Abstract
The ability to engineer genetic programs governing cell fate will permit new safeguards for engineered organisms and will further the biological understanding of differentiation and aging. Here, we have designed, built and implemented a genetic device in the budding yeast Saccharomyces cerevisiae that controls cell-cycle progression selectively in daughter cells. The synthetic device was built in a modular fashion by combining timing elements that are coupled to the cell cycle, i.e. cell-cycle specific promoters and protein degradation domains, and an enzymatic domain which conditionally confers cell arrest. Thus, in the presence of a drug, the device is designed to arrest growth of only newly-divided daughter cells in the population. Indeed, while the engineered cells grow normally in the absence of drug, with the drug the engineered cells display reduced, linear growth on the population level. Fluorescence microscopy of single cells shows that the device induces cell arrest exclusively in daughter cells and radically shifts the age distribution of the resulting population towards older cells. This device, termed the ‘daughter arrester’, provides a blueprint for more advanced devices that mimic developmental processes by having control over cell growth and death.
INTRODUCTION
Control over cell fate is a biological process ubiquitous in nature. In unicellular organisms it is used to control cell division and in multi-cellular organisms it regulates developmental processes. External control of cell fate has been demonstrated using a variety of chemical, genetic and small molecule methods. Small molecules that perturb signaling pathways of stem cells are used to modulate cell fate decisions (1,2) such as differentiation, cell survival and proliferation (3). Alternatively, suicide gene therapy is used to selectively kill cells that have taken up an exogeneous gene, which converts a prodrug to an active drug (4). In bacteria, a more complex genetic feedback circuit based on quorum sensing similarly controls cell fate to enable programmed population control (5,6). However, the described systems above act conditionally depending on population density and/or affect all genetically identical cells, rather than exerting selective control over certain members of a genetically identical population at a specific cell cycle or developmental stage. The ability to read the cell-cycle state and act upon it could allow for more fine-tuned devices that implement conditional programs or tightly time-regulated behaviors.
Cells undergo highly regulated transcriptional, translational and degradation programs to progress through the cell-cycle, and these programs are regulated by modular elements (7,8). We reasoned that we could use rational design of modular elements to design a genetic device to selectively arrest newly formed cells upon a conditional signal in a cell density independent fashion. Living systems are, in principle, organized from building blocks that through their interactions give rise to their inherent complexity. Modularity in nature has led to the idea of re-using genetic parts to form a synthetic biology toolbox comprising different functions that can be interfaced and recombined (9). This toolbox encompasses genetic components such as promoters, functional protein domains, operator regions as well as non-coding regulatory regions such as miRNAs. Parts can then be pieced together to generate circuits encoding commands or tasks that can be carried out in series or in parallel. For example, Chen et al. (10) integrated exogenous synthesis and receptor modules from Arabidopsis thaliana into Saccharomyces cerevisiae’s endogenous protein phosphorylation pathway to implement artificial cell-cell communication in yeast. Also using a modular approach, Ajo-Franklin et al. (11) built a positive feedback loop by assembling novel proteins based on modular protein domains to achieve new functionality. Other efforts in modular designs have been extensively described elsewhere (9,12–15). Despite this explosion of genetic devices, the integration of engineered genetic circuits into native cell-cycle progression remains an unexplored area.
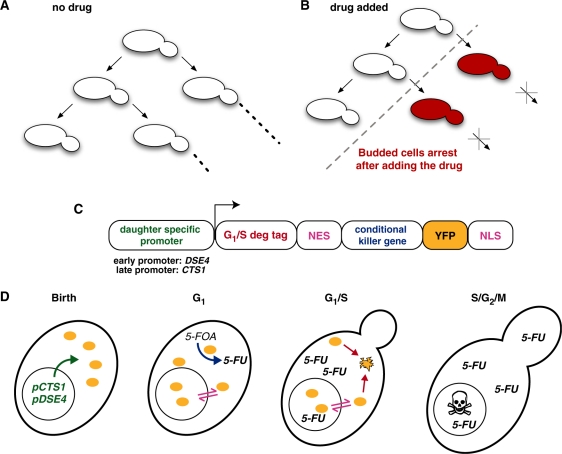
Here we report construction of a ‘daughter arrester’, a device that arrests the growth of daughters while allowing mother cells to grow. To accomplish this, we engineered a drug-sensitive enzyme that is transiently present in yeast daughters during the normal cell-cycle progression. In the absence of the drug, all cells divide normally (Figure 1A), while in the presence of the drug, the enzyme selectively arrests daughter cell growth (Figure 1B). This device constitutes a first successful attempt in building a modular genetic device that integrates information from the endogenous cell-cycle and an external stimulus. Devices that follow this design philosophy have potential application as tools to study aging and as fail-safe devices that are activated based on proliferation rates.
