Abstract
De novo gene and genome synthesis enables the design of any sequence without the requirement of a pre-existing template as in traditional genetic engineering methods. The ability to mass produce synthetic genes holds great potential for biological research, but widespread availability of de novo DNA constructs is currently hampered by their high cost. In this work, we describe a microfluidic platform for parallel solid phase synthesis of oligonucleotides that can greatly reduce the cost of gene synthesis by reducing reagent consumption (by 100-fold) while maintaining a ∼100 pmol synthesis scale so there is no need for amplification before assembly. Sixteen oligonucleotides were synthesized in parallel on this platform and then successfully used in a ligation-mediated assembly method to generate DNA constructs ∼200 bp in length.
INTRODUCTION
The ability to synthesize genes and genomes is a powerful tool with a broad cross-section of applications, including the study of large sets of genes, the design of genetic circuitry, and the engineering of metabolic pathways for the production of small molecules. The basic concept of de novo gene synthesis involves taking a pool of short oligonucleotides and assembling them into larger constructs using some form of polymerase chain assembly or ligase chain reaction approach (1,2). Traditional methods of solid-phase oligonucleotide synthesis (3,4), where each oligonucleotide is synthesized individually on a separate column or in a multi-well plate, produce high yields but are costly. For the assembly of long genes or even whole genomic sequences, the cost of the starting oligonucleotides alone can be prohibitive. Smaller-scale synthesis strategies are needed to bring down the cost of starting oligonucleotides before de novo gene synthesis will be widely adopted.
The development of optical and electro-chemical deprotection strategies have led to commercially available DNA microarrays (5). Depending on the chip platform being used, several thousand to several hundred thousand distinct oligonucleotides can be synthesized on a single chip. In principle, these massive parallel microarrays can reduce the cost of oligonucleotides by orders of magnitude. However, microarrays produce very small amounts of oligonucleotides (107–108 molecules per sequence) (6). Such low yields of oligonucleotides are insufficient to drive a gene assembly reaction and require additional polymerase chain reaction (PCR) steps, which add complexity. Another problem associated with microarray synthesis technology is that all the oligonucleotides must be cleaved off the chip as a single mixture. While the ability to synthesize thousands or even hundreds of thousands of oligonucleotides on a single chip is beneficial, the sheer number of different strands synthesized on a chip requires careful planning to handle the complexity of all the possible interactions from the various sequences in the mixture (7). Finally, in some cases oligonucleotides obtained from microarray technologies can be of a lower quality compared to existing solid-phase strategies due to a deblocking step that is inefficient.
Here, we report a programmable microfluidic synthesis platform that addresses many of the limitations inherent with current microarray DNA synthesis technologies. Using conventional solid-phase chemistry, this device is capable of synthesizing ∼100 pmol of an oligonucleotide sequence per column while consuming <250 nl of 0.1 M phosphoramidite solution per column in each reaction cycle. This is a substantial reduction (∼100-fold) in reagent consumption compared to other solid-phase synthesis technologies while keeping the product on a scale large enough to not require subsequent amplification steps. The microfluidic architecture with integrated valves allows one to individually manipulate and collect products from each column. Sets of oligonucleotides with potentially disruptive interactions can be separated into different pools to avoid problems in gene assembly. Furthermore, this platform lends itself to further integration with other processes including gene assembly, cloning, or purification on chip (7–9). As a proof of principle, we synthesized 16 oligonucleotides on a single device; without further purification, we successfully used these sequences to assemble a DNA construct ∼200 bp long.
MATERIALS AND METHODS
Microfluidic oligonucleotide synthesizer
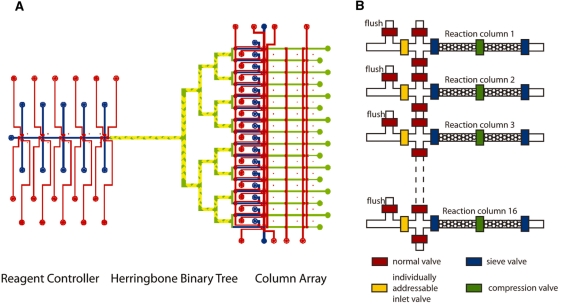
The parallel microfuidic DNA synthesizer, shown in Figure 1, was fabricated using a standard multilayer soft lithography (MSL) method (10,11). The device can be roughly divided into three components. The front component handles all the reagents. In each synthesis step, the valve for the desired reagent opens while others remain closed, delivering the appropriate reagent by means of back pressure. The middle component consists of a binary tree which produces a uniform flow rate across all reaction columns. The herringbone topology within the binary tree facilitates proper mixing of reagents as shown in Supplementary Movie M1. The total volume of the binary tree is 1.49 µl. The end component consists of a 16-column array. The column array was designed to allow parallel loading of solid supports. Purge lines were implemented to avoid cross contamination between synthesis steps by properly purging residual reagent within the binary tree. Each reaction column measures 4500 µm long, 200 µm wide and 30 µm tall to give a column volume of 27 nl. Additional sieve valves (denoted as compression valves) were added to the middle of the columns to prevent rearrangement of closely packed spherical beads when a pressure drop occurs between synthesis steps, minimizing bead loss. Over 95% of resin was retained after 40 cycles (20 h) of synthesis.
Figure 1.
(A) Schematic diagram of a 16-column microfluidic DNA synthesizer. The control lines are shown in red, the fluidic lines in blue, the herringbone mixers in yellow, and the square profiled binary tree and reactor columns in green. (B) Close up schematic of the column array. The red and yellow valves close fully when activated; the blue and green valves are sieve valves that allow fluid flow while trapping CPG beads. Each column has a flush valve that opens and closes in unison to allow residual reagent in the binary tree to be washed away between steps to avoid contamination. The addressable inlet valve controls which columns the reagents enter. The compression valves prevent rearrangement of closely packed spherical beads when a pressure drop occurs between steps.
Device preparation and operation
Microfluidic devices were stored in a desiccator overnight before use. Glass vials filled with molecular sieves and steel tubes (New England Small Tube Co., Litchfield, NH) were dried in a 180°C oven overnight and allowed to cool to room temperature 15 min prior to use. The control lines were interfaced with computer-controled solenoid valves (Pneumadyne, Plymouth, MN) through steel tubes connected to Tygon tubing. The control lines were primed with Krytox oil through dead-end filling. All reagents were prepared fresh prior to experiments. Glass vials filled with reagent were connected to the device via microbore PTFE tubing through the septa. Control porous silica beads (5 µm, 500 Å) functionalized with universal linker (SynGen) were used to construct the bead columns as described in Supplementary Figure S1. Both reagent delivery (10 psi for acetonitrile, 5 psi for everything else) and valve actuation (35 psi) were controled using argon gas.
Microfluidic oligonucleotide synthesis
The synthesis used standard phosphoramidite chemistry (12,13) except for the deprotection step, where 10% trifluoroacetic acid in acetonitrile was used to ensure solvent compatibility with polydimethylsiloxane (PDMS). Short oligonucleotides ∼15–25-nt in length can be synthesized without any capping step. Oligonucleotides >40 nt in length required a capping step to achieve high yield of full-length product. Each synthesis cycle contained seven reaction steps and four washing steps. The step-sequence was deblocking (1.8 min), washing (4 min), coupling T (2.5 min), coupling G (2.5 min), coupling A (2.5 min), coupling C (2.5 min), washing (1 min), capping (2.5 min), washing (2 min), oxidizing (2 min), washing (2 min). An extended deblocking condition of 2.5 min instead of the normal 1.8 min was used for the initial removal of the dimethoxytrityl (DMT) group from the secondary hydroxyl group of the universal linker. The initial coupling step to the universal linker was run for 3 min instead of the normal 2.5 min. During the coupling step, the two coupling reagents (phosphoramidite and activator) were sent through the reaction columns alternately with one reagent flowed continuously for 0.27 s followed by the other for 0.27 s before cycling. The final deblocking of the finished oligonucleotides was done by flowing deblocking reagent into reaction columns for 2 min. After an extended acetonitrile wash, we released the ‘column valves’ and flushed the beads out of the chip and into micro-centrifuge tubes. The acetonitrile was evaporated using centri-vap. In the final cleavage step, 75 µl concentrated ammonium hydroxide was added into the tubes and then incubated at 55°C for 12 h. The beads were centrifuged and then the liquid phase was transferred into new tubes and lyophilized to yield oligonucleotides as pellets (with salts).
