Abstract
The engineering of biological components has been facilitated by de novo synthesis of gene-length DNA. Biological engineering at the level of pathways and genomes, however, requires a scalable and cost-effective assembly of DNA molecules that are longer than ∼10 kb, and this remains a challenge. Here we present the development of pairwise selection assembly (PSA), a process that involves hierarchical construction of long-length DNA through the use of a standard set of components and operations. In PSA, activation tags at the termini of assembly sub-fragments are reused throughout the assembly process to activate vector-encoded selectable markers. Marker activation enables stringent selection for a correctly assembled product in vivo, often obviating the need for clonal isolation. Importantly, construction via PSA is sequence-independent, and does not require primary sequence modification (e.g. the addition or removal of restriction sites). The utility of PSA is demonstrated in the construction of a completely synthetic 91-kb chromosome arm from Saccharomyces cerevisiae.
INTRODUCTION
De novo synthesis of DNA is becoming an increasingly valuable resource for a broad range of applications (1). While synthesis of gene-length DNA (1–3 kb) is common, the ability to quickly and cost-effectively assemble longer-length DNA (>10 kb) remains a challenge. The process of synthetic DNA construction typically involves the assembly of overlapping oligonucleotides into contiguous fragments of dsDNA using PCR-based and/or ligation-based methods (2–5). The products of these assemblies generally require extensive clonal screening and sequence-verification to avoid mis-assembled products or products containing errors due to imperfect oligonucleotide fidelity. Coupling oligonucleotide assembly with error-filtration methods (5–7) aids in the production of higher fidelity synthetic DNA, but there remains a need for scalable technologies for subsequent assembly of sub-fragments to create longer-length DNA.
Several methods have been developed for downstream assembly of synthetic DNA to create multigene pathways or chromosome-sized molecules. PCR-based approaches have been used to assemble fragments up to ∼20 kb (8), but have been shown to have limited utility for construction of larger DNA fragments (8), and are generally less practical for construction of sequences with high GC content or repeat regions. Ligation-based methods avoid some of the sequence-dependent issues associated with PCR-based assembly, and have been used to construct large viral genomes (9) and gene clusters up to 32 kb (10). Some ligation-based methods have been developed to use common sets of restriction sites and vectors for modular (11) or hierarchical (12,13) assembly of DNA sub-fragments. All of these ligation-based assembly methods require the insertion and/or removal of restriction sites that are used in the assembly process from the primary sequence being assembled. The necessity for sequence modification is non-ideal, as the phenotypic effects of such modifications are difficult to predict, and can be detrimental (14).
Recently, recombination-based methods were used to construct a 134-kb fragment in Bacillus subtilis (15), and a 583-kb fragment in Saccharomyces cerevisiae (16,17). While these assemblies demonstrate the feasibility of constructing fragments as large as a bacterial genome (16,17), they either involve stepwise assembly (15), or rely on the absence of a particular restriction site (16,17), potentially limiting their scalability. A one-step, in vitro recombination method was also recently developed and used to assemble DNA products as large as 900 kb in vitro (18). This simple and rapid method was used to join two ∼150-kb fragments with an 8-kb vector, and, after transformation of Escherichia coli with an assembly product, approximately 50% of the colonies screened contained the correct insert size (18).
The difficulty in scaling conventional methods, together with the high cost of screening and validation of construct intermediates, hinders the reliable and cost-effective construction of long-length DNA. Here we present a general, fundamentally scalable method for the construction of long DNA sequences. This method, termed pairwise selection assembly (PSA), is a ligation-based assembly method that uses a standard set of components and operations for hierarchical DNA assembly that is ‘sequence-independent’. Importantly, PSA does not require sequence modification to accommodate the construction method (e.g. through the addition or removal of restriction sites) and uses stringent selection to minimize the screening of assembly intermediates.
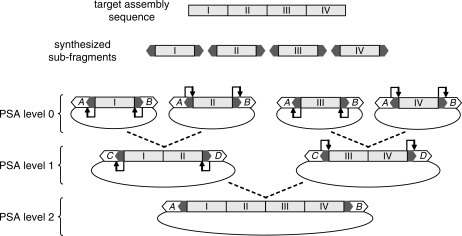
The basic components of PSA include two cassettes that encode divergently oriented, non-functional antibiotic resistance markers and a corresponding set of two recyclable activation tags. Activation tags flank all initial sub-fragments and serve two functions: (i) they enable positive selection of a paired ligation product through activation of cassette-encoded antibiotic resistance markers, and (ii) they contain sites for type IIS restriction enzymes that allow sub-fragment excision throughout the assembly process (because type IIS restriction enzymes cut outside of their recognition sequence, one activation tag from each sub-fragment is recycled for use in the next assembly round). Figure 1 illustrates the assembly of four sub-fragments over two PSA levels. At each assembly level, each sub-fragment is digested with one of two type IIS restriction enzymes (whose cognate sites are located within the activation tags) such that only one activation tag is retained (indicated by U-shaped arrows in Figure 1). This ensures that both sub-fragment members of a pair are necessary for activation of the selectable markers in the destination assembly vector. Sub-fragments from one vector type (e.g. containing selectable markers A and B) become a joined product in the second vector type (e.g. containing selectable markers C and D) (Figure 1). This enables a stringent selection of correctly ligated pairs, obviating the need for sub-fragