Abstract
One of the foundations of synthetic biology is the project to develop libraries of standardized genetic parts that could be assembled quickly and cheaply into large systems. The limitations of the initial BioBrick standard have prompted the development of multiple new standards proposing different avenues to overcome these shortcomings. The lack of compatibility between standards, the compliance of parts with only some of the standards or even the type of constructs that each standard supports have significantly increased the complexity of assembling constructs from standardized parts. Here, we describe computer tools to facilitate the rigorous description of part compositions in the context of a rapidly changing landscape of physical construction methods and standards. A context-free grammar has been developed to model the structure of constructs compliant with six popular assembly standards. Its implementation in GenoCAD makes it possible for users to quickly assemble from a rich library of genetic parts, constructs compliant with any of six existing standards.
INTRODUCTION
The compelling vision of libraries of biological components with standardized interfaces enabling a fast and cheap assembly of large biological systems is one of the foundations of synthetic biology (1,2). The BioBrick Foundation (BBF) has been instrumental in promoting the BioBrick standard. A BioBrick compliant part is a DNA fragment flanked by a prefix and a suffix sequence having specific restriction sites (3,4). Two BioBrick parts can be assembled by using a specific series of restriction digestions and ligations independent of the parts sequences. The different restriction sites used by the prefix and suffix result in complementary overhangs that can be ligated without recreating any of the prefix and suffix restriction sites. The legacy sequence between two adjoining parts is called the scar. BioBrick parts are physically composable in the sense that the assembly of two BioBricks results in a new part compliant with the same standard. The first BioBrick assembly standard, BBa1.0, was proposed by Knight in a BBF Request For Comments (BBF RFC 10). It uses EcoRI, NotI and Xbal in the prefix, and SpeI, NotI and PstI in the suffix. Later on, it has been proposed to replace PstI with SbfI, an enzyme with a longer restriction site less likely to be found in parts sequences (BBF RFC 11). Both standards have been well received by the community and widely used by teams enrolled in the international Genetically Engineered Machine (iGEM) competition (5,6). However, both BBa1.0 and BBa2.0 create an eight-base scar (TACTAGAG), which results in a frame shift when assembling two protein-coding sequences. To address this problem, several new standards have been proposed (BBF RFCs 12, 21, 23 and 25) to allow protein fusion by introducing six-base scars. These standards are summarized in Table 1.
Table 1.
Summary of prefix, suffix and scar groups used in different BioBrick assembly standards
| Standard | Reference | Prefix | Suffix | Scar | Compatible parts |
||||
|---|---|---|---|---|---|---|---|---|---|
| Prom. | RBS | Gene | Ter. | PB | |||||
| BBa1.0 | RFC 10 | EcoRI | SpeI | TACTAGAG | 2166 | ||||
| NotI | NotI | 761 | 149 | 1149 | 98 | 9 | |||
| XbaI | PstI | ||||||||
| BBa2.0 | RFC 11 | EcoRI | SpeI | TACTAGAG | 2166 | ||||
| NotI | NotI | ||||||||
| XbaI | SbfI/PstI | 761 | 149 | 1149 | 98 | 9 | |||
| Biofusion | RFC 23 | EcoRI | SpeI | ACTAGA | 2166 | ||||
| NotI | NotI | ||||||||
| XbaI | PstI | 761 | 149 | 1149 | 98 | 9 | |||
| Freiburg | RFC 25 | EcoRI | AgeI | ACCGGC | 1969 | ||||
| NotI | SpeI | ||||||||
| XbaI | NotI | ||||||||
| Met | PstI | 743 | 148 | 973 | 96 | 9 | |||
| NgoMIV | |||||||||
| BBb | RFC 21 | EcoRI | BamHI | GGATCT | 2019 | ||||
| BglII | XhoI | 636 | 149 | 1112 | 83 | 39 | |||
| Knight | RFC 12 | EcoRI | NheI | GCTAGT | 2140 | ||||
| SpeI | PstI | 724 | 150 | 1159 | 97 | 10 | |||
‘The best thing about standards is that there are so many to choose from!’ summarizes well the difficulty of navigating this increasingly complicated technical landscape. The multiplication of assembly standards creates a number of new difficulties. Most parts are only compliant with some of the assembly standards due to the presence of reserved restriction sites in their sequence. A design framework that could automatically manage the constraints associated with the different standards could help the community better leverage ongoing standardization efforts. Here, we introduce a context-free grammar (CFG) (7) to model the structure of genetic constructs compliant with any of the existing assembly standards. A CFG is a set of rewriting rules, which defines the set of all designs that can be derived by the grammar. A context-free rule can be written as χ→γ, where χ is a single non-terminal and γ is any string of terminals and/or non-terminals (possibly empty). In the case of the BioBrick grammar presented in this article, non-terminals include parts categories (e.g. promoter) and categories of composite parts (e.g. cistron), while terminals are specific BioBricks (e.g. BBa_R0040) and standard-specific prefixes, suffixes and scars. For instance, a rule “Cass1 → Prom1 C1 Cist1 C1 Term1” is interpreted as an expression cassette (Cass1) can be transformed into a DNA sequence comprising a promoter (Prom1), a BioBrick scar (C1), a cistron (Cist1), a BioBrick scar (C1) and a terminator (Term1).