Polyacrylamide gel electrophoresis
The synthesized oligonucleotides were re-suspended in water and normalized to a concentration of 50 ng/µl using a NanoDrop 1000 (Thermo Scientific). Before gel loading, the oligonucleotides were mixed with TBE–Urea sample loading buffer (Invitrogen) and heated to 70°C for 3 min. Samples of 1.5 µl were loaded onto polyacrylamide gel (15% Novex® TBE–Urea gel, Invitrogen). Samples ∼20 nt in length were run at room temperature at 175 V for 75 min. Samples ∼40-nt in length were run under the same condition but for 100 min. The gels were stained with SYBR Gold dye and visualized using Typhoon 9410 (Amersham Biosciences). Final images were analyzed using ImageQuant (Amersham Biosciences).
Design of oligonucleotides for gene synthesis
A gene fragment from Bacillus cereus was selected for synthesis through a ligation mediated assembly method. Oligonucleotide sequences were derived from a custom-developed program that first divides the sequence into oligonucleotides with a desired melting temperature range while keeping track of the melting temperature variation between the pieces. The set of pieces with the least variation in melting temperature was chosen. The program then generated the sequence of the sense strand by stitching every two pieces together, and the anti-sense strand using the same method but with an offset of a single piece. A summary of the oligonucleotide set is provided in Table 1.
Table 1.
Oligonucleotide sequences used for the ligation-mediated assembly of a gene fragment of B. cereus
| Forward sequence | Reverse sequence | ||
|---|---|---|---|
| F1 | GTATACTTCCAATCCAATGCA | R1 | TCGCTTTCATTGCATTGGAT |
| F2 | ATGAAAGCGAAGAAAAAAGATAAA | R2 | ATATTCAGAAGTTTTATCTTTTTTCT |
| F3 | ACTTCTGAATATCAGTACGTTG | R3 | ACCATGCGTCAACGTACTG |
| F4 | ACGCATGGTACTCTAATCAG | R4 | CGGCCGTTCTGATTAGAGT |
| F5 | AACGGCCGTAGCATCCC | R5 | ACGTTTCCACGGGATGCTA |
| F6 | GTGGAAACGTATCCCGTCC | R6 | CACTTCGCTGGACGGGAT |
| F7 | AGCGAAGTGAAACAGTTCCA | R7 | CACCGGTCTGGAACTGTTT |
| F8 | GACCGGTGAGGCGTTCA | R8 | ACAGTTGAAGTTGAACGCCT |
| F9 | ACTTCAACTGTTTCGCAACTGTTCAGCGTTTC |
The similar starting sequences of F3 and F9 that led to side products during the assembly are in bold.
Ligation assembly of DNA constructs
Each oligonucleotide strand (8 µl, 5 µM) was mixed with 1 µl of T4 polynucleotide kinase reaction buffer (New England Biolabs) and 1 µl of 10 mM ATP, then cooled to 4°C. T4 polynucleotide kinase (0.7 µl) was then added. The mixture was allowed to react for 1 h at 37°C and then terminated by heating the reaction mixture to 65°C for 15 min. The phosphorylated oligonucleotides were then added together and diluted into a 75 nM solution. One microliter of Taq DNA ligase reaction buffer and 0.75 µl of Taq DNA ligase were added to 9 µl of phosphorylated oligonucleotide solution. The mixture was heated to 65°C for 30 s to ensure all the oligonucleotides were melted off. The ligation reaction was then allowed to take place at 42°C for 4 h.
PCR of ligation product
Polymerase chain reaction was performed with 100 µl of reaction mixture including 10 µl PCR buffer, 3 µl of 50 mM MgCl2, 2 µl of Primer A1 (10 µM), 2 µl of Primer B1 (10 µM), 1 µl of dNTPs, 1 µl of ligation mixture. The PCR was conducted using the following conditions: 2 min initial denaturation at 94°C, then 20 cycles of 94°C for 45 s, 55°C for 30 s, 72°C for 1.5 min.
DNA sequencing
PCR products were sequenced to check for errors. The DNA construct was cloned using plasmid vector pCR®4-TOPO (Invitrogen) and transformed into chemically competent OneShot® TOP10 chemically competent Escherichia coli cells. The cells were spread on prewarmed LB plates containing 50–100 µg/ml ampicillin and incubated at 37°C. After overnight growth, the plates were sent to Molecular Cloning Laboratories (MCLab) where individual colonies were amplified and sequenced.