Figure 1.
Design of the daughter arrester. (A) Mother and daughter yeast cells of the daughter arrester strain both divide normally in the absence of drug. In the presence of drug (B), mother cells continue to undergo cell divisions, but daughter cells arrest (red) and fail to divide. (C) The daughter arrester is a gene comprised of a daughter specific promoter (pCTS1 or pDSE4 for the early or late daughter-specific arrester, respectively), a G1/S degradation tag (the N-terminal of Sic1p) fused to the PKI nuclear export sequence (NES), the conditional killer protein (Ura3p), the Venus yellow fluorescent protein (YFP), and the SV40 nuclear localization sequence (NLS). (D) Detail of the engineered strain throughout the cell cycle. The fluorescent conditional kill protein (yellow ovals) is expressed due to daughter specific promoters at birth (panel 1), shuttles between the nucleus and cytoplasm, and converts the pro-drug (5-FOA) to the toxic 5-FU throughout G1 (panel 2). At the G1/S transition (panel 3), this protein is targeted for proteasomal degradation due to the degradation tag (C, in red). As the cell progresses through the cell cycle (panel 4), the daughter cell will arrest due to the incorporation of 5-FU in RNA and DNA.
MATERIALS AND METHODS
Yeast strains
The constructs used to create the early (PSY3651) and late (PSY3652) daughter arrester strains were assembled in sequential fashion using the Biobrick assembly method (16). All promoters or proteins sequences are from S. cerevisiae unless otherwise noted and were subcloned from PSY580 genomic DNA. The CTS1 and DSE4 promoters were subcloned from region −762 to −3 and from region −1233 to −3, respectively, relative to the start codon of their downstream coding sequence. Both promoters were fused to a Kozak sequence containing the start codon, and the coding sequences of the G1/S degradation tag (amino acids 2–160 of Sic1p), PKI nuclear export sequence (NES; amino acid sequence ELALKLAGLDIN), Ura3p (amino acids 2–267), yeast codon optimized Venus yellow fluorescent protein (17,18) and the SV40 NLS (amino acid sequence PKKKRKV), followed by the ADH1 terminator. The final Biobrick constructs were confirmed by sequencing and then subcloned into pRS integrating vectors (19). The pRS vectors were then linearized and integrated into the URA3 mutant PSY580 strain (MATa, ura3-52, trp1Δ63, leu2Δ1) at the TRP1 locus. Integrations were confirmed by colony PCR.
Spectroscopy
Yeast cultures were grown overnight in synthetic complete media, back diluted to an OD600 0.2, grown until OD600 of 0.6, and back diluted again to OD600 0.2. Two milliliters of culture were then transferred to two plastic cuvettes containing magnetic stir bars. The drug 5-fluoroorotic acid (5-FOA) (100 mg/ml in DMSO) (Sigma Aldrich, St. Louis, MO) was added to a final concentration of 1 mg/ml to one of the cultures, an equal volume of DMSO was added to the second, and the OD600 of both cultures was measured every 5 min for 7 h at a constant 30°C with stirring using a Cary 300 spectrophotometer.
Growth assays
Yeast cultures were grown overnight in synthetic complete media, back diluted to an OD600 0.2, grown until OD600 of 0.6, and back diluted again to OD600 0.2. These cultures were serially diluted by factors of 10 and spotted from the leftmost side to the right on plates containing different types of media.
Cell imaging
Time-lapse movies of yeast growth were obtained by seeding and growing cells within a microfluidic device (CellASIC Corporation, Berkeley, CA) according to the manufacturer’s instructions. Time-lapse microscopy acquisition was obtained using Metamorph software using a Nikon TE-2000 inverted microscope with automated stage inside a temperature controlled chamber set to 30°C. The JP2 filter set from Chroma was used for YFP acquisition every 3 min. Image analysis was performed using Metamorph and NIH ImageJ software (20).
