monitored by 31P NMR (121 Hz, 90% H2O + 10% D2O). Simultaneously with disappearance of the starting material (–20.79 ppm, s), formation of three news peaks [–0.39 ppm (d, JP–P = 20.06 Hz), –4.97 ppm (d, JP–P =17.58 Hz), –19.65 ppm (t, JP–P = 18.99 Hz)], assigned to triphosphoro iminodiacetic acid (TPI), was observed (80% yield, from 31P peak integration). The mixture was lyophilized; solvent (DMF, DMSO, hexamethylphosphoramide or mixture thereof with water) (1 ml), deoxyadenosine (16 mg, 0.0625 mmol) and magnesium chloride (127 mg, 0.625 mmol) were added. The resulting mixture was stirred at r.t. for 1 h and monitored by 31P NMR. Disappearance of TPI signals was simultaneous with appearance of two singulets (0.38 ppm and –9.24 ppm), attributed to iminodiacetic monophosphate and to inorganic pyrophosphate, respectively.
Oligodeoxyribonucleotides preparation
Oligodeoxyribonucleotides P1, T1, T2 and T3 were purchased from Sigma Genosys or Eurogentec. The concentrations were determined with a Varian Cary-300-Bio UV Spectrophotometer.
The lyophilized oligonucleotides were dissolved in diethylpyrocarbonate (DEPC)-treated water and stored at –20°C. The primer oligonucleotides were 5′-33P-labeled with 5′-[γ33P]-ATP (Perkin Elmer) using T4 polynucleotide kinase (New England Biolabs), according to standard procedures. The labelled oligonucleotide was further purified using IllustraTM MicrospinTM G-25 columns (GE Healthcare).
DNA polymerase reactions using IDA-dAMP as substrate
End-labelled primer was annealed to its template by combining primer and template in a molar ratio of 1:2 and heating the mixture to 70°C for 10 min, followed by slow cooling to r.t. over a period of 1.5 h. For the incorporation of IDA-dAMP a series of 20-µl-batch reactions was performed with the enzyme HIV-1 RT (Ambion, 10 U/µl stock solution, specific activity 8.095 U/mg). The final mixture contained 125 nM primer template complex, RT buffer [250 mM Tris–HCl, 250 mM KCl, 50 mM MgCl2, 2.5 mM spermidine, 50 mM dithiothreitol (DTT); pH 8.3], 0.025 U/µl HIV-1 RT, and different concentrations of IDA-dAMP building blocks (1 mM, 500 µM, 200 µM and 100 µM). In the control reaction with the natural nucleotide, a 10-µM dATP concentration was used. The mixture was incubated at 37°C and aliquots were quenched after 5, 10, 20, 30 and 60 min. For elongation experiments, the same mixture with appropriate primer:template hybrid (Table 1) was incubated at 37°C and aliquots were quenched after 15, 30, 60, 90, 120 min. In the reaction with the natural nucleotide, a 50-µM dATP concentration was used.
Table 1.
Overview of the primer-template complexes used in the DNA polymerase reactions
| Single-nucleotide incorporation and kinetic experiments | |
|---|---|
| P1 | 5′-CAGGAAACAGCTATGAC-3′ |
| T1 | 3′-GTCCTTTGTCGATACTGTCCC-5′ |
| Elongation experiments | |
| P1 | 5′-CAGGAAACAGCTATGAC-3′ |
| T2 | 3′-GTCCTTTGTCGATACTGTTTTTTT-5′ |
| T3 | 3′-GTCCTTTGTCGATACTGTTTTTTTGGAC-5′ |
Bold letters indicate the template overhang in the hybridized primer–template duplex.
Steady-state kinetics of single-nucleotide incorporation
The steady-state kinetics of single-nucleotide incorporation of the iminodiacetate phosphoramidate and of the natural nucleoside triphosphate (dATP) was determined by gel-based polymerase assay. In all the experiments, the template T1 and the primer P1 were used. The primer and template in a 1:2 molar ratio were hybridized in a buffer containing 20 mM Tris–HCl, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.3 and used in an amount to provide 125 nM concentration of the primer in each 20-µl reaction. A range of building block concentrations between 10 µM and 1 mM for the phosphoramidate and between 0.1 and 10 µM for the natural building block was used. The final concentrations of primer–template complex and HIV-1 RT were 125 nM, and 0.0063 U/µl, respectively. Reaction mixtures were incubated at 37°C and aliquoted at six different time intervals. The reactions were quenched by addition of a buffer containing 80% formamide, 2 mM EDTA and 1X TBE buffer. The analysis of polymerase reaction was performed by polyacrylamide gel electrophoresis (see detailed protocol below). The incorporation rates (V) were calculated based on the percentage of the extended oligonucleotide in the mixture (P+1 band). The kinetic parameters (VMax and KM were determined by plotting V (nM/min–1) versus substrate concentration (µM) and fitting the data to a non-linear Michaelis–Menten regression using GraphPad Prism Software version 5.0.
Electrophoresis
All polymerase reaction aliquots (2.5 µl) were quenched by the addition of 10 µl of loading buffer [90% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol and 50 mM ethylenediaminetetraacetic acid (EDTA)]. Samples were heated at 85°C for 3 min prior to analysis by electrophoresis for 2.5 h at 2000 V on a 30 cm × 40 cm × 0.4 mm 20% (19:1 mono:bis) denaturing gel in the presence of a 100 mM Tris–borate, 2.5 mM EDTA buffer; pH 8.3. Products were visualized by phosphorimaging. The amount of radioactivity in the bands corresponding to the products of enzymatic reactions was determined by using the imaging device Cyclone® and the Optiquant image analysis software (Perkin Elmer).
Molecular modelling
Modelling of the iminodiacetate-dAMP in the RT active site was prepared and performed as previously described (12–14). Solvated molecular dynamics was used to verify the stability. The complex was solvated and simulated for 1 ns. The presence of the base pair hydrogen bonds between primer and template strand was taken as a valid indicator for the stability of the structures: nearly all H-bonds were still present in the final dynamic structures. An average structure of the last 50 ps was generated and analysed.
RESULTS AND DISCUSSION
Synthesis of IDA-dAMP
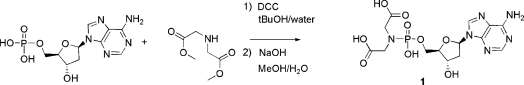
The synthesis of the methyl esters of the IDA-dAMP 1 was accomplished according to the method described by Wagner and colleagues (15) starting from the nucleoside monophosphate. Deprotection of the methyl ester was carried out with sodium hydroxide in methanol–water solution. Scheme 1 shows the synthetic route, which is an easy-to-perform two-step reaction.
Scheme 1.
Obtention of IDA-dAMP 1.
Single-Nucleotide Incorporation
HIV-1 reverse transcriptase serves as a catalyst in the HIV-1 viral replication process and uses deoxynucleotides as substrates. This polymerase is error-prone and thus has a high mutation rate (16,17). Previous experiments carried out with the l-aspartic acid phosphoramidate of dAMP demonstrated that this amino acid was an acceptable leaving group for the polymerase in the nucleotidyl condensation process (5,6). In the present study, we evaluated the capacity of HIV-1 RT to incorporate IDA-dAMP as a substrate in the primer- template complex P1:T1. The initial experiments were carried out using a template with an overhang of one thymidine nucleotide followed by three non-pyrimidine bases (Table 1). Incorporation efficiency was analysed by the polyacrylamide-gel-based single-nucleotide incorporation assay (18,19).
