Abstract
Circadian clocks have long been known to be essential for the maintenance of physiological and behavioral processes in a variety of organisms ranging from plants to humans. Dysfunctions that subvert gene expression of oscillatory circadian-clock components may result in severe pathologies, including tumors and metabolic disorders. While the underlying molecular mechanisms and dynamics of complex gene behavior are not fully understood, synthetic approaches have provided substantial insight into the operation of complex control circuits, including that of oscillatory networks. Using iterative cycles of mathematical model-guided design and experimental analyses, we have developed a novel low-frequency mammalian oscillator. It incorporates intronically encoded siRNA-based silencing of the tetracycline-dependent transactivator to enable the autonomous and robust expression of a fluorescent transgene with periods of 26 h, a circadian clock-like oscillatory behavior. Using fluorescence-based time-lapse microscopy of engineered CHO-K1 cells, we profiled expression dynamics of a destabilized yellow fluorescent protein variant in single cells and real time. The novel oscillator design may enable further insights into the system dynamics of natural periodic processes as well as into siRNA-mediated transcription silencing. It may foster advances in design, analysis and application of complex synthetic systems in future gene therapy initiatives.
INTRODUCTION
The rapidly developing field of synthetic biology has emerged as a discipline capable of linking theoretical model predictions to experimental implementations consisting of minimal modular building blocks arranged to form synthetic networks (1) with different levels of complexity (2–9). Substantial information on such minimal units and circuitry originated from prokaryotes (10–18) and the relatively recent development of heterologous mammalian gene regulation technologies (19–21) enabled subsequent applications in complex mammalian systems (2,3,22,23).
In synthetic biology, oscillatory behavior has gained substantial attention due to its impact on repair (24), metabolic (25) and signaling pathways (26–27). Such pathways, like the p53-mdm2-triggered apoptosis cascade (28), NF-κB-regulated immune responses (29) or Hes1-mediated developmental control (30), are based on periodic induction of negative feedback loops controlling expression of key pathway factors (31,32).
Utilizing antisense-mediated silencing of a transactivator, we have recently created a synthetic gene network capable of oscillatory expression of the green fluorescent protein (22). Future applications, however, are expected to require fine-tuning of oscillatory properties, such as frequency and amplitude. This could be achieved by alternative selection and reconfiguration of the underlying modular building blocks. Here, we present an alternative oscillatory network that displays robust low-frequency fluctuations in target gene expression. Capitalizing on recent progress showing that RNA interference may be a powerful component of synthetic gene networks (33,34), we have designed a synthetic mammalian clock that combines siRNA-mediated, time-delayed negative feedback loop with autoregulated expression of the tetracycline-dependent transactivator (tTA). The experimental data thus obtained were in good agreement with a mathematical model based on ordinary differential equations. Information gained from such combined dynamic studies should facilitate advances in our understanding of natural circadian clocks and rhythms (35).
MATERIALS AND METHODS
Expression vector design
pBP283 (PETR3→d2EYFP), harboring a destabilized enhanced yellow fluorescent protein (d2EYFP) under control of the erythromycin-responsive promoter (PETR3), was constructed by excising d2EYFP from pd2EYFP (Clontech) using EcoRI/NotI and cloning it into the corresponding sites (EcoRI/NotI) of pBP90 (36). pDG156 (PhCMV*−1-ET1), containing the erythromycin-dependent transactivator [ET1, E-VP16; (37)] driven by the tetracycline-responsive promoter (PhCMV*−1) was designed by excising PhCMV*−1 from pMF111 (38) by SspI/EcoRI and cloning it into the corresponding sites (SspI/EcoRI) of pWW35 (19). pDG157 (PhCMV*−1→E-siRNALUC-VP16) expresses the ET1, E-VP16; (37) containing an intronically encoded siRNA specific for firefly luciferase mRNAs (siRNALUC; nt 62–80). pDG157 was obtained by replacing PSV40 of pDG17 (39) with PhCMV*−1 (40) via SspI/EcoRI. pDG178 (PhCMV*−1→tTA) harboring an autoregulated expression unit for tetracycline-responsive control of the tTA was obtained by excising tTA from pSAM200 (38) using EcoRI/BssHII into pDG156, thereby replacing ET1. pND10 (PhCMV*−1→TAGLUC-tTA) contains an expression unit for PhCMV*−1-driven expression of the tTA preceded by a noncoding sequence tag derived from the firefly luciferase coding sequence (nt 62–80). pND10 was constructed in two steps, first by replacing PhCMV*−1 of pDG178 with PhEF1α-TAGLUC (PhEF1α, human elongation factor 1α promoter) of pDG54 via SspI/EcoRI and then by replacing PhEF1α with PhCMV*−1 of pDG178 via SspI/KpnI.
Cell culture, transfection and gene regulation
Chinese hamster ovary cells (CHO-K1, ATCC CCL61) were cultivated in ChoMaster® HTS (Cell Culture Technologies, Gravesano, Switzerland) supplemented with 5% fetal calf serum (Pan Biotech GmbH, Aidenbach, Germany; cat. no. 3302-P231902, lot no. P231902). CHO-K1 were cultivated at 37°C in a humidified atmosphere containing 5% CO2, and 35 000 cells were transfected using the FuGENE®6 transfection reagent (Roche Molecular Biochemicals, lot. no. 93535720) and a network-encoding plasmid mixture calibrated to a total of 1.2 µg DNA using the extender vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). Transfection experiments were standardized to provide efficiencies of up to 35 ± 5%. The number of oscillating cells was indicated as percent relative to the total number of transfected cells (100%). Transfected cells were arrested in the G1 phase by a down-shift of the cultivation temperature to 32°C (41), and monitoring of the fluorescence dynamics by time-lapse microscopy was started 18 h posttransfection (t = 0).
Fluorescence imaging
Time-lapse fluorescence microscopy was performed using an inverted Leica fluorescence microscope (Leica Microsystems DMI 6000B, 11888107) equipped with an incubation chamber, a Leica DFC350FX R2 digital camera (Leica, cat. no: 112730043), an 10× objective (Leica, Obj. HC PL FL 10×/0.30 PH1 -/D 11.0, cat. no. 11506507) and the following excitation/emission filter set: d2EYFP (513/527 nm; C/Y/R, cat. no. 11513897). Time-lapse movies were produced with the LAS AF imaging software (Leica FW4000-TZ, cat. no. 12723979) set to exposure times of 610 ms every 20 min. Fluorescence was quantified for single cells using a custom-designed software implemented in MATLAB® (Mathworks, Nantucket/MA, USA) All details on software development are available as Supplementary Data.
Computational modeling
All details on mathematical models and computational methods are provided as Supplementary Data. Simulations were performed with MATLAB®, Version R2007b.
RESULTS
Design of the low-frequency mammalian oscillator
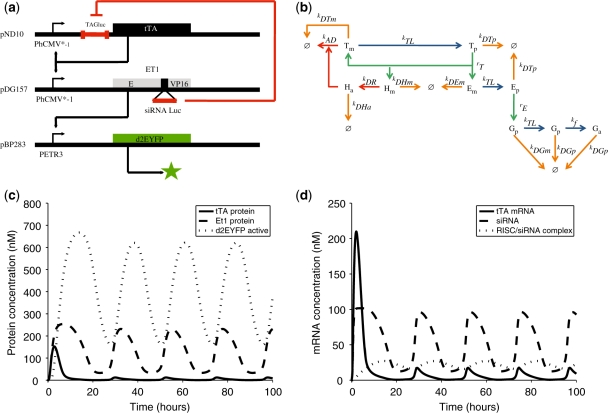
The low-frequency mammalian oscillator (Figure 1a) consists of: (i) an autoregulated expression unit harboring the tTA and an untranslated Photinus pyralis firefly luciferase-derived sequence tag (TAGLUC; 42, 43) targeted by a luciferase-specific siRNA (siRNALUC; 42) under control of the tTA-specific tetracycline-responsive promoter (PhCMV*−1; PhCMV*−1→TAGLUC-tTA); (ii) a PhCMV*−1-driven erythromycin-dependent transactivator [ET1; E-VP16; (37)] harboring an intronic siRNALUC specific for LUC-tagged tTA (TAGLUC-tTA) between the macrolide-dependent repressor (E) and the Herpes simplex-derived transactivation domain (VP16) (PhCMV*−1→E-siRNALUC-VP16); and (iii) a destabilized yellow fluorescent protein variant with a half-life of 2 h (d2EYFP) (44, 45) controlled by the ET1-specific macrolide-responsive promoter (PETR3; PETR3-d2EYFP) (Figure 1a).
Figure 1.
Mammalian clock components and predicted oscillation dynamics. (a) The mammalian oscillator consists of an autoregulated expression unit encoding the tTA under control of the tetracycline-responsive promoter (PhCMV*−1). A luciferase-specific sequence TAG engineered into the noncoding 5′-region of the tTA transcript enables interference-based silencing of TAGLUC-tTA mRNAs using a luciferase-specific siRNA (siRNALUC). siRNALUC is intronically encoded within the macrolide-dependent transactivator (ET1; between the macrolide-responsive repressor (E) and the Herpes simplex-derived transactivation domain VP16; E-siRNALUC-VP16), which is driven by the tTA-specific PhCMV*−1. Splicing of E-siRNALUC-VP16 transcripts produces both, siRNALUC, which triggers interference-based elimination of TAGLUC-tTA mRNAs and ET1, which transactivates PETR3, thereby controlling expression of the destabilized d2EYFP and enabling visualization of the oscillatory network dynamics. Tetracycline and erythromycin can be used to modulate tTA- and ET1-mediated induction of PhCMV*−1 and PETR3, respectively. (b) Intracellular processes considered in the mathematical model. Transcription (green arrow), translation and protein folding (blue arrow), degradation (orange arrow) and RNA interference (red arrow). (c and d) Model predictions for the reference parameter set (all plasmids transfected at a 1:1:1 ratio, no antibiotics) (c) with protein concentrations (tTA, solid line; ET1, dashed line; active fluorescent d2EYFP, dotted line) and (d) with mRNA concentrations (tTA, solid line; ET1, dashed line), and RISC/siRNALUC complex (dotted line).
When co-transfected into mammalian cells, leaky PhCMV*−1-mediated TAGLUC-tTA transcription is expected to initiate an autoregulated positive feedback loop sustaining high-level expression of tTA and consequently of PhCMV*−1-driven E-siRNALUC-VP16 expression. Constitutive splicing of E-siRNALUC-VP16 produces equimolar amounts of ET1 (E-VP16) and siRNALUC, which results in concomitantly increasing ET1-triggered PETR3-driven d2EYFP expression and siRNALUC-mediated breakdown of LUC-tagged tTA transcripts. siRNALUC-based depletion of TAGLUC-tTA transcripts will reduce tTA levels and temporarily shut down the network. This results in low d2EYFP expression and low siRNALUC production, which eventually allows the circuit to start over again. The gene network can be tuned by the addition of tetracycline and erythromycin to regulate PhCMV*−1 and PETR3 promoter activity, respectively.
Model-based analysis of circuit behavior
To evaluate the general capability of the network to generate oscillatory behavior, we developed a mathematical model based on ordinary differential equations (see Figure 1b and Supplementary Data). This deterministic model describes the circuitry behavior in a mechanistic fashion. A first model prediction based on experimentally determined, as well as literature-based, parameter sets (see Supplementary Data) demonstrated the general capacity of the circuit to display oscillatory gene expression with periods similar to those observed for natural circadian clocks (Figure 1c and d).
Evaluation of circuit behavior in CHO-K1 cells by monitoring d2EYFP expression
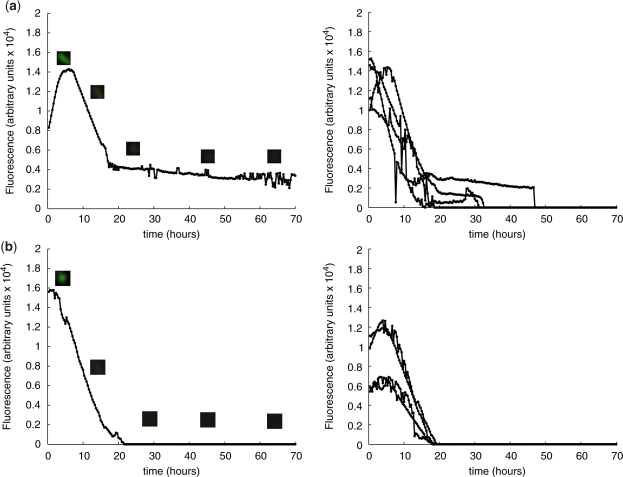
According to model simulations, any inactivation of transactivators tTA and/or ET1 by the addition of antibiotics would disrupt the rhythmic expression pattern (Supplementary Figure 1). The impact of regulating antibiotics on the system was therefore first analyzed to validate the functionality of the single components. For this purpose, pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) were co-transfected at equimolar ratios (1 : 1 : 1), followed by subsequent addition of (i) 200 ng/500 µl tetracycline (Figure 2a) or (ii) 200 ng/500 µl erythromycin (Figure 2b). As expected, addition of either antibiotic disrupted the oscillatory behavior, leading to decreasing levels of d2EYFP fluorescence overtime.
Figure 2.
Impact of regulating antibiotics on the mammalian oscillator components and systems behavior using single cell time-lapse fluorescence microscopy of transfected CHO-K1. (a) CHO-K1 co-transfected with pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP); 200 ng tetracycline was added to the culture, (b) CHO-K1 transfected with pND10, pDG157 and pBP283; 200 ng erythromycin was added to the culture. Left panels show fluorescence dynamics and micrographs of a representative individual cell, and right panels show the fluorescence profiles of several cells with t = 0 corresponding to the onset of fluorescence.
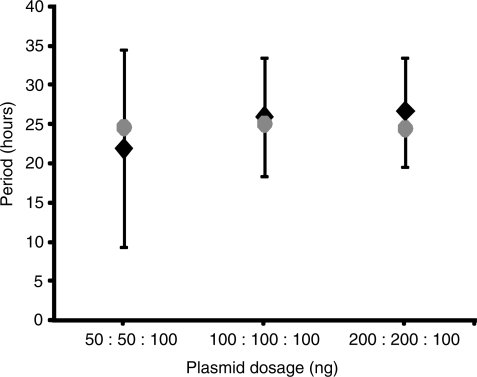
In contrast, model predictions suggested that the system would be insensitive to variations in the amount of transfected single components (Figure 3 and Supplementary Data). Therefore, we next evaluated the effect of plasmid dosage variations by co-transfecting pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PETR3→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) at differing amounts, namely 1:1:1 (100 ng each, Figure 4a), 0.5:0.5:1 (Figure 4b), 2:2:1 (Figure 4c), and varying ratios 2:1:1 (Figure 5a) and 1:2:1 (Figure 5b). All variations displayed oscillatory behavior with similar periods of ∼26 h and with substantial amplitudes. Hence, the experimental data confirmed the theoretically predicted robustness of the network to moderate changes in the amounts of single components. This contrasts with the observed and predicted sensitivity of the oscillatory behavior to plasmid dosage in our earlier synthetic oscillator based on antisense-mediated silencing of the tTA (22). There, changes in ratio and/or amount of system components influence the period and amplitude of oscillatory gene expression significantly.
Figure 3.
Impact of plasmid dosage on oscillation frequency. Alignment of mathematically simulated and predicted period (gray disc) with the experimental data (black diamonds with error bars) for the conditions (i) pND10 (PhCMV*−1→TAGLUC-tTA):pDG157 (PhCMV*−1→E-siRNALUC-VP16):pBP283 (PETR3→d2EYFP); 50 ng:50 ng:100 ng, (ii) 100 ng:100 ng:100 ng, (iii) 200 ng:200 ng:100 ng.
Figure 4.