RESULTS
Microfluidic oligonucleotide synthesis
A PDMS-based microfluidic device was used as a miniaturized synthesizer for solid-phase parallel synthesis of oligonucleotides. This device has 16 nl-scale reaction chambers as illustrated in Figure 1B. Smaller, more compact reaction columns are also possible using the same fabrication method. The 100 pmol synthesis scale is chosen so that sufficient products are produced for several ligation assembly reactions after standard analysis methods. This device allows more than an order of magnitude higher synthesis throughput than the single reactor chemical DNA synthesizer we described previously (14).
In a typical experiment, oligonucleotide synthesis starts on universal linker-derivatized controlled porous glass beads (15). During operation, appropriate reagents are driven by pressure into binary trees that precede the reaction columns. Each reaction column is gated by an individually addressable valve that not only controls which phosphoramidite solution enters the column but also allows varied exposure time of solutions to that column if necessary. Because each reaction column is physically separated from the others, the reaction cross-talk between adjacent columns is minimized. Potential contamination between synthesis steps when residual reagents might enter reactors is eliminated by adequate flushing of the binary tree after each step. In addition, by physically separating the columns, the contents of each column can be selectively eluted, unlike microarray technologies where all the products are cleaved off into a single mixture.
Each reaction column consumes ∼250 nl of 0.1 M phosphoramidite solution in each reaction cycle, a substantial savings (>100-fold) over conventional DNA synthesizers on a per sequence basis. More importantly, unlike other microarray technologies where only femtomoles of each oligonucleotide are synthesized, the 100 pmol synthesis scale allows direct gene assembly without any need for amplification which increases cost and oligonucleotide length to accommodate adapters before assembly.
Synthesis of oligonucleotide using trifluoroacetic acid deblocking
Because dichloromethane is incompatible with PDMS and leads to swelling and possible delamination of multilayer devices (16), acetonitrile is the preferred solvent. However, the reagent traditionally used for deblocking (dichloroacetic acid) forms a complex with acetonitrile that drastically slow detritylations (17). Therefore, a new deblocking solution that is more suitable for PDMS devices had to be formulated.
Heterogeneous phase detritylation processes are complex with many chromatography effects such as the retardation of detritylation due to acid binding with the oligonucleotides and recapture of free DMT cation by 5′-OH group. In addition, the interplay of incomplete deblocking and losses of oligonucleotide bases due to depurination make rigorous kinetic modeling and analysis difficult. We analyzed the end product of the synthesis and used the ratio of full-length product to truncated byproducts as a benchmark to evaluate the success of the deblocking step.
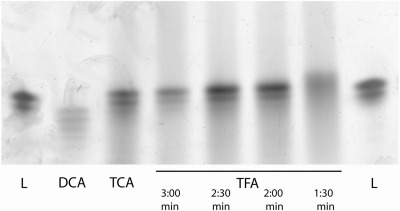
We surveyed several deblocking solutions composed of different organic acids in acetonitrile using spectroscopy to detect the release of the colored DMT cation when beads are exposed to these solutions. The deblocking solutions that release the most DMT cation were used in the synthesis of the homopolymer A20. Adenine was chosen because it is the base most sensitive to depurination (18). We found that trifluoroacetic acid was most suitable (Figure 2). Initially, we established that 5% TFA in acetonitrile with a deblocking time of 2.5 min was sufficient for synthesis of short oligonucleotides. However, in experiments where long oligonucleotides (longer that 35mers) were synthesized, we observed that the orange color of DMT cation persists well over the 2.5 min deblocking time at later synthesis cycles. We believe that acid binding to the growing oligonucleotide depletes free acid from the deblocking solution at higher oligonucleotide mass densities due to the increased length. We addressed this problem by increasing the deblocking acid concentration to 10%. The use of higher acid concentration allows quicker saturation and gives faster and more complete detritylation. In subsequent experiments where 10% TFA in acetonitrile was chosen as the deblocking solution, the optimum deblocking time was found to be between 1 and 2 min (Table 2). At longer deblocking times, losses of full-length nucleotide product through depurination exceeded gains from increased deblocking efficiency.
Figure 2.