The grammar was implemented in GenoCAD (www.genocad.org), a web-based application to design synthetic genetic constructs (8). GenoCAD is built upon a solid computational linguistic foundation. Yet, its point-and-click graphical user interface enables users to design complex constructs in a matter of minutes. GenoCAD captures design strategies of synthetic genetic constructs in the form of grammatical models. The linguistic models can be used in two ways: a user can design a synthetic construct by successively selecting design rules to transform the structure of the design; or a user can upload a DNA sequence designed outside GenoCAD to validate its consistency with the grammatical model. GenoCAD provides a central parts database with each grammar, and the BioBrick grammar comes with a library of 2312 basic genetic parts available in the Registry of Standard Biological Parts in May 2009. Users, who elect to create a GenoCAD personal account, can log in the system to create project-specific parts libraries, upload new parts into their workspace and save designs for later use.
MATERIALS AND METHODS
A static snapshot of the Registry content is available as a FASTA file at http://partsregistry.org/fasta/parts/All_Parts. For each part, the file includes its identifier, category, a short description and the part sequence. The version of this file published in May 2009 included 9526 parts. A Perl script was developed to parse out the content of this file into structured data format, which could be imported into a MySQL database.
RESULTS
Compliance with different BioBrick standards
The Registry includes both basic parts (e.g. promoter and RBS) and composed parts, which include multiple basic parts (e.g. device, project and composite). As the set of composed parts can be regenerated from the basic parts (9), we only focused on the basic parts which include categories of Regulatory, RBS, Coding, Terminator and, Plasmid Backbone. By querying the MySQL database, we extracted a set of 2312 basic parts with DNA sequences. Because a part is compatible with a BioBrick standard if its sequence does not include any of the restriction sites used by the assembly standard, we developed SQL queries to check for the presence of the restriction sites listed in Table 1.
Interestingly, there are 2166 parts compliant with the BBa1.0, BBa2.0 and Biofusion standards. This observation is not surprising because these three standards use almost identical restriction sites. There are slightly fewer parts available for newly proposed standards like the BBb standard.
Grammar design
The general methodology of developing grammars to model the structure of synthetic genetic constructs has been described elsewhere (7). Here, we highlight the introduction of new rewriting rules and non-terminals that augment the previously described grammars. The full grammar is described in Table 2.
Table 2.
A CFG describing different BioBrick assembly standards
| Rule | Comments | Left term | Right term |
|---|---|---|---|
| 1 | Select a standard (BBa1.0) | S | BBa1.0 |
| 2 | Similar to rule 1 | S | BBa2.0 |
| 3 | Similar to rule 1 | S | Biofusion |
| 4 | Similar to rule 1 | S | Freiburg |
| 5 | Similar to rule 1 | S | BBb |
| 6 | Similar to rule 1 | S | Knight |
| 7 | Transform a standard (BBa1.0) into a plasmid backbone (PB1), a prefix (P1), a cassette (Cass1) and a suffix (S1) | BBa1.0 | PB1 P1 Cass1 S1 |
| 8 | Transform a cassette (Cass1) into two cassettes (Cass1) with a scar (C1) in between | Cass1 | Cass1 C1 Cass1 |
| 9 | Reverse the sequence orientation of a cassette (Cass1) | Cass1 | [Cass1] |
| 10 | Transform a cassette (Cass1) into a promoter (Prom1), a scar (C1), a cistron (Cist1), a scar (C1) and a terminator (Term1) | Cass1 | Prom1 C1 Cist1 C1 Term1 |
| 11 | Transform a cistron (Cist1) into two cistrons (Cist1) with a scar (C1) in between | Cist1 | Cist1 C1 Cist1 |
| 12 | Transform a cistron (Cist1) into a RBS (RBS1), a scar (C1) and a gene (Gene1) | Cist1 | RBS1 C1 Gene1 |
| 13 | Transform a terminator (Term1) into two terminators (Term1) with a scar (C1) in between | Term1 | Term1 C1 Term1 |
| 14 | Similar to rule 7 | BBa2.0 | PB2 P1 Cass2 S2 |
| 15 | Similar to rule 8 | Cass2 | Cass2 C1 Cass2 |
| 16 | Similar to rule 9 | Cass2 | [Cass2] |
| 17 | Similar to rule 10 | Cass2 | Prom2 C1 Cist2 C1 Term2 |
| 18 | Similar to rule 11 | Cist2 | Cist2 C1 Cist2 |
| 19 | Similar to rule 12 | Cist2 | RBS2 C1 Gene2 |
| 20 | Similar to rule 13 | Term2 | Term2 C1 Term2 |
| 21 | Similar to rule 7 | Biofusion | PB3 P3 Cass3 S3 |
| 22 | Similar to rule 8 | Cass3 | Cass3 C3 Cass3 |
| 23 | Similar to rule 9 | Cass3 | [Cass3] |
| 24 | Similar to rule 10 | Cass3 | Prom3 C3 Cist3 C3 Term3 |
| 25 | Similar to rule 11 | Cist3 | Cist3 C3 Cist3 |
| 26 | Similar to rule 12 | Cist3 | RBS3 C3 Gene3 |
| 27 | Transform a gene (Gene3) into two genes (Gene3) with a scar (C3) in between, i.e. protein fusion | Gene3 | Gene3 C3 Gene3 |
| 28 | Similar to rule 13 | Term3 | Term3 C3 Term3 |
| 29 | Similar to rule 7 | Freiburg | PB4 P4 Cass4 S4 |
| 30 | Similar to rule 8 | Cass4 | Cass4 C4 Cass4 |
| 31 | Similar to rule 9 | Cass4 | [Cass4] |
| 32 | Similar to rule 10 | Cass4 | Prom4 C4 Cist4 C4 Term4 |
| 33 | Similar to rule 11 | Cist4 | Cist4 C4 Cist4 |
| 34 | Similar to rule 12 | Cist4 | RBS4 C4 Gene4 |
| 35 | Similar to rule 27 | Gene4 | Gene4 C4 Gene4 |
| 36 | Similar to rule 13 | Term4 | Term4 C4 Term4 |
| 37 | Similar to rule 7 | BBb | PB5 P5 Cass5 S5 |
| 38 | Similar to rule 8 | Cass5 | Cass5 C5 Cass5 |
| 39 | Similar to rule 9 | Cass5 | [Cass5] |
| 40 | Similar to rule 10 | Cass5 | Prom5 C5 Cist5 C5 Term5 |
| 41 | Similar to rule 11 | Cist5 | Cist5 C5 Cist5 |
| 42 | Similar to rule 12 | Cist5 | RBS5 C5 Gene5 |
| 43 | Similar to rule 27 | Gene5 | Gene5 C5 Gene5 |
| 44 | Similar to rule 13 | Term5 | Term5 C5 Term5 |
| 45 | Similar to rule 7 | Knight | PB6 P6 Cass6 S6 |
| 46 | Similar to rule 8 | Cass6 | Cass6 C6 Cass6 |
| 47 | Similar to rule 9 | Cass6 | [Cass6] |
| 48 | Similar to rule 10 | Cass6 | Prom6 C6 Cist6 C6 Term6 |
| 49 | Similar to rule 11 | Cist6 | Cist6 C6 Cist6 |
| 50 | Similar to rule 12 | Cist6 | RBS6 C6 Gene6 |
| 51 | Similar to rule 27 | Gene6 | Gene6 C6 Gene6 |
| 52 | Similar to rule 13 | Term6 | Term6 C6 Term6 |
Terminals are italicized. P, C and S are terminals representing prefix, scar and suffix, respectively. As BBa2.0 uses the same prefix and scar as BBa1.0, there is no P2 and C2 in the grammar.