Characterization of oscillator dynamics using single-cell time-lapse fluorescence microscopy of CHO-K1 transfected with different amounts of oscillator components. (a–c) CHO-K1 co-transfected with pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP): (a) plasmid ratio of 1:1:1, no antibiotics; (b) 0.5:0.5:1, no antibiotics; and (c) 2:2:1, no antibiotics. Left panels show fluorescence dynamics and micrographs of a representative individual cell, and right panels show the fluorescence profiles of several cells with t = 0 corresponding to the onset of fluorescence.
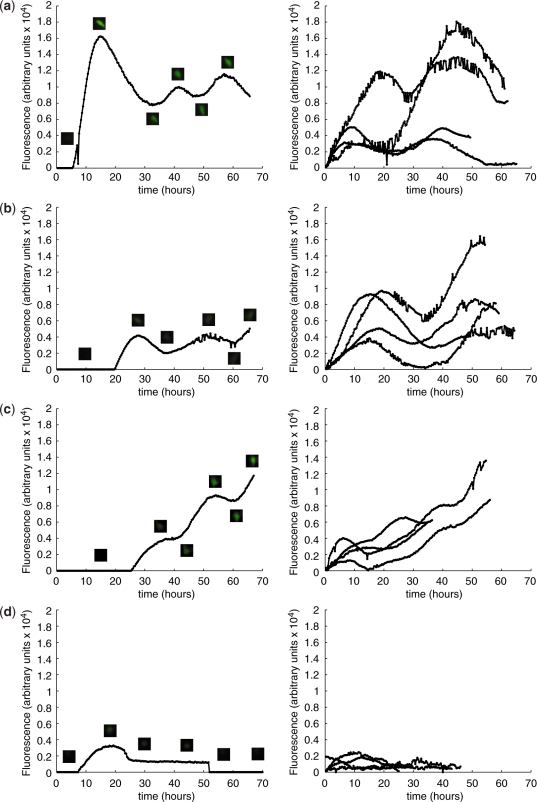
Figure 5.
Characterization of oscillator dynamics using single-cell time-lapse fluorescence microscopy of CHO-K1 transfected with different ratios of oscillator components. (a–d) Single-cell time-lapse fluorescence microscopy of CHO-K1 co-transfected with pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP): (a) plasmid ratio of 2:1:1, no antibiotics; (b) 1:2:1, no antibiotics; (c) 10:1:1, no antibiotics; and (d) 1:10:1, no antibiotics. Left panels show fluorescence dynamics and micrographs of a representative individual cell, and right panels show the fluorescence profiles of several cells with t = 0 corresponding to the onset of fluorescence.
When expressing excessive amounts of either pND10 (PhCMV*−1→TAGLUC-tTA) or pDG157 (PETR3→E-siRNALUC-VP16) using ratios of 10:1:1 (Figure 5c) and 1:10:1 (Figure 5d), the oscillatory circuit was substantially repressed. Interestingly, even with a 10-fold excess in tTA gene dosage, silencing induced visible fluctuations in reporter gene expression (Figure 5c). In contrast, a 10-fold excess of siRNALUC, which removes TAGLUC-tTA transcripts from the system, completely eliminated such behavior (Figure 5d). These results point to a major role of the amplifying and long-lasting siRNA-mediated silencing mechanism in generating the observed network behavior.
To further characterize the impact of siRNALUC activity and specificity, we co-transfected CHO-K1 cells with (i) pDG178 (PhCMV*−1→tTA) lacking the 5′-TAGLUC, together with pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) and (ii) pND10 (PhCMV*-1→TAGLUC-tTA), pDG156 (PETR3→E-VP16) lacking the intronically encoded siRNALUC and pBP283 (PETR3→d2EYFP) (Figure 6a and b). For both conditions, we detected no oscillations but a constitutive increase in fluorescent intensity over time. This was comparable with conditions in which only pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) were co-transfected (Figure 6c). The observed oscillatory behavior is therefore specific and directly linked to the siRNALUC-mediated elimination of TAGLUC-tTA transcripts that leads to silencing of tTA expression. These observations confirm functional siRNA-mediated breakdown of target mRNAs and corroborate modeling results based on a specific set of known RNA-interference parameters (see Supplementary Data). Owing to the strong amplification of the mRNA destruction capacity by RNA interference components such as RISC and DICER (46), even residual siRNA levels were predicted to completely eliminate leaky tTA transcripts and, therefore, prevent tTA production (Figure 5c and d).