Effect of deblocking acids and deblocking times on the quality of A20 homopolymer. A selection of organic acids is broadly surveyed as candidates to be used in an acetonitrile based deblocking solution. Five percent TFA is found to be more suitable than 10% DCA and 10% TCA with an optimal deblocking time between 2:00 and 2:30 min.
Table 2.
Effect of deblocking time on the length distribution of A40 homopolymer using 10% TFA in acetonitrile
| Length |
Deblocking time |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 s | 30 s | 1 min | 1.5 min | 2.0 min | 2.5 min | 3.0 min | 3.5 min | |
| N | 0 | 0.351 | 0.671 | 0.651 | 0.656 | 0.644 | 0.644 | 0.598 |
| N−1 | 0 | 0.284 | 0.329 | 0.349 | 0.344 | 0.356 | 0.356 | 0.402 |
| N−2 | 0 | 0.214 | ||||||
| N−3 | 0 | 0.151 | ||||||
A minimum of 1 min deblocking time is needed for successful deblock. The quality of the homopolymer starts to drop with deblocking time above 2.5 min. This suggests that increase deblocking efficiency at longer time is offset by losses of full-length product through depurination.
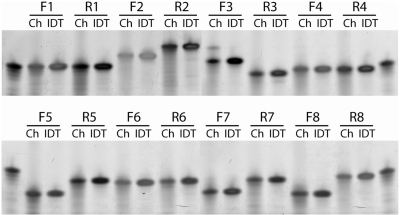
Using the established synthesis protocol with our deblocking conditions, we proceeded to synthesize the oligonucleotides needed to assemble a gene fragment from B. cereus. Each oligonucleotide product was verified though gel electrophoresis. The gel image (Figure 3) shows that the chip products had the same motility as commercially ordered standards from Integrated DNA Technologies (IDT). The lanes with chip products were partially smeared because no desalting step was taken; however, the major products all corresponded with full-length oligonucleotides.
Figure 3.
Polyacrylamide gel electrophoresis comparing oligonucleotides synthesized from the microfluidic platform (Ch) to oligonucleotides purchased from IDT. The chip products have the same motility as the commercially-prepared standards. However, the chip products are also partially smeared because of excess salt as no desalting step was performed after cleavage from the beads.
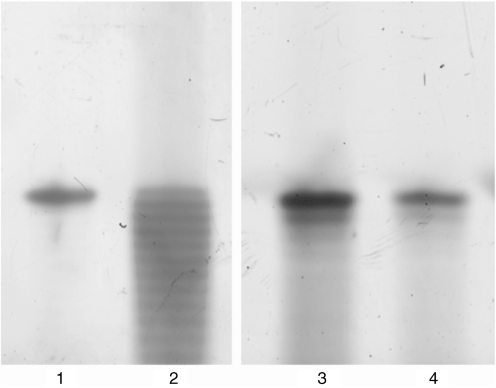
To test the limits of this synthesis method, we also synthesized oligonucleotides up to 40 bases in length. We found that at these lengths, the resulting oligonucleotides were not of sufficiently high quality for assembly. We therefore added an additional capping step that was not used in our original synthesis of shorter oligonucleotides. By capping unreacted bases, we prevented truncated sequences from extending further. This eliminated most of the N-1 and N-2 oligonucleotides observed in the uncapped synthesis, which are particularly problematic because they can lead to deletion errors in the gene assembly reaction. PAGE analysis of the reactions (Figure 4) shows that the dominant bands in the capped synthesis are from full-length product as opposed to truncated products from the uncapped synthesis. By comparing the ratio of full-length product to the N-1-mers, we calculated our deblocking efficiency to be >98.6%.
Figure 4.
Polyacrylamide gel electrophoresis showing oligonucleotides 40 bases in length synthesized under different conditions. Lane 1 is a purified standard purchased from IDT. Lane 2 is product synthesized using the microfluidic chip using 5% TFA deblocking solution without any capping during the synthesis. Lanes 3 and 4 are products from different reaction columns synthesized using 10% TFA deblocking solution with capping after each synthesis cycle. The gel image shows that the dominant bands in the capped synthesis are from full-length products whereas there are a large number of truncated products in the uncapped synthesis.