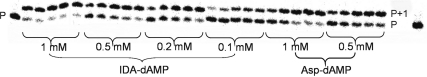
The phosphoramidate analogue IDA-dAMP was correctly processed by HIV-1 RT converting a primer to an (P+1) strand up to 86% in 60 min, at 500 µM deoxynucleotide concentration (Figure 2). So as for L-Asp-dAMP, efficient substrate incorporation was also detected with a 5-fold lower substrate concentration (40% P+1 product at 100 µM).
Figure 2.
Incorporation of IDA-dAMP and L-Asp-dAMP into P1:T1 by HIV-1 reverse transcriptase. Aliquots were taken at 5, 10, 20, 30 and 60 min.
Elongation experiments
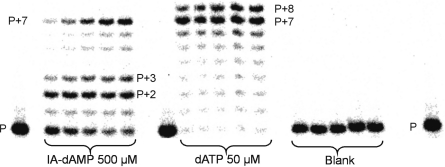
Previous studies carried out with L-Asp-dAMP and HIV-1 RT showed its ability to serve as a substrate for template-dependent incorporation of more than one phosphoramidate nucleotide. Primer elongation of up to 6 nt was observed but only to a limited extent, and with prevalence of the (P+2) and (P+3) products. Such a stalling represents a significant hindrance for future use of phosphoramidate nucleotides for enzymatic DNA synthesis in vivo. For the evaluation of the strand elongation capacity of IDA-dAMP using the same enzyme, the primer–template duplexes P1:T2 and P1:T3 were used (Table 1).
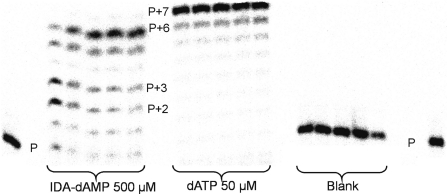
A range of concentrations of the building block was incubated at the appropriate temperature with the primer–template complex and 0.025 U/µl of enzyme. Samples were taken between 30 and 120 min and analysed by 20% polyacrylamide gel electrophoresis. In the P1:T2 experiment, the template has an overhang of seven thymidine nucleotides. With IDA-dAMP, we observed a prevalence of the (P+6) product, disclosing an excellent elongation capacity (Figure 3). The initial stalling at the (P+2) and (P+3) products disappears over time and is not so pronounced as with L-Asp as substrate (Table 2). However, we did not observe a complete chain extension to (P+7) products when using the P1:T2 system.
Figure 3.
Elongation of P1:T2 with IDA-dAMP by HIV-1 reverse transcriptase. Aliquots were taken at 15, 30, 60, 90 and 120 min. Blank: no triphosphate analogue was added in the reaction.
Table 2.
L-Asp-dAMP versus IDA-dAMP in the elongation of P1 directed by template T2: % of P+n product after a 120-min reaction
| Product | % substratea L-Asp-dAMP (6) | % substratea IA-dAMP |
|---|---|---|
| P+6 | Traces | 40 (major) |
| P+5 | Traces | 19 |
| P+4 | Traces | 3 |
| P+3 | 34 | 14 |
| P+2 | 64 (major) | 12 |
aPercentage of the total amount of radio-emitting oligonucleotides in the mixture.
The major product of L-Asp-dAMP incorporation is the (P+2) resulting oligonucleotide, whereas the (P+6) product is only present in trace amounts and barely quantifiable. In contrast, with IDA-dAMP, the major product is the (P+6) oligonucleotide (Table 2).
In the subsequent P1:T3 experiment using a P1:T3 duplex, where the seven thymidine bases overhang of the template is flanked by four non-thymidine units, the primer extension resulted in formation of a (P+7) product (Figure 4).
Figure 4.
Elongation of P1:T3 with IDA-dAMP by HIV-1 reverse transcriptase. Aliquots were taken at 15, 30, 60, 90 and 120 min. Blank: no triphosphate analogue was added in the reaction.
The observed full-length elongation obtained when using the purine-elongated template T3, could be due to the increased affinity of the longer template overhang for the enzyme and the formation of a more stable tertiary complex. As was suggested by Kohlstaedt in 1992, it has been shown that the template strand binds to the fingers subdomain of the polymerase active site (20–22). Besides, the binding affinity of RT with DNA duplexes is increased when template overhang is extended to six additional bases (23). From models based on crystal structures of the complex (23,24), the β3–β4 loop, situated in this domain, contacts the template 3 nt upstream from the primer terminus (near or at Leu74). Kew et al. (25) postulated that a stronger complex between the fingers subdomain of a HIV-RT and the template overhang may be responsible for higher processivity by the enzyme.
Although it seems that binding and incorporation of a natural triphosphate substrate dATP is not dependent on the length of the template overhang (24) (Figure 3), it appears to be the case with the triphosphate mimic IDA-dAMP. The presence of a four base 5′-overhang at the template’s 5′-end could favour a more stable dsDNA–protein complex. The template length also seems to influence the ability of HIV-1 RT to recognise and incorporate an incoming dNTP carrying a modified leaving group.
The superior substrate properties of IDA-dAMP as compared to l-Asp-dAMP might also be arising from the increased chelating ability of the N-diacetate group of IDA-dAMP when compared with the aspartate group of l-Asp-dAMP, from structural differences of the transition state or from modified conformational changes of the enzyme. In the case of T7 RNA polymerase, Yin and Steitz proposed a model where cross-linking between the DNA helix and the enzyme depends on the presence of pyrophosphate. They demonstrated that the nature of the electrostatic interactions of this complex undergo changes between the pre- and post-translocation state of nucleotide incorporation (26). In a previous model postulated by Steitz, one magnesium ion is coordinated by the 3′ end of the primer and the α-phosphate, while the second magnesium ion is thought to facilitate the leaving group properties of the pyrophosphate moiety by chelating with the β- and γ-phosphates of the nucleotide (8).
Both iminodiacetic acid and aspartic acid have good chelating properties. These dicarboxylic acids display dissociation constants (KD,25°C) for the divalent magnesium ion of 2.98 and 2.43, respectively, while the dissociation constant of pyrophosphate is 5.4 (27). The observed elongation demonstrates that iminodiacetic acid has functioned as a leaving group. If the reaction mechanism of the chemical step mimics that of the natural substrate, one magnesium ion should leave the catalytic site together with the leaving group. If this happens, given that the amino group participates in a phosphoramidate bond, the contribution of the lone pair on the nitrogen atom to the chelating properties of the leaving group is reduced before phosphoryl transfer, but not when the P–N bond has been cleaved (coordination of iminodiacetate).
Kinetic experiments
Further investigation of IDA-dAMP was focused on kinetics of incorporation. Kinetic parameters for the incorporation of both the natural and the modified substrate by HIV-1 RT were determined on the basis of the single completed hit model (19,28); in the study P1 and T1 (Table 1) were used as the primer and template DNA strands, respectively. The steady-state kinetic values KM and VMax corresponding to the substrate efficiency of the nucleotide triphosphate analogues are reported in Table 3.
Table 3.
Determination of the kinetic parameters of the incorporation of dAMP with the natural pyrophosphate leaving group (dATP) and the iminodiacetate leaving group (IDA-dAMP) into P1T1 by HIV-1 RT DNA polymerase at 0.025 U/µl
| KM (µM) | Vmax (nM min−1) | Vmax/KM (min−1) | |
|---|---|---|---|
| dATP | 29 ± 5 | 65.7 ± 4.3 | 2.26 × 10−3 |
| IDA-dAMP | 312.5 ± 70.5 | 49.0 ± 4.5 | 0.16 × 10−3 |
Along with showing a similar single incorporation after 60 min and far better elongation results, IDA-dAMP also demonstrates better incorporation kinetics than L-Asp-dAMP for HIV-1 RT (5). The measured VMax/KM ratio towards HIV-1 RT is 1 order of magnitude lower than that of dATP. Similarly to the L-Asp phosphoramidate analogue, the KM of IDA-dAMP is much higher than for the natural substrate, whereas the VMax is 1.3-fold lower. The increased capacity for chain elongation is paralleled by a similar increase in incorporation efficiency. It is clear that enzyme evolution should focus on the selection of a polymerase enhancing KM (rather than VMax) which is easier to achieve than the inverse.