Figure 6.
Evaluation of siRNA specificity on the oscillatory behavior using single-cell time-lapse fluorescence microscopy of transfected CHO-K1. (a) CHO-K1 co-transfected with pDG178 (PhCMV*−1→tTA), pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) at a plasmid ratio of 1:1:1, no antibiotics, (b) co-transfection of pND10 (PhCMV*−1→TAGLUC-tTA), pDG156 (PhCMV*−1→ET1; ET1, E-VP16) and pBP283 (PETR3→d2EYFP) at a plasmid ratio of 1:1:1, no antibiotics and (c) co-transfection of pDG157 (PhCMV*−1→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) at a plasmid ratio of 1:1, no antibiotics. Left panels show fluorescence dynamics and micrographs of a representative individual cell and right panels show the fluorescence profiles of several cells with t = 0 corresponding to the onset of fluorescence.
Quantitative analysis of network behavior using automated image tracking and quantification
To obtain statistically relevant information on the network dynamics, the experimental data were systematically analyzed using customized tracking and image analysis software. Using high-throughput processing of acquired image series (for detailed information see Supplementary Data), we determined a consistent period for all analyzed plasmid ratio variations. In addition, we obtained significant relative amplitudes of >30% for total cellular fluorescence (Table 1).
Table 1.
Overview on oscillation characteristics resulting from different conditions
| Transfected plasmid | Amount (ng) | Antibiotic | No. of cells analyzed | Oscillating cells (%) | Period (h) | Relative amplitude (%) | Figure |
|---|---|---|---|---|---|---|---|
| pND10:pDG157:pBP283 | 100:100:100 | Tetracycline | 109 | 0 (0.0) | − | − | 2a |
| pND10:pDG157:pBP283 | 100:100:100 | Erythromycin | 23 | 0 (0.0) | − | − | 2b |
| pND10:pDG157:pBP283 | 100:100:100 | None | 336 | 39 (11.6) | 25.8 ± 7.6 | 34.8 ± 22.5 | 4a |
| pND10:pDG157:pBP283 | 50:50:100 | None | 34 | 6 (17.6) | 23.1 ± 11.5 | 28.3 ± 17.5 | 4b |
| pND10:pDG157:pBP283 | 200:200:100 | None | 588 | 42 (7.1) | 26.7 ± 7.0 | 41.7 ± 21.4 | 4c |
| pND10:pDG157:pBP283 | 200:100:100 | None | 318 | 31 (9.7) | 26.0 ± 8.5 | 29.7 ± 16.6 | 5a |
| pND10:pDG157:pBP283 | 100:200:100 | None | 425 | 35 (8.2) | 25.3 ± 7.5 | 33.1 ± 19.1 | 5b |
| pND10:pDG157:pBP283 | 1000:100:100 | None | 192 | 9 (4.7) | 26.7 ± 10.5 | 33.3 ± 19.7 | 5c |
| pND10:pDG157:pBP283 | 100:1000:100 | None | 282 | 0 (0.0) | - | - | 5d |
| pDG178:pDG157:pBP283 (no TAGLUC) | 100:100:100 | None | 93 | 0 (0.0) | - | - | 6a |
| pND010:pDG156:pBP283 (no siRNALUC) | 100:100:100 | None | 152 | 0 (0.0) | - | - | 6b |
| pDG157:pBP283 (no tTA) | 100:100 | None | 50 | 0 (0.0) | - | - | 6c |
ET1, macrolide-dependent transactivator (E-VP16); pBP283 (PETR3-d2EYFP), expression vector encoding a d2EYFP under control of the erythromycin-responsive promoter; PETR3, macrolide-responsive promoter (ETR-PhCMVmin); PhCMV, promoter of the human cytomegalovirus immediate early promoter; PhCMVmin: minimal version of PhCMV; PhCMV*−1, tetracycline-responsive promoter (tetO7-PhCMVmin); pDG156 (PhCMV*−1-ET1), expression vector encoding the erythromycin-dependent transactivator (ET1, E-VP16); pDG157 (PhCMV*−1-E-siRNALUC-VP16), expression vector encoding ET1 containing an intronically encoded siRNA specific for firefly luciferase mRNA (siRNALUC; nt 62–80); pDG178 (PhCMV*−1-tTA), autoregulated tTA expression vector; pND10 (PhCMV*−1-TAGLUC-tTA), tTA expression vector containing an siRNALUC-specific tag (TAGLUC) in the 5′ untranslated region derived from the firefly luciferase (nt 62–80); siRNALUC, firefly luciferase-specific siRNA; TAGLUC, firefly luciferase-specific sequence tag; tTA, tetracycline-dependent transactivator (TetR-VP16); VP16, transactivation domain of Herpes simplex virus.