Ligation assembly of DNA constructs
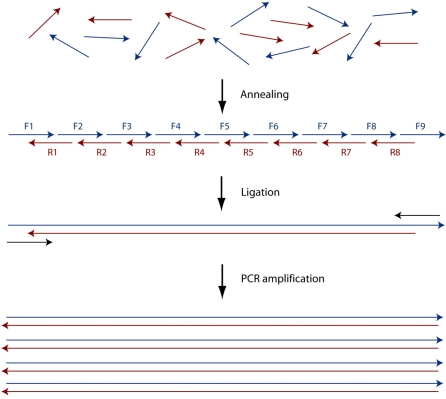
To confirm the identity of synthesized sequences and to determine whether the eluted oligonucleotides were functional without further amplification or purification, a gene fragment from B. cereus was assembled from a mixture of oligonucleotide 17–24-mers that was obtained using the microfluidic synthesis method described earlier. The general strategy of gene assembly used involves ligation of oligonucleotides into a complete construct followed by PCR amplification of the full-length product (Figure 5). The 230-bp gene fragment sequence and its component oligonucleotides are given in the Supplementary Data. The oligonucleotides were each collected in a 10 µl volume; the material produced from one synthesis is sufficient for 50–100 ligation reactions.
Figure 5.
Schematic of gene assembly by ligation. The starting oligos are placed in a single mixture and allowed to anneal and ligate into complete constructs. Primers are then added to amplify the full-length product by PCR.
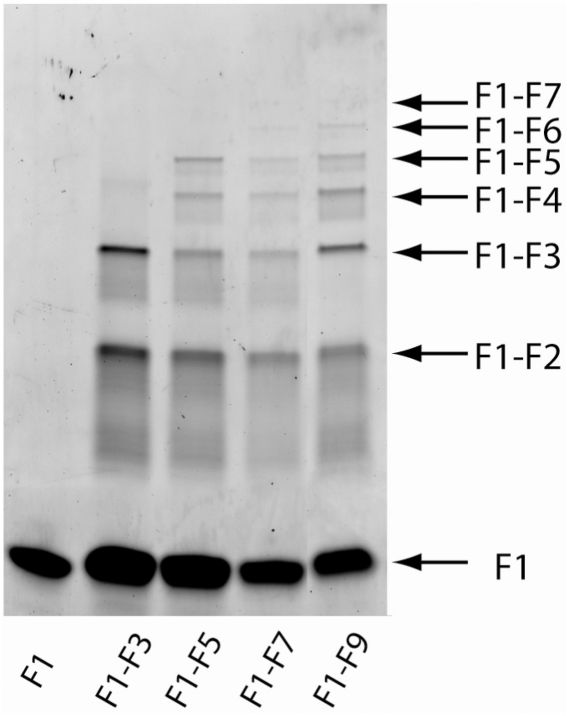
A special set of ligation reactions with Cy3-labeled F1 strand was performed to examine the ligation reaction in detail by gel electrophoresis without the need of amplification (Figure 6). Reaction pots with increasing numbers of strands that form products of increasing length were prepared. We expected species of various starting and ending strands to be ligated in each reaction. However, only species that included the Cy3-labeled F1 strand could be visualized after electrophoresis.
Figure 6.
Polyacrylamide gel electrophoresis for a series of ligation reactions with a Cy3-labeled F1 strand in combination with different numbers of starting oligonucleotides. No products longer than the expected length are present in each reaction mixture. For example, in the mixture containing F1 through F5, only F1, F1–F2, F1–F3, F1–F4 and F1–F5 are observed. No products longer than F1–F7 can be visualized without further amplification. In general, the amount of ligation product decreases as the number of ligation steps needed to form a particular product increases.
Our results show that no products longer than the expected length are present in each reaction mixture. In general, the amount of ligation product decreases as the number of ligation steps needed to form a particular product increases. The majority of the Cy3-labeled F1 strands failed to take part in any ligation reaction to form longer products. The longest detectable ligation product without further amplification is F1–F7. The yield of this ligation reaction was not optimized.
PCR of ligation product
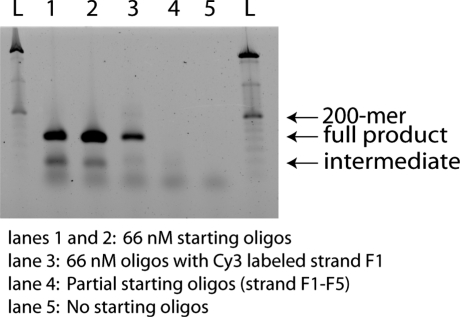
We performed PCR of the ligation reactions and analyzed the results on an agarose gel. Strong bands were evident for the desired product in lanes with the full set of starting oligonucleotides. ‘Primer-only’ and incomplete oligonucleotide sets used as controls yielded no product-length bands, confirming that the presence of desired-length species was not a consequence of amplification of a contaminating species. We observed a weak band in the main reaction lanes that indicated there were lower molecular weight species present that could be assembly intermediates present in the ligation product (Figure 7).