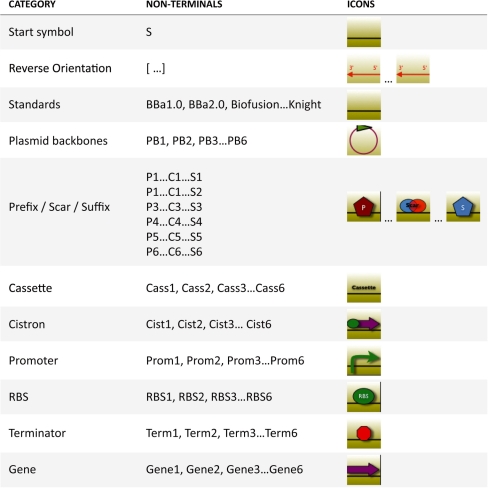
Figure 1 lists the non-terminals along with the icons used for their graphical representation. S is the start symbol used to initiate the design process. In order to ensure the consistency of a design with a specific standard, it is necessary to introduce for each category of parts a different non-terminal for each standard. For instance, instead of having a single non-terminal for genes, we defined the non-terminal Gene1 to represent genes compliant with the BBa1.0 standard, Gene2 for genes compliant with BBa2.0 standards and so on. Non-terminals P, C and S were introduced to represent the prefixes, scars and suffixes of different standards. Non-terminals PB1–PB6 represent the plasmid backbone. Finally, we used non-terminals that do not correspond to specific DNA sequences. A class of non-terminals is used to represent the different assembly standards. Square brackets are introduced to represent that part of a construct is coded on the reverse strand of the DNA molecule, as illustrated in Figure 2.
Figure 1.
Correspondence between parts categories, non-terminals and icons used to graphically represent construct structures.
Figure 2.
Three different representations of a BBa2.0 design. (A) This design includes two gene expression cassettes in opposite orientations. The first icon represents the construct backbone. The icons P1 (second to the left) and S2 (last) represent the construct prefix and suffix. The brackets [and] indicate the reverse orientation of the first cassette. Because BBa1.0 and BBa2.0 share the same prefix and scars, the design includes P1, C1 and S2. (B) The sequence generated by the grammar includes the special characters [ and ] to indicate the fragment in reverse orientation in bold characters. (C) The sequence of the construct is generated by replacing the sequence in bold character by its reverse complement.
Table 2 lists all the production rules of the six standards. Rules P1–P6 specify the assembly standard the design complies with. Rules P7, P14, P21, P29, P37 and P45 specify that a design is composed of a plasmid backbone and a gene expression cassette flanked by the standard prefix and suffix. P8, P15, P22, P30, P38 and P46 allow a single cassette to be transformed into two cassettes with a scar in the middle. Applying these rules n times will create n + 1 cassettes in the design. P9, P16, P23, P31, P39 and P47 can be used to reverse the orientation of a cassette. P10, P17, P24, P32, P40 and P48 define the structure of a cassette to be a promoter, a cistron and a terminator, separated by scars. P11, P18, P25, P33, P41 and P49 allow multiple cistrons in a cassette. P12, P19, P26, P34, P42 and P50 specify that a cistron is composed of a RBS, a scar and a gene. P13, P20, P28, P36, P44 and P52 allow introducing multiple terminators. As BBa1.0 and BBa2.0 both use an eight-base scar (TACTAGAG), which results in the frame shift, protein fusion is not permissible. However, the other standards use six-base scars (such as ACTAGA for the Biofusion standard) compatible with in-frame fusion of protein-coding parts. The grammar reflects this fact by having rules P27, P35, P43 and P51 for protein fusion while using those standards.
GenoCAD implementation
We imported all the basic parts present in the Registry of Standard Biological Parts into the GenoCAD-backend database. We also implemented the BioBrick grammar in GenoCAD.
The large number of parts included in the BioBrick parts library may be difficult to navigate when working on a specific project. After they have logged into the system, users can customize their workspace by adding new parts and creating new parts libraries. When starting a project, it is suggested to first create a parts library for the project. This parts library should contain all the parts needed for the project. Most parts will be imported from the general BioBrick library. However, if there is a need for additional parts, it is possible to define new parts and include them in the project parts library.