Model of IDA-dAMP (1) bound to HIV-1 reverse transcriptase
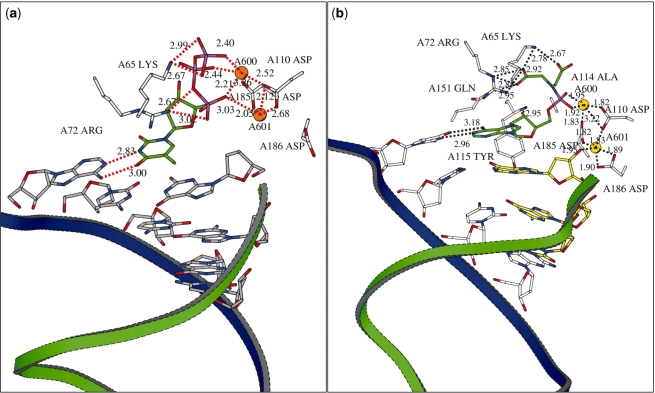
With the IDA-dAMP molecule bound to HIV-1 RT, stable molecular dynamics (MD) trajectories were obtained. In the calculated ground-state complex (Figure 5b), the spatial arrangement of two Mg2+ ions is comparable to their situation in the original TTP complex (Figure 5a) (21). Although in the ground state the carboxylate groups of IDA-dAMP (mimicking the β and γ phosphate groups) are not yet involved in an ionic bond with the Mg2+ ions, they are stabilized by ionic interactions with Lys65A, Arg72A and Gln151A. The difference in binding affinity between PPi and IDA could also contributes to a difference in kinetics or to the lower processivity of the enzyme when challenged with IDA-dAMP in place of dATP (29). In the crystal structure obtained with a dNTP, Gln151 only contacts the dTTP, without forming an actual ionic bond (21).
Figure 5.
(a) The thymidine triphosphate (TTP) in the RT dNTP pocket [X-ray structure from Huang 1998 (18)]. The primer strand is shown as a green ribbon; the template strand is in blue. The stabilizing interactions of the TTP within the complex are shown: hydrogen bond distances between the bases are indicated in angstroms. The ligation of the 2 Mg2+ ions to the complex is also visualized by dashes and distances in angstroms. (b) Average structure of IDA-dAMP in the RT dNTP binding pocket. This figure was generated using Bobscript, Molscript and Raster3d (37–39).
The divalent magnesium ions are located in a very strong network of ionic bonds that involves the non-bridging oxygen of the phosphoramidate group and the three aspartate residues Asp185A, Asp110A and Asp186A, which are highly conserved in the polymerase family (22,23). The ionic bond interaction observed between the divalent cation and residue Asp186A in the ground state is specifically noted in the complex of HIV-1 RT with IDA-dAMP and Asp-dAMP (6,30).