RESULTS
Design of daughter arrester
Our goal was to design a gene that would selectively prevent daughter cells from dividing. Thus the protein product of this gene needed to meet design specifications: it would (i) arrest cell growth if and only if the drug were present (otherwise the strain could never propagate), (ii) be expressed only in daughter cells after cytokinesis, but be degraded before the daughter cell completed its first cell division, (iii) be in the appropriate cellular compartments to carry out the above functions, and (iv) be readily detectable. To meet these criteria, we combined genetic modules for a conditionally lethal enzyme with cell-cycle dependent synthesis, cell-cycle dependent degradation, visualization and subcellular localization (Figure 1C) as described in detail below.
Our first major consideration was to use a gene encoding an enzyme capable of interacting with certain substrates, generating a toxic chemical and inducing cell death—we term this the ‘conditional killer gene’. We selected URA3 as the conditional killer gene; it codes for the protein orotidine-5′-phosphate (OMP) decarboxylase which converts 5-FOA into the cytotoxic pyramidine analog 5-fluorouracil (5-FU) (21). 5-FU misincorporates into RNA and DNA in place of uracil or thymine, hampering normal RNA and DNA processing, and leads to cell arrest (22). In strains lacking the URA3 gene, 5-FOA is not converted into 5-FU, and cell growth is not affected (23).
The second key design requirement was to restrict this conditional killer protein to daughter cells. Therefore, we paired a cell-cycle dependent promoter with a cell-cycle dependent protein degradation sequence to limit this protein to daughter cells between G1- and S-phase. To prevent the conditional kill protein from being present past G1 (24,25), we fused the N-terminal region of Sic1p in frame to the conditional kill gene coding sequence. This protein region, a so-called G1/S degradation tag, causes protein fusions (26) to be specifically degraded at the G1/S transition with the same timing as the native Sic1p (27). Since our engineered protein must be present long enough to convert 5-FOA into 5-FU, yet should not be present before G1, we chose two different promoters with slight different temporal patterns of expression to create two versions of our device. While both the CTS1 and DSE4 promoters (28,29) specifically drive gene expression in newly budded cells, CTS1 lags slightly behind DSE4 in driving the expression of any gene coupled to it (29). Hence we term the DSE4 promoter-variant of our device the ‘early daughter arrester’ and the CTS1-variant the ‘late daughter arrester’.
Lastly, we included genetic modules for subcellular localization and visualization in our device. Since OMP decarboxylase is usually a cytosolic protein, yet RNA and DNA processing occur in the nucleus, we reasoned that we could increase the probability of arrest by targeting our fusion protein to both compartments. A partial nuclear localization is also necessitated by our G1/S degradation tag: the degradation of endogenous Sic1 is triggered in the nucleus, where it is phosphorylated by B-type cyclins (27). Thus, the coding sequence for both a nuclear localization signal (NLS) and a nuclear export signal (NES) were fused to the URA3 module. A protein that contains both signals continuously shuttles (30) between the cytoplasm and the nucleus. As a final component, a yeast codon optimized YFP DNA sequence was fused to the Sic1 module in order to quantify the presence of the engineered protein with spatial and temporal resolution.
Assuming these components maintain their modularity, we can build a timeline predicting how our device should selectively arrest daughter cells (Figure 1D). Starting at birth (panel 1), the daughter-specific promoters are up-regulated and result in expression of the fluorescent conditional kill protein (yellow ovals). Throughout G1 (panel 2), this protein will continue to be expressed, will shuttle continuously between the cytoplasm and the nucleus, and will convert 5-FOA (if present) into 5-FU. At the G1/S transition (panel 3), the daughter-specific promoters will no longer be active and the conditional kill protein will be phosphorylated in the nucleus, ubiquitinated, and rapidly degraded by the proteasome. Subsequently (panel 4), the daughter cell will arrest due to incorporation of 5-FU in RNA and DNA. In contrast, mother cells should not express the conditional kill protein and should divide normally.
The daughter arrester conditionally inhibits growth of bulk cultures
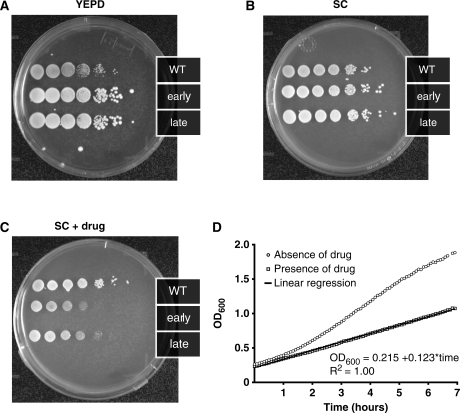
To test whether this genetic device could conditionally arrest cell growth on the population level, the wild-type, early daughter arrester and late daughter arrester strains were grown in liquid culture, serially diluted starting from the same initial cell density, and plated on rich media and synthetic complete plates with and without the added drug 5-FOA (Figure 2A–C). All three strains cells grew to a similar extent when grown on rich YEPD (Figure 2A) or synthetic complete media (Figure 2B), suggesting that the device does not affect growth in the absence of a drug. Likewise, cell growth of the wild-type strain in the presence of drug was not affected (Figure 2C), indicating that the drug does not impact its growth. In comparison, both the early and late daughter arrester strains displayed reduced growth, as demonstrated by the growth of fewer colonies in the serial dilution (Figure 2C). Additionally, the early daughter arrester strain displayed a slower growth rate compared to late daughter arrester strain. Based on these results, we used only the early daughter arrester strain in the subsequent experiments.
Figure 2.
The daughter arrester strain shows reduced growth on the population level only in the presence of drug. Serial dilutions of the parent strain (PSY580), the early daughter arrester (pCTS1, PSY3652) and the late daughter arrester strain (pDSE4, PSY3651) on (A) rich media (YEPD), (B) synthetic complete media (SC) and (C) synthetic complete media containing the drug 5-FOA show that only in the presence of drug, cell growth of the daughter arrester strains is reduced. (D) Biomass growth as measured by OD600 as a function of time for the early daughter arrester strain in the absence (circles) and presence of drug (squares) indicates that normal exponential growth is observed without drug while the predicted linear growth is observed in the presence of drug.
Under normal growth conditions, the biomass and cell density of a yeast cell culture grow exponentially over time after the lag-phase and before stationary-phase. In a population where the newly budded cells arrest their transit through the cell cycle, biomass will be generated in a linear fashion since current mother cells will bud new cells, but these new cells will fail to bud (Figure 1B). The plot of cell growth versus time can be qualitatively described by one of the following behaviors: (i) concave up, typical of a wild-type population during exponential growth; (ii) concave down, expected for a growing population where mother and daughter cells arrest or die and cease to bud; (iii) linear, expected for a growing population where mother cells continuously bud new daughter cells that get arrested. In order to assay growth rate over time we indirectly quantified biomass by measuring light scattering at 600 nm using a spectrophotometer. Measurements were obtained while growing yeast cell cultures in a temperature controlled well-stirred mini-reactor. Figure 2D shows a representative time course of the early daughter arrester strain growing in the mini-reactor over time in the presence and absence of the stimulus. Cells grown in the absence of drug (circles) display exponential growth similar to wild-type cells (data not shown), whereas cells grown in the presence of drug (squares) display reduced growth that fits perfectly to a linear regression [OD600 = 0.215 + 0.123 × time (h), R2 = 1.00]. These observations indicate that engineered genetic device reduces the rate of cell division in a manner that is qualitatively consistent with our design.
The daughter arrester conditionally inhibits growth of daughter cells
In order to determine whether the reduced growth of the populations expressing our genetic device was due to the arrest of the newly budded daughter cells, we followed individual cells in a microfluidic device by differential interference contrast and fluorescence microscopy. The microfluidic format conferred two important advantages: the media composition could be changed within less than 1 min without altering the field of view and growing cells remained in the same focal plane because they are trapped in a 4 µm high region. In a typical experiment, the microfluidic chamber was seeded with cells, the cells were grown for 1 h in synthetic complete media, and then the media was switched to either drug-containing media (t = 0 in Figure 3), and individual cells were tracked over many generations.
Figure 3.
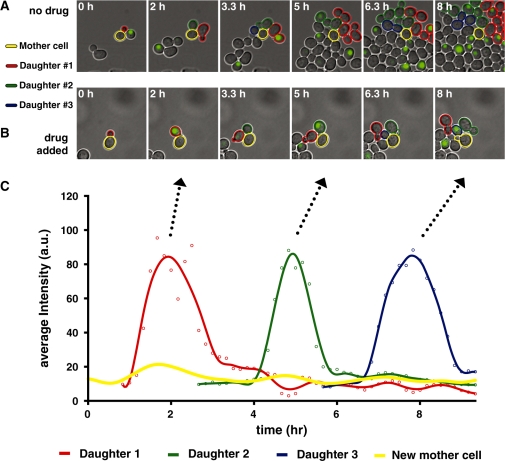
Time-lapse microscopy and single-cell analysis of daughter arrester cells growing in a microfluidic device shows specific arrest of daughter cells only in the presence of drug. Cells were seeded into a microfluidic device and grown in the absence of drug for 1 h before switching to either identical or drug-containing media. (A) When grown in the absence of drug, newly budded cells (outlined in red, green and blue) are fluorescent from G1- to S-phase, and both mothers (outlined in yellow) and daughters progress through the cell cycle. When grown in the presence of drug (B), a mother cell (outlined in yellow) divides multiple times, but its daughter cells (outlined in red, green and blue) do not complete a cell division. (C) Average fluorescence intensity of single cells shown in (B) as a function of time. The peaks of fluorescence intensity have remarkably similar durations as well as magnitudes.
We analyzed representative cell trajectories for the early daughter arrester strain grown in the presence and absence of drug (Figure 3). Figure 3A shows a mother cell (outlined in yellow), its first, second and third daughter cells (outlined in red, green and blue, respectively) and their progeny (outlined in red, green and blue) when grown in the absence of drug. While the mother cell divides several times over the course of the experiment, it never shows significant fluorescence. In contrast, newly budded cells (e.g. first daughter cell outlined in red) rapidly accumulate bright fluorescence centered in the nucleus (Figure 3A, panel 1), which persists until the cell enters into S-phase and then it rapidly disappears and does not reappear (Figure 3A, panels 2–6). This pattern of expression and degradation indicates that the CTS1 daughter-specific promoter and G1/S degradation tag can be combined in a modular fashion to tightly restrict an arbitrary protein to a particular phase of the cell cycle. The predominately nuclear localization of the conditional killer protein is in accordance with previous observations in human fibroblasts that a fusion protein carrying both the PKI NES and the SV40 NLS leads to predominately nuclear localization (31), indicating that in this configuration the NLS prevails. Additionally, both mother and daughter cells of the daughter arrester strain divided many times over the course of the experiment and displayed a growth rate inside the microfluidic device comparable to their wild-type counterparts (∼90 min).
When grown in the presence of the drug, both mother cells and post-G1 daughter cells of the daughter arrester strain are not significantly fluorescent (Figure 3B) and divide at slightly slower rate compared to wild-type cells in the absence of drug (140 versus 90 min, respectively). We speculate that this slower growth could be due to very low expression of conditional killer protein or a bystander effect originating from nearby arrested daughter cells. As in the absence of drug, fluorescence appeared only in newly budded cells, e.g. red cell in panel 2, and then precipitously disappeared as the cells entered into S-phase. However, these cells subsequently arrested as they started to bud new cells, e.g. red cell in panels 3–6, and never completed a cell division over the next ∼14 h, i.e. until the end of the experiment.
In order to quantitatively assess the function of our daughter arrester strain on the population level and to identify possible modes of failure, we analyzed the trajectories of many cells grown and imaged under identical conditions as Figure 3B. Of the daughter cells born in the presence of drug (n = 185), 86% arrested after the G1/S transition and did not complete a cell division during the remainder of the experiment (>30 min). Additionally, a very small subset of the daughter cells died before the G1/S transition. Interestingly, the daughter cells that completed a division displayed noticeably slower growth and in all cases that we observed, their daughters do arrest. Thus we suggest that these cells have divided in spite of significant accumulated damage rather than mutating in such a way that 5-FOA is no longer converted into toxic 5-FU.