With respect to the efficiency of oscillator behavior in the cell population, we found the highest number of cells that showed oscillating d2EYFP fluorescence for transfection with equimolar plasmid ratios (1:1:1). A movie showing the oscillatory behavior of the system after transfection of pND10 (PhCMV*−1→TAGLUC-tTA), pDG157 (PETR3→E-siRNALUC-VP16) and pBP283 (PETR3→d2EYFP) at equimolar ratios 1:1:1 (100 ng each) can be found at http://mf-229-serv.ethz.ch/fussi_download/marcel/suppmov.mov. The share of oscillating cells was 11.7% compared to 9.7% (2:1:1), 8.2% (1:2:1), 7.1% (2:2:1) and 17.6% (0.5:0.5:1) (Table 1). In contrast to the first-generation mammalian oscillator (22), which was tunable by varying plasmid ratio and amount (i.e. it exhibited frequency and amplitude changes in response to variations in these parameters), the alternative low-frequency mammalian oscillator is largely insensitive to component fluctuations.
DISCUSSION
The design of functional replicas of natural circadian clocks using native circadian clock components has been unsuccessful, suggesting that engineering of oscillatory behavior remains a non-trivial challenge (47). Synthetic biology at this point seems to be a valuable alternative, as several synthetic networks in prokaryotic backgrounds have efficiently produced functional circuits displaying damped (48), self-sustained (11), tunable (49,50) or metabolically (51) controlled oscillations. By incorporating posttranscriptional antisense-based control mechanisms, together with transcriptional regulation, we have previously created self-sustained and tunable oscillatory behavior in mammalian cells (22). However, the understanding and availability of single building blocks for complex network reconstruction is still relatively low. Any successful future initiative for gene therapeutic applications will require a broad spectrum of single components that can be interconnected to generate tunable and predictable oscillatory behavior in terms of frequency and amplitude of target gene expression levels. Our intronic-siRNA-based low frequency and robust oscillator therefore not only provides another tool for oscillatory gene expression. Due to its characteristics (siRNA-based feedback loops, long period, robustness to variations of DNA amounts and ratios), it serves as a valuable addition as well as counterpart to established systems. Besides synchronization of oscillators across a population of cells which was recently achieved in bacteria (17,18), the design of synthetic clocks with increased robustness of oscillatory behavior to variations in relative expression of network components and intrinsic noise remain major challenges on the way to use rhythmic transgene expression dosing in future therapeutic applications. Based on extensive simulations of novel oscillator architectures using a mathematical model, we have chosen to combine a two-level transcription cascade (autoregulated tTA expression triggers expression of the ET1 transactivator which induces d2EYFP expression) with an intronic siRNA-based negative feedback loop eliminating tTA-encoding transcripts by RNA interference. This network design enabled oscillating d2EYFP expression with a fixed frequency and amplitude that was almost insensitive to changes in plasmid ratio and dosage. Such extensions will make custom-tailored designs of regulatory feedback loops for specific applications possible. Furthermore, new insights into intronic siRNA-mediated silencing impact and dynamics may advance the understanding of naturally occurring silencing-mediated feedback and feed-forward circuits.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (31003A0-126022); EC Framework 6 (COBIOS). Funding for open access charge: Swiss Federal Institute of Technology, Zurich (ETH Zurich).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wilfried Weber for critical comments on the article.