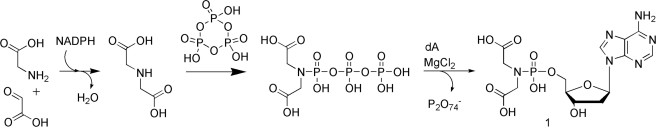
Metabolic accessibility
In order to investigate the metabolic accessibility and recycling potential of a novel triphosphate mimic, we synthesized triphospho-iminodiacetate (TPI) and tested the possibility of displacement of a pyrophosphate moiety by deoxyadenosine (Scheme 2). The triphosphate was formed by the reaction of trimetaphosphate with iminodiacetic acid, by adapting the method described by Feldmann et al. (31). Pyrophosphate displacement reaction with deoxyadenosine in the presence of magnesium ions with release of pyrophosphate was, however, not successful due to poor nucleophilicity of the 5′-hydroxyl group of deoxyadenosine and solubility problems. On the other hand, cleavage of cyclic trimetaphosphate by adenosine in the presence of MgCl2 in neutral conditions was previously observed by Yamagata (32). The presence of a divalent cation that would facilitate the pyrophosphate displacement implies the use of a non-nucleophilic polar solvent. Therefore further experiments are needed to be done to circumvent the solubility problems. Another approach to solve the aforementioned difficulty with pyrophosphate displacement is to develop aptamers/aptazymes to catalyse the reaction.
Scheme 2.
Hypothesized metabolic pathway towards iminodiacetic acid deoxyadenosine phosphoramidate.
CONCLUSION AND PERSPECTIVES
Deoxyadenosine triphosphate analogues in which the pyrophosphate moiety is replaced by L-aspartic acid had previously been shown to act as substrates for DNA polymerisation catalysed by HIV-1 RT. Using iminodiacetate dAMP as a possible substrate improved incorporation and elongation results are obtained. The contribution of the pyrophosphate leaving group to DNA synthesis, in terms of catalytic efficiency and fidelity of DNA polymerase β, was studied recently (33). These data suggest a leaving-group-induced change in the rate-limiting step due to stabilization of pyrophosphate group release. It is clear that the substitution of pyrophosphate group by L-Asp and IDA moieties would be likely to have a major impact on this process. Furthermore, chemical modification of the leaving group influences active-site structural differences between correct and incorrect base-paired transition state (34).
Elongation results with IDA-dAMP allowed us to confirm the positive influence of a longer template overhang for the recognition and incorporation of several successive units carrying a modified leaving group. Therefore, our results validated the importance of interactions between finger subdomain residues and the template’s upstream nucleotides during the HIV-1 RT directed DNA synthesis.
Modelling experiments showed us that in the ground state, the carboxylate groups are most likely to be bound to an intricate network of amino acid residues Lys65, Arg72 and Gln151 located in the enzyme’s active site via divalent cations. Our study suggests that in the ground state the magnesium ion could form an ionic bond with Asp110, Asp185 and Asp186. The last interaction mentioned (Asp186) is observed only in the case of IDA-dAMP and L-Asp-dAMP. This model could suggest that in the ground state, the stabilization of IDA-dAMP is enhanced, involving six amino acid residues instead of four with the natural substrate and five with Asp-dAMP (6). The increase in amino acids interactions observed in the complex formation with IDA-dAMP is not reflected in a change in KM. The electrostatic involvement of residues Gln155A and Asp186A has not been observed in previous studies (6). The difference in binding affinity between pyrophosphate and iminodiacetic acid is likely to contribute to a difference in kinetics (29). In addition, the difference in intrinsic energy of the released leaving group could be a possible ‘brake’ for further conformational changes of the enzyme (26,29).