Quantifying the average fluorescence intensity of the YFP reporter, which is indicative of the protein concentration, allows a fine-grained examination of device function over multiple cell divisions (Figure 3C). The fluorescence intensity as a function of time for a mother cell and three of its daughter cells is plotted in Figure 3B and C, in where mother cell is circled in yellow and daughter cells are indicated by red, green, and blue outlines, respectively. The mother cell only displays a very small fluorescent signal above its own time averaged background level some minutes before the newly budded cell shows a fluorescent signal. Newly budded cells display a fluorescent signal with a remarkable features that are reproduced over several generations (Figure 3C), such as the peak protein level (∼85 A.U. at hours 2, 5 and 8), the period that the fluorescent protein is present (∼2 h), the rate of protein production and the precipitous degradation as cells go through the G1/S transition (Figure 3C). Additionally, the peaks centered around hours 2, 5 and 8 lack a plateau, indicating that the system never reaches a steady-state before the protein is degraded.
The daughter arrester enriches the population for oldest cells
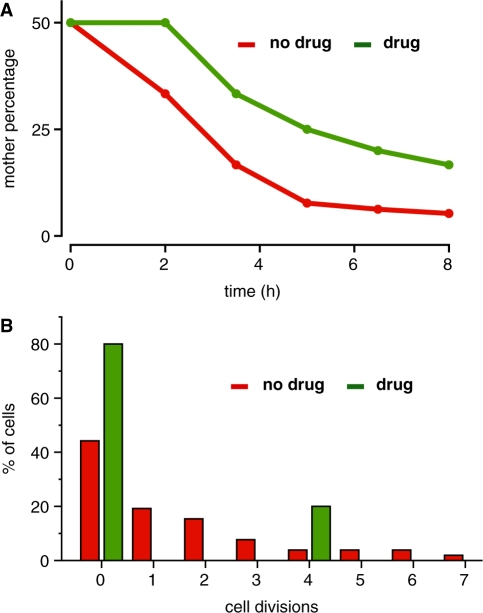
During exponential phase, a wild-type culture of budding yeast will roughly contain 50% new daughter cells, 25% cells that have divided once, 12.5% that have budded twice, etc., so that the oldest n-generational cells should be 100/2n% of the population. While the population distribution of the daughter arrester strain should be unchanged relative to the wild-type strain in absence of drug, it should be significantly altered in the presence of the drug since newly budded daughters do not divide. To explore to what degree our device can enrich for this oldest population of cells, we identified mother–daughter lineages present at the beginning of the microfluidic imaging experiment in either the presence or absence of drug, mapped the number of divisions undergone by these cells and the descendents, and calculated the percentage of the population corresponding to the oldest mother cell. Using the mother–daughter lineage shown in Figure 3A and B, we calculated and plotted in the percentage of the population corresponding to the oldest mother cell as a function of time for representative trajectories. In the absence of drug (Figure 4A, red line), the percentage of oldest cells drops sharply as would be expected. However, in the presence of drug (green line), the fraction of oldest cells is much higher, reflecting the much greater number of oldest cells in the total population. This trend is reproduced over many manually tracked mother–daughter lineages (n = 15 mother cells), confirming that the daughter arrester strain can be used to create populations enriched in old cells.
Figure 4.
The daughter arrester strain can enrich a yeast cell population in older cells. (A) Starting populations of two cells—one mother and daughter—were tracked over time and the percentage of original mother cells (highest replicative age) in the total population was calculated. In the absence of drug (red), the percentage sharply drops over time, while in the presence of drug (green) it decreases significantly less over time. (B) The replicative age distribution after 8 h of cell growth in the absence (green) and presence of drug (red) shows that the age distribution is radically skewed in the presence of drug.
Mapping cellular divisions also reveals that the replicative age distribution of the daughter arrester strain (Figure 4B, analysis of Figure 3A and B) is radically altered in the presence of drug (red) relative to without drug (green). In the absence of drug, the population distribution of the daughter arrester strain closely resembles the expected wild-type distribution. In the presence of drug (Figure 4B, red), the age distribution is bimodal. While most of the cells in the population are arrested daughter cells, the mother cells that budded four times are highly enriched (20%) compared with the population in the absence of drug (4%). Again, these results were qualitatively reproduced over several mother–daughter lineages (n = 15 mother cells), and we never observed a mother cell arrest or die. Thus growing the daughter arrester strain in the presence of drug radically changes the population replicative age distribution, creating one peak population of newly born daughter cells and a second peak corresponding to cells with n number of cell divisions. The number of cell divisions—n—can be manipulated by growing the cells for longer times, e.g. different stages of the time course in Figure 3B.
DISCUSSION AND CONCLUSIONS
Here we have designed and implemented a genetically encoded biological device—the daughter arrester—that can incorporate information from an external stimulus and from the endogenous cell cycle. In the presence of a drug, the daughter arrester strain shows reduced, linear growth on the population level. Furthermore, time-lapse fluorescence microscopy shows that the device reduces the overall growth rate by arresting daughter cells if and only if drug is present. The arrest of daughter cells leads to an enrichment of older mother cells in the population and a time-dependent shift in the replicative age distribution within a population.
Close examination of our device function in the absence of drug reveals new details on the timing and specificity of cell-cycle regulatory elements. We observed very weak, but measurable, expression of the conditional killer protein in mother cells, which we attribute to slight expression from the DSE4 promoter in mother cells. This weak expression most likely arises from imperfect segregation of the transcriptional activator Ace2 or imperfect regulation by the kinase Cbk1 (32,33). In daughters, the conditional killer protein expressed by the DSE4 promoter does not reach a steady-state concentration, indicating that the DSE4 promoter is still active when the G1/S degradation tag triggers degradation of the protein. This information on the relative timing of cell-cycle events in S. cerevisiae is an example of new biology that can be learned from building synthetic devices. The rates of protein production and degradation, and the peak protein concentration (Figure 3C) appear remarkably consistent across single cells. However, a rigorous analysis of this behavior is outside the scope of the paper. We speculate that this apparent lower variability is due to the higher regulation of cell-cycle elements and that with more regular behavior, cell cycle modules may have special utility in synthetic biology.
This genetic device also reveals additional information on the action of URA3 and 5-FU. This device provides a first example that even transiently expressed Ura3p can convert sufficient 5-FOA into 5-FU to cause cell arrest. However, the increased ability of the DSE4-based early daughter arrester to restrict growth relative to the CTS1 late daughter arrester may suggest that the longer residence time of the conditional killer protein translates into a higher toxic effect on cells’ transcriptional and translational mechanism. That the conditional killer protein induces arrest after it has been degraded is also somewhat unexpected. In particular, despite 5-FU toxic-effects on both DNA and RNA processing, it is striking that the cells still past the G1/S checkpoint and enter S-phase, as confirmed by the appearance of a small bud. The cell-cycle program progresses until cells eventually arrest and do not finish the cell cycle. We speculate that the cells are arresting in S/G2 or G2/M, where genome duplication becomes critical to go through the checkpoint.
While our device does indeed arrest growth of daughter cells, it would be desirable to reduce expression of the killer protein in mother cells and simultaneously increase expression of the killer protein in daughter cells. The most straightforward way to achieve this goal would be to make the daughter-specific promoter less leaky in mother cells and a stronger promoter in daughter cells. Since there are several putative daughter-specific promoters in addition to DSE4 and CTS1, creating variants of our device with these alternative promoters might allow us to identify an even more selective device. Alternatively, future experimental approaches might entail using directed evolution coupled with flow cytometry analysis to generate and select artificial promoter variants that have higher expression in daughters and simultaneously lower expression in mothers.
Successful design and construction of this new genetic device expands the limits of modularity used in synthetic biology. Specifically, we show for the first time that cell-cycle specific promoters and degradation sequences can be used as modular timing elements in genetic devices. Importantly, they also accomplish these functions without significantly affecting the overall cell-cycle progression, suggesting that they do not overload the endogenous cell cycle machinery. We also show that these timing devices can be used with exogenous signals, i.e. a drug, to give fine control over device activity. We anticipate that the daughter arrester and related variants will have uses in basic research and biotechnology applications. For example, the daughter specific arrester can be used to study aging in multiple ways. Since the daughter arrester allows mother cells to be tracked by microscopy over many more generations than its parent strain, it will permit examination of the time-evolution of age-related changes to thus elucidate mechanisms underlying aging. Additionally, cell populations enriched in mother cells that have undergone many divisions can be generated in much smaller cell cultures than the current state of the art methods, eliminating a key bottleneck to proteomic analysis of old cells. These novel tools would enable simplified and closer analysis of replicative aging processes. In biotechnology applications, variations of this device could be used as a fail-safe device to prevent cells from proliferating under certain conditions.
FUNDING
PhD fellowship from Fundação para a Ciência e a Tecnologia and GABBA Program (grant number SFRH/BD/15237/2004 to B.A.); Director, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, of the US Department of Energy (Contract No. DE-AC02-05CH11231 to C.A-F.); Experimental design, data analysis and manuscript preparation performed by B.A. Data analysis and manuscript preparation performed by C.A-F. at the Molecular Foundry. Funding for open access charge: Director, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering, of the U.S. Department of Energy [Contract No. DE-AC02-05CH11231 to C.A-F.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Michael Moore for bringing URA3 to our attention and Dirk Landgraf for experimental suggestions and discussions throughout the project.
REFERENCES
- 1.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat. Biotechnol. 2004;22:833–840. doi: 10.1038/nbt987. [DOI] [PubMed] [Google Scholar]
- 2.Hwang K-C, Kim JY, Chang W, Kim D-S, Lim S, Kang S-M, Song B-W, Ha H-Y, Huh YJ, Choi I-G, et al. Chemicals that modulate stem cell differentiation. Proc. Natl Acad. Sci. USA. 2008;105:7467–7471. doi: 10.1073/pnas.0802825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 4.Lal S, Lauer UM, Niethammer D, Beck JF, Schlegel PG. Suicide genes: past, present and future perspectives. Immunol Today. 2000;21:48–54. doi: 10.1016/s0167-5699(99)01550-9. [DOI] [PubMed] [Google Scholar]
- 5.Balagaddé FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 6.You L, Cox RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 7.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 9.Voigt CA. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 2005;23:1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 11.Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EPS, Landgraf D, Phillips I, Silver PA. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle PM, Silver PA. Harnessing nature's; toolbox: regulatory elements for synthetic biology. J. Roy. Soc. Interface / the Royal Soc. 2009;6(Suppl. 4):S535–S546. doi: 10.1098/rsif.2008.0521.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 15.Agapakis CM, Silver PA. Synthetic biology: exploring and exploiting genetic modularity through the design of novel biological networks. Mol. Biosyst. 2009;5:704–713. doi: 10.1039/b901484e. [DOI] [PubMed] [Google Scholar]
- 16.Phillips I, Silver PA. MIT Artificial Intelligence Laboratory, MIT Synthetic Biology Working Group. Cambridge, MA.: Massachusetts Institute of Technology; 2006. [4, February, 2010, date last acessed]. A new biobrick assembly strategy designed for facile protein engineering DSpace. http://hdl. handle. net/1721.1/32535. [Google Scholar]
- 17.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2001;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 18.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 19.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–43. [Google Scholar]
- 21.Jones EW, Fink GR. Regulation of amino acid and nucleotide biosynthesis in yeast. Cold Spring Harbor Monograph Archive. 1982;11:181. [Google Scholar]
- 22.Ko N, Nishihama R, Pringle JR. Control of 5-FOA and 5-FU resistance by Saccharomyces cerevisiae YJL055W. Yeast. 2008;25:155–160. doi: 10.1002/yea.1554. [DOI] [PubMed] [Google Scholar]
- 23.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 24.Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 25.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 26.Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 27.Cross FR, Schroeder L, Bean JM. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics. 2007;176:1541–1555. doi: 10.1534/genetics.107.073494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'C;onallain C, Doolin MT, Taggart C, Thornton F, Butler G. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 1999;262:275–282. doi: 10.1007/s004380051084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 30.Taura T, Schlenstedt G, Silver PA. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J. Biol. Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 31.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 32.Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourens M, Racki W, Becam AM, Panozzo C, Boulon S, Bertrand E, Herbert CJ. Mutations in a small region of the exportin Crm1p disrupt the daughter cell-specific nuclear localization of the transcription factor Ace2p in Saccharomyces cerevisiae. Biol. Cell. 2008;100:343–354. doi: 10.1042/BC20070077. [DOI] [PubMed] [Google Scholar]