REFERENCES
- 1.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 2.Weber W, Daoud-El Baba M, Fussenegger M. From the Cover: synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc. Natl Acad. Sci. USA. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 5.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 6.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 7.Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Curr. Opin. Biotechnol. 2008;19:454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Weber W, Kramer BP, Fussenegger M. A genetic time-delay circuitry in mammalian cells. Biotechnol. Bioeng. 2007;98:894–902. doi: 10.1002/bit.21463. [DOI] [PubMed] [Google Scholar]
- 9.Voigt C. Genetic parts to program bacteria. Curr. Opin. Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMillen D, Kopell N, Hasty J, Collins JJ. Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl Acad. Sci. USA. 2002;99:679–684. doi: 10.1073/pnas.022642299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 16.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 17.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fussenegger M. Synthetic biology: synchronized bacterial clocks. Nature. 2010;463:301–302. doi: 10.1038/463301a. [DOI] [PubMed] [Google Scholar]
- 19.Weber W, Kramer BP, Fux C, Keller B, Fussenegger M. Novel promoter/transactivator configurations for macrolide- and streptogramin-responsive transgene expression in mammalian cells. J. Gene Med. 2002;4:676–686. doi: 10.1002/jgm.314. [DOI] [PubMed] [Google Scholar]
- 20.Weber W, Marty RR, Keller B, Rimann M, Kramer BP, Fussenegger M. Versatile macrolide-responsive mammalian expression vectors for multiregulated multigene metabolic engineering. Biotechnol. Bioeng. 2002;80:691–705. doi: 10.1002/bit.10461. [DOI] [PubMed] [Google Scholar]
- 21.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 22.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 23.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahav G. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci. STKE. 2004;264:pe55. doi: 10.1126/stke.2642004pe55. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Kaasik K, Blackburn MR, Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature. 2006;439:340–343. doi: 10.1038/nature04368. [DOI] [PubMed] [Google Scholar]
- 26.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 27.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 28.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 29.Krishna S, Jensen MH, Sneppen K. Minimal model of spiky oscillations in NF-kappaB signaling. Proc. Natl Acad. Sci. USA. 2006;103:10840–10845. doi: 10.1073/pnas.0604085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl Acad. Sci. USA. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen MH, Sneppen K, Tiana G. Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 2003;541:176–177. doi: 10.1016/s0014-5793(03)00279-5. [DOI] [PubMed] [Google Scholar]
- 32.Tiana G, Krishna S, Pigolotti S, Jensen MH, Sneppen K. Oscillations and temporal signalling in cells. Phys. Biol. 2007;4:R1–17. doi: 10.1088/1478-3975/4/2/R01. [DOI] [PubMed] [Google Scholar]
- 33.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 34.Greber D, El-Baba MD, Fussenegger M. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic Acids Res. 2008;36:e101. doi: 10.1093/nar/gkn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benner SA, Sismour AM. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer BP, Weber W, Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- 37.Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 38.Fussenegger M, Moser S, Mazur X, Bailey JE. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;13:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]
- 39.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J. Biotechnol. 2007;130:329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufmann H, Mazur X, Fussenegger M, Bailey JE. Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol. Bioeng. 1999;63:573–582. doi: 10.1002/(sici)1097-0290(19990605)63:5<573::aid-bit7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 42.Greber D, Fussenegger M. Multi-gene engineering: simultaneous expression and knockdown of six genes off a single platform. Biotechnol. Bioeng. 2007;96:821–834. doi: 10.1002/bit.21303. [DOI] [PubMed] [Google Scholar]
- 43.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 45.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 46.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chilov D, Fussenegger M. Toward construction of a self-sustained clock-like expression system based on the mammalian circadian clock. Biotechnol. Bioeng. 2004;87:234–242. doi: 10.1002/bit.20143. [DOI] [PubMed] [Google Scholar]
- 48.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 49.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swinburne IA, Miguez DG, Landgraf D, Silver PA. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 2008;22:2342–2346. doi: 10.1101/gad.1696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fung E, Wong WW, Suen JK, Bulter T, Lee SG, Liao JC. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.