The nucleoside phosphoramidates developed in this study lend themselves to metabolic implementation, as described in Scheme 2. With the help of the divalent magnesium cation, a further nucleophilic attack of the α-phosphate by the 5′ hydroxyl group of adenosine should form the phosphoramidate 1, with the energetically favoured release of a pyrophosphate. Biosynthesis would start from the two common metabolites glycine and glyoxylate, that is its corresponding 2-oxo-acid. A specific transaminase (serine-glyoxylate aminotransferase) performs interconversion of glycine and glyoxylate in certain methylotrophic bacteria (35). Reductive condensation to glycine with glyoxylate resulting in iminodiacetate is reminiscent to octopin biosynthesis, and enzymes in charge of the production of these nutrient shuttles could be harnessed for our purpose (36,37). Furthermore, ammonia reacts in neutral aqueous media with cyclotriphosphate to yield triphosphoramidates and equilibrium constant of this reaction is about 1.52 min–1 (31). We propose that this reaction could be readily observed in the case of the secondary amine IDA, spontaneously providing TPI in good yield (80% yield after 24 h at room temperature). Putative enzymes capable of accelerating this spontaneous reaction could possibly be found in the class of hydrolases that act on acid anhydrides, such as trimetaphosphatase (EC 3.6.1.2) (38). Our study though showed further condensation of TPI with the 5′ hydroxyl of the nucleoside deoxyadenosine did not proceed spontaneously under the various conditions tested. Nevertheless, since this reaction features the nucleophilic attack of a phosphoanhydride by a primary alcohol as performed by many kinases, the systematic assay of such enzymes can unveil a biocatalyst marginally endowed with the desired activity.
FUNDING
The European Commission Orthosome Project (FP6-NEST-PATHFINDER) and by the K.U. Leuven, Bijzonder Onderzoeksfonds, Geconcerteerde Onderzoeksacties (GOA/10/013). Funding for open access charges: Departmental account.
ACKNOWLEDGMENTS
We would like to thank Prof. Jef Rozenski for providing MS and HRMS analytical data, Prof. Arthur Van Aerschot, Dr Natalia Dyubankova and Dr Marleen Renders for technical and scientific advice, Luc Baudemprez for recording 125 MHz 13C spectra. We are also grateful to Dr. Olga Adelfinskaya and Prof. Susan Cure for careful reading of the manuscript.
REFERENCES
- 1.Herdewijn P, Marliere P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009;6:791–808. doi: 10.1002/cbdv.200900083. [DOI] [PubMed] [Google Scholar]
- 2.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 3.Pochet S, Kaminski A, Van Aerschot A, Herdewijn P, Marlière P. Replication of hexitol oligonucleotides as a prelude to the propagation of a third type of nucleic acid in vivo. C.R. Biol. 2003;326:1175–1184. doi: 10.1016/j.crvi.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Renders M, Lievrouw R, Krecmerova M, Holy A, Herdewijn P. Enzymatic polymerization of phosphonate nucleosides. Chembiochem. 2008;9:2883–2888. doi: 10.1002/cbic.200800494. [DOI] [PubMed] [Google Scholar]
- 5.Adelfinskaya O, Herdewijn P. Amino acid phosphoramidate nucleotides as alternative substrates for HIV-1 reverse transcriptase. Angew. Chem. Int. Ed. 2007;46:4356–4358. doi: 10.1002/anie.200605016. [DOI] [PubMed] [Google Scholar]
- 6.Adelfinskaya O, Terrazas M, Froeyen M, Marliere P, Nauwelaerts K, Herdewijn P. Polymerase-catalyzed synthesis of DNA from phosphoramidate conjugates of deoxynucleotides and amino acids. Nucleic Acids Res. 2007;35:5060–5072. doi: 10.1093/nar/gkm498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrazas M, Marlière P, Herdewijn P. Enzymatically catalyzed DNA synthesis using L-Asp-dGMP, L-Asp-dCMP, and L-Asp-dTMP. Chem. Biodivers. 2008;5:31–39. doi: 10.1002/cbdv.200890013. [DOI] [PubMed] [Google Scholar]
- 8.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 9.Garforth SJ, Parniak MA, Prasad VR. Utilization of a deoxynucleoside dphosphate substrate by HIV reverse transcriptase. PLos One. 2008;3:4, e2074. doi: 10.1371/journal.pone.0002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraut A, Dyubankova N, Song XP, Herdewijn P. Phosphodiester substrates for incorporation of nucleotides in DNA using HIV-1 reverse transcriptase. Chembiochem. 2009;10:2246–2252. doi: 10.1002/cbic.200900270. [DOI] [PubMed] [Google Scholar]
- 11.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis PJ. Molscript – a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 15.Drontle DP, Wagner CR. Designing a pronucleotide stratagem: lessons from amino acid phosphoramidates of anticancer and antiviral pyrimidines. J. Med. Chem. 2004;4:409–419. doi: 10.2174/1389557043403945. [DOI] [PubMed] [Google Scholar]
- 16.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse-transcriptase – contribution of template–primer misalignment, miscoding, and termination probability to mutational hot-spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 17.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 18.Goodman MF, Creighton S, Bloom FB, Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 19.Creighton S, Bloom FB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 20.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal-structure at 3.5 angstrom resolution of HIV-1 reverse-transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Chopra R, VErdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 22.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 23.Boyer PL, Tantillo C, Jacobo-Molina A, Nanni RG, Ding J, Arnold E, Hughes S. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc. Natl Acad. Sci. USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel PH, Jacobo-Molina A, Ding J, Tantillo C, Clark AD, Jr, Raag R, Nanni RG, Hughes SH, Arnold E. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 25.Kew Y, Olsen LR, Japour AJ, Prasad VR. Insertions into the [beta]3-[beta]4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J. Biol. Chem. 1998;273:7529–7537. doi: 10.1074/jbc.273.13.7529. [DOI] [PubMed] [Google Scholar]
- 26.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 27.Martell AE, Schmitt RM. Critical Stability Constants. Vol. 1. New-York, London: Plenum; 1974. [Google Scholar]
- 28.Creighton S, Goodman MF. Gel kinetic-analysis of DNA-polymerase fidelity in the presence of proofreading using bacteriophage-T4 DNA-polymerase. J. Biol. Chem. 1995;270:4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- 29.Götte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SJ. 2009. Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions, Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics, 7 August 2009 (doi:10.1016/j.bbapap.2009.07.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarafianos SG, Das K, Ding JP, Boyer PL, Hughes SH, Arnold E. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem. Biol. 1999;6:R137–R146. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 31.Feldmann W, Thilo E. Zur chemie der kondensierten phosphate und arsenate. (XXXVIII) Amidotriphosphat. Zeit. Anorg. allg. Chem. 1964;328:113–216. [Google Scholar]
- 32.Yamagata Y, Inoue H, Inomata K. Specific effect of magnesium ion on 2',3'-cyclic dAMP synthesis from adenosine and trimeta phosphate in aqueous solution. Origins Life Evol. B. 1995;25:47–52. doi: 10.1007/BF01581572. [DOI] [PubMed] [Google Scholar]
- 33.Sucato C, Goodman MF. Modifying the [beta, gamma] leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase [beta] Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 34.Sucato C, Goodman MF. DNA polymerase [beta] fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry. 2008;47:870–879. doi: 10.1021/bi7014162. [DOI] [PubMed] [Google Scholar]
- 35.Karsten WE, Ohshiro T, Izumi Y, Cook PF. Initial velocity, spectral, and pH studies of the serine-glyoxylate aminotransferase from Hyphomicrobiuim methylovorum. Arch. Biochem. Biophys. 2001;388:267–275. doi: 10.1006/abbi.2001.2294. [DOI] [PubMed] [Google Scholar]
- 36.Thompson J, Donkersloot JA. N-(carboxyalkyl)amino acids – occurrence, synthesis, and functions. Annu. Rev. Biochem. 1992;61:517–557. doi: 10.1146/annurev.bi.61.070192.002505. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Louie TM, Payne J, Bohuslavek J, Bolton H, Xun LY. Identification, purification, and characterization of iminodiacetate oxidase from the EDTA-degrading bacterium BNC1. Appl. Environ. Microbiol. 2001;67:696–701. doi: 10.1128/AEM.67.2.696-701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornberg SR. Tripolyphosphate and trimetaphosphate in yeast extracts. J. Biol. Chem. 1956;218:23–31. [PubMed] [Google Scholar]