Figure 7.
Agarose gel electrophoresis showing full-length assembly of a gene fragment from B. cereus. In the presence of the full set of starting oligos, full-length products are observed following amplification. ‘Primer-only’ and incomplete oligonucleotide sets used as controls yielded no product-length bands. Weaker bands with lower molecular weight indicate assembly intermediates present in the ligation product.
DNA sequencing of assembled sequence
Sequencing of microfluidic gene synthesis products unambiguously confirmed the identity of the gene construct. Even though such sequencing does not directly report on the quality of the starting synthesized oligonucleotides, errors present in the starting oligonucleotides would be a source of deletions and other errors in the assembled sequences. The fidelity of the polymerase used in amplification step may also contribute to any observed errors (8).
Gel purification of the resulting assembled sequence was omitted to prevent the masking of errors. The clones were also not screened prior to sequencing other than to confirm successful insertion into the cloning vector. Thus, gene assembly products including both full-length species along with other incomplete, intermediate products should be detected in the clones.
Ten randomly selected clones of DNA constructs assembled from purified oligonucleotides purchased from IDT were sequenced. Eighteen randomly selected clones of DNA constructs assembled from oligonucleotides synthesized on the microfluidic synthesis chip (Chip) were also sequenced. The results of this sequencing are summarized in Table 3.
Table 3.
Summary of errors in the assembly of a gene fragment of Bacillus cereus using purified oligonucleotides purchased commercially (IDT clones) versus the synthesis products from the microfluidic device (Chip clones)
| Error type | IDT assembly of oligos | Chip assembly of oligos |
|---|---|---|
| None | 8/10 | 4/18 |
| Single deletion | 2/10 | 5/18 |
| Single mutation | 2/18 | |
| Single insertion and deletion | 1/18 | |
| Single insertion and mutation | 1/18 | |
| Sequence truncation (missing oligos) | 5/18 | |
| Total bases sequenced | 1300 | 1690 (without truncation) |
| Overall assembly success rate | 80% | 22% |
| Total number of errors | 2 | 11 |
| Per base error rate | 1 in 650 | 1 in 153 |
Out of the 10 IDT clones, eight have perfect sequence while two had single deletions. Out of the 18 Chip clones, four had perfect sequences, five had single deletions, two had single point mutations, one had a single deletion and insertion, and one had single point mutation and a single insertion. More interestingly, three of the chip clones were missing a stretch of sequence from F3 to F8. Looking more closely at the sequences of the starting oligonucleotides, we found that the five starting bases for F3 were identical to F9. Thus, F9 can incorrectly ligate to F2 instead of F3. Such a problem could be avoided with more careful design of the starting oligonucleotide pool.
For the IDT clones, a total of two errors were found out of 1300 bases sequenced, an error rate of 1 per 650 bp or 0.15%. Excluding clones with incompletely assembled sequences, the Chip clones had 11 errors out of 1690 bases sequenced. That is an error rate of 1 per 153 bp or 0.65%. This error rate is very similar to the error of 1 in 160 bp given by Tian (19).
DISCUSSION
In this study we have developed a microfluidic synthesis platform capable of generating a number of oligonucleotides in parallel for gene assembly. This system addresses many of the limitations associated with microarray technology that may lead to reduced product purity such as deprotection, insufficient synthesis scale, and adverse interactions in large oligonucleotide pools while keeping many of its advantages such as reduced cost. Our chip is programmable for synthesis of any desired sequence, easy to operate and consumes minute quantities of reagent and solvent on a per sequence basis.
The physical separation of individual reaction chambers eliminates the potential problem of cross contamination. Having individually addressable valves gating each reaction chamber also allows for easy recovery of specific sequences synthesized for direct use in the subsequent gene assembly. This is especially useful for multiplex reactions where sequences with adverse interactions with each other can be eluted into different mixtures. Such precise fluid handling is not available in microarray technologies and will help allow the integration of this chip to other processes in the future.