Once the project library is complete, the design phase can start. After selecting the BioBrick grammar, the project-specific parts library can be selected. The construct design proceeds through a series of rewriting operations corresponding to the selection of specific grammar rules. The BioBrick grammar first prompts the user to select a particular assembly standard and then a cloning vector. The design then proceeds through a series of steps to specify the structure of the constructs and specific parts to implement this structure. A more detailed description of the design workflow and GenoCAD various features have been published recently (8).
The recent multiplication of assembly standards has led to new design challenges. When all parts complied with a single standard, it was very straightforward to combine them. Now, it becomes necessary to verify that all parts used in a project are consistent with the standard selected for the project. GenoCAD structured approach to the design of genetic constructs makes it possible to gracefully navigate complex libraries of genetic parts compliant with multiple assembly standards. Once a standard has been selected, only the parts compatible with this standard are available to the designer. The construct prefix and suffix along with the scars are properly represented along with the sequence of the cloning vector used to propagate the design.
Designing an iGEM project using GenoCAD
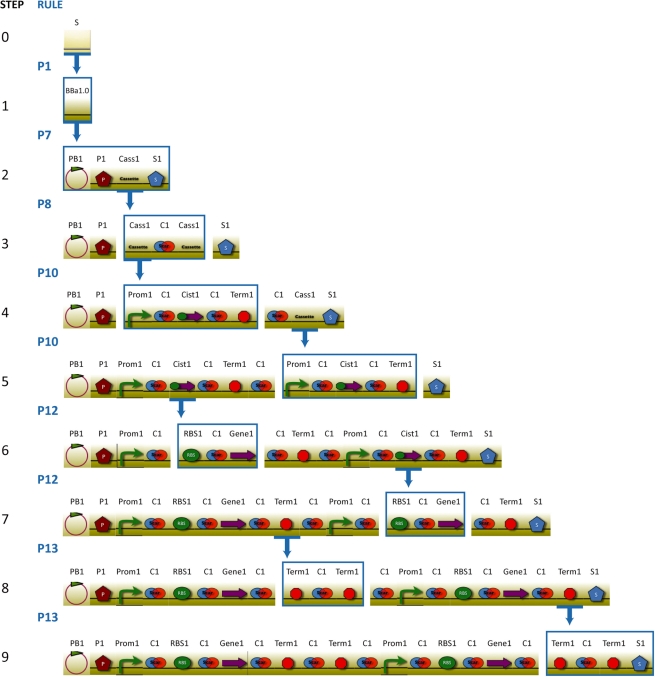
To demonstrate how to use GenoCAD and the BioBrick grammar to quickly design an iGEM project, we selected the wintergreen odor biosynthetic system (http://bit.ly/85Hhgd) designed and implemented by the MIT iGEM team in 2006. The system contains two expression cassettes: one produces salicylate acid from the cellular metabolite, and the second one converts the salicylic acid to methyl salicylate that produces the wintergreen odor. We designed this system with the BBa1.0 assembly standard using GenoCAD. The step-by-step design process is depicted in Figure 3. The design process starts with selecting the BBa1.0 assembly standard (P1); P7 is used to transform the design into a plasmid backbone, a prefix, a cassette and a suffix; as there are two cassettes needed in the wintergreen odor biosynthetic system, P8 is applied to allow two cassettes in the design; by applying P10 to both cassettes, we specified the structure of each cassette to be a promoter, a scar, a cistron, a scar and a terminator; by applying P12 to each cistron, the structure of a cistron is expanded to a RBS, a scar and a gene; finally, we used P13 to allow the usage of a double terminator in each cassette, which ensures a tight transcription termination. After specifying the structure of the design, the last step is to select a specific part for each category, and the DNA sequence of the design is ready for export as a text file.
Figure 3.
Step-by-step design process of a wintergreen odor system using GenoCAD. The construct is designed in nine steps. For each step, the rewriting rule used is indicated in blue in the second column using the same number as in Table 2. The rewriting resulting from the rule selection is indicated in the graphical representation of the construct. The icon underlined by the base of the arrow indicates the left term of the rule. The icons enclosed in a blue rectangle correspond to the right term of the rule. For instance, applying the rule P8 to Cass1 in step 2 transforms this element into Cass1 C1 Cass1 in step 3.
DISCUSSION
Data exchange
The method used to import in GenoCAD data from the Registry suffers from a number of obvious limitations. A live connection between parts registries has been envisioned for some time. The recently launched BioBrick Parts Catalog (www.biobrickparts.org) provides an API to read its content using the JavaScript Object Notation (JSON). We are also working on the development of web services allowing other clients to communicate with the GenoCAD database.
However, setting up web services to access databases solves only part of the data exchange challenge. As data are available easily, it will become apparent that the nature of the data exchanged needs to be documented. It is safe to assume that all registries will associate a unique identifier, a DNA sequence and a description with the parts. The description of the nature of parts is a more difficult issue. The Registry of Standard Biological Parts, the BioBrick Parts Catalog and GenoCAD use their own system of categories, but these categorization systems are developed independently of each other making it difficult to map categories of one resource into categories used by another system. This problem can be solved by the development of an ontology giving the community a common controlled vocabulary to describe genetic parts. Early efforts to develop the Synthetic Biology Open Language have been somewhat hampered by the magnitude of the task. In particular, it is difficult to properly appreciate the scope of what needs to be described by this language. It is also challenging to evaluate the possibility of using existing ontologies like the Sequence Ontology (10) for this new application.
Ensuring that parts are properly delimited at the DNA sequence level is another challenge. The BioBrick grammar presented in this article carefully handles the fusion of coding sequences when using assembly standards allowing this type of construct. However, the possible inclusion of a stop codon in the sequence of genes may prevent the actual fusion of two adjacent proteins. It is, therefore, necessary to set standards to delimit the DNA sequences of different categories of parts (BBF RFC 13).
Advantages and limitations of the BioBrick grammar
The syntactic model proposed in this article constrains the design space of BioBrick-based systems. The point-and-click approach to the design process makes it easy for someone to quickly design constructs compliant with any of the proposed standards. The design strategy embedded in the grammar is very conservative to maximize the chances of designing functional systems. However, GenoCAD currently excludes some ‘out of the box’ designs, e.g. an expression cassette with multiple promoters. Advanced users can overcome this limitation by creating new parts in their personal workspace. For instance, it is possible to use a sequence editor to combine the sequence of two promoters and then save it in GenoCAD as a regular promoter.
Domain-specific languages like Eugene (http://sourceforge.net/projects/eugene) or GEC (11) provide users with richer frameworks and greater design flexibility, but these programming environments may have steeper learning curves than GenoCAD. The Registry of Standard Biological Parts or Gene Designer (12) provides the ultimate design flexibility by allowing users to combine any parts in any order, but the lack of verification or guidance creates more opportunities for design errors that will manifest only when the part is fabricated or characterized.
Fabrication
GenoCAD and the BioBrick grammar described in this article do not provide users with a path to fabrication, but it generates the theoretical DNA sequence of a design that can be used to analyze sequencing data collected to verify physical implementations of a design.
It is fairly common for expression vectors to include expression cassettes in opposite orientations (13) as this configuration limits interferences between promoters. The BioBrick grammar allows users to select the orientation of gene expression cassettes. The reverse complementation operation, necessary to generate the final sequence, includes a reverse complementation of the scar sequences between parts. The final sequence is identical to the sequence of a cassette first assembled in a direct orientation and then flipped before its insertion into cloning vector. Because most parts are defined in the same orientation in the various registries, this scenario is the most likely assembly strategy, but other strategies leading to different DNA sequences can be imagined.
Choosing an assembly standard is only one element in the development of an assembly strategy. The availability of clones, representing physical implementations of various elements of the design, guides a fabrication process that often includes de novo synthesis steps and assembly of existing DNA fragments using various cloning techniques (14). Note that the choice of a particular assembly standard does not automatically restrict the user to a specific assembly process. As they do not rely on restriction enzymes, USER fusion (BBF RFC 39) (15,16) or In-Fusion cloning (BBF RFC 26) (17) are compatible with any assembly standard. The determination of an optimal assembly process can be solved by dynamic programming algorithms (18).
GenoCAD for iGEM
GenoCAD provides users having limited domain expertise with a user-friendly environment to quickly design structurally valid BioBrick constructs compliant with different assembly standards. Students enrolled in the iGEM competition represent an important group of potential users, and the BioBrick grammar has been developed with this group in mind. By importing parts available in the Registry, reusing the system of categories used by the Registry, capturing physical and basic functional composition rules, the BioBrick grammar customizes the GenoCAD environment for the needs of iGEM participants. As a result, any curation of the data imported from the registry has been avoided.
GenoCAD is part of a quickly growing arsenal of software tools for synthetic biology (19–22). It has been recently proposed to use attribute grammars, an extension of the CFG formalism used in this report, to develop semantic models of DNA sequences (23). Embedding this formalism in GenoCAD will enable users to translate their designs into SBML files describing their expected behavior. This capability will make it possible to investigate the possible influence on gene expression of the scars associated with the different assembly standards. It was initially assumed that scars would not significantly influence the phenotype coded in a genetic design, but rapid progress in the characterization of the relations between structure and functions of ribosome-binding sites (24,25) and promoters (26,27) may contribute to re-evaluate this hypothesis.
FUNDING
National Science Foundation Award EF-0850100; Genetics, Bioinformatics and Computational Biology interdisciplinary graduate program at Virginia Tech (a graduate fellowship to Y.C.). Funding for open access charge: National Science Foundation Award EF-0850100.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Authors acknowledge Jenny Wang for assisting in the preparation of the figures and the BBF and the iGEM community for contributing the RFCs and BioBricks parts upon which this project was developed.
REFERENCES
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R. Engineering life: building a fab for biology. Sci. Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 3.Arkin A. Setting the standard in synthetic biology. Nat. Biotechnol. 2008;26:771–774. doi: 10.1038/nbt0708-771. [DOI] [PubMed] [Google Scholar]
- 4.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 5.Smolke CD. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009;27:1099–1102. doi: 10.1038/nbt1209-1099. [DOI] [PubMed] [Google Scholar]
- 6.Goodman C. Engineering ingenuity at iGEM. Nat. Chem. Biol. 2008;4:13. doi: 10.1038/nchembio0108-13. [DOI] [PubMed] [Google Scholar]
- 7.Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23:2760–2767. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- 8.Czar MJ, Cai Y, Peccoud J. Writing DNA with GenoCAD. Nucleic Acids Res. 2009;37:W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peccoud J, Blauvelt MF, Cai Y, Cooper KL, Crasta O, DeLalla EC, Evans C, Folkerts O, Lyons BM, Mane SP, et al. Targeted development of registries of biological parts. PLoS ONE. 2008;3:e2671. doi: 10.1371/journal.pone.0002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen M, Philipps A. Toward programming languages for synthetic biology. J. Roy. Soc. Interface. 2009;6:S437–S450. doi: 10.1098/rsif.2008.0516.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 14.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Smith C, Day PJ, Walker MR. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and application for cloning into restriction digest-linearized vectors. PCR Methods Appl. 1993;2:328–332. doi: 10.1101/gr.2.4.328. [DOI] [PubMed] [Google Scholar]
- 16.Rashtchian A, Buchman GW, Schuster DM, Berninger MS. Uracil DNA glycosylase-mediated cloning of polymerase chain reaction-amplified DNA: application to genomic and cDNA cloning. Anal. Biochem. 1992;206:91–97. doi: 10.1016/s0003-2697(05)80015-6. [DOI] [PubMed] [Google Scholar]
- 17.Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Densmore D, Hsiau THC, Batten C, Kittleson JT, DeLoache W, Anderson JC. Algorithms for automated DNA assembly. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq165. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandran D, Bergmann FT, Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J. Biol. Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchisio MA, Stelling J. Computational design of synthetic gene circuits with composable parts. Bioinformatics. 2008;24:1903–1910. doi: 10.1093/bioinformatics/btn330. [DOI] [PubMed] [Google Scholar]
- 21.Marchisio MA, Stelling J. Computational design tools for synthetic biology. Curr. Opin. Biotechnol. 2009;20:479–485. doi: 10.1016/j.copbio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Hill AD, Tomshine JR, Weeding EM, Sotiropoulos V, Kaznessis YN. SynBioSS: the synthetic biology modeling suite. Bioinformatics. 2008;24:2551–2553. doi: 10.1093/bioinformatics/btn468. [DOI] [PubMed] [Google Scholar]
- 23.Cai Y, Lux MW, Adam L, Peccoud J. Modeling structure-function relationships in synthetic DNA sequences using attribute grammars. PLoS Comput. Biol. 2009;5:e1000529. doi: 10.1371/journal.pcbi.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]