The oligonucleotides synthesized from our device were directly used for ligation to give DNA constructs ∼200-bp long. Employing such direct synthesis without concentration or an initial amplification step not only reduces the time and cost of the overall synthesis protocol, but also eliminates the possibility that additional errors will be generated during the amplification procedure. While the error rate of 1 in 153 bp from chip product is higher than that observed for a product assembled from purified oligonucleotides ordered from IDT, it is consistent with the 1 in 160 bp error rate given by Tian for assembly of DNA constructs using unpurified oligonucleotides. The 17–24-mer synthesis also lacked capping; given the improvement in purity for longer syntheses (Figure 3), performing capping during the synthesis of the oligonucleotides used for ligation should also improve the error rate. Optimized ligation conditions, including varied temperature, may also be able to minimize the incorporation of errors into the full-length product.
These results demonstrate significant progress towards high-throughput gene synthesis and could help meet the growing demands of this emergent field of synthetic biology. Substantial improvements are still needed but many can still be achieved through further optimization of reaction conditions. Systems integration of other processes that have already been demonstrated in stand-alone units, such as purification, cloning and protein expression (20) will greatly increase the range of suitable applications for this technology.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
Funding for open access charge: National Science Foundation under grant DMI-0328162.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Au LC, Yang FY, Yang WJ, Lo SH, Kao CF. Gene synthesis by a LCR-based approach: High-level production of leptin-L54 using synthetic gene in Escherichia coli. Biochem. Biophys. Res. Commun. 1998;248:200–203. doi: 10.1006/bbrc.1998.8929. [DOI] [PubMed] [Google Scholar]
- 2.Stemmer WPC, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 3.Letsinger RL, Lunsford WB. Synthesis of thymidine oligonucleotides by phosphite triester intermediates. J. Am. Chem. Soc. 1976;98:3655–3661. doi: 10.1021/ja00428a045. [DOI] [PubMed] [Google Scholar]
- 4.Matteucci MD, Caruthers MH. Nucleotide chemistry. 4. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981;103:3185–3191. [Google Scholar]
- 5.Gao XL, Gulari E, Zhou XC. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–596. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- 6.Mueller S, Coleman JR, Wimmer E. Putting synthesis into biology: a viral view of genetic engineering through de novo gene and genome synthesis. Chem. Biol. 2009;16:337–347. doi: 10.1016/j.chembiol.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong DS, Carr PA, Chen L, Zhang SG, Jacobson JM. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007;35:e61. doi: 10.1093/nar/gkm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang MC, Ye H, Kuan YK, Li MH, Ying JY. Integrated two-step gene synthesis in a microfluidic device. Lab. Chip. 2009;9:276–285. doi: 10.1039/b807688j. [DOI] [PubMed] [Google Scholar]
- 9.Ye HY, Huang MC, Li MH, Ying JY. Experimental analysis of gene assembly with TopDown one-step real-time gene synthesis. Nucleic Acids Res. 2009;37:e51. doi: 10.1093/nar/gkp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 11.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 12.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 13.McBride LJ, Caruthers MH. An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 1983;24:245–248. [Google Scholar]
- 14.Huang YY, Castrataro P, Lee CC, Quake SR. Solvent resistant microfluidic DNA synthesizer. Lab. Chip. 2007;7:24–26. doi: 10.1039/b613923j. [DOI] [PubMed] [Google Scholar]
- 15.Wang ZW, Olsen P, Ravikumar VT. A novel universal linker for efficient synthesis of phosphorothioate oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2007;26:259–269. doi: 10.1080/15257770701257277. [DOI] [PubMed] [Google Scholar]
- 16.Lee JN, Park C, Whitesides GM. Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Anal. Chem. 2003;75:6544–6554. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- 17.Paul CH, Royappa AT. Acid binding and detritylation during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3048–3052. doi: 10.1093/nar/24.15.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3053–3058. doi: 10.1093/nar/24.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian JD, Gong H, Sheng NJ, Zhou XC, Gulari E, Gao XL, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Ran R, Zhao JL, Xu YS. Design, simulation, and optimization of a miniaturized device for size-fractioned DNA extraction. Electrophoresis. 2007;28:4661–4667. doi: 10.1002/elps.200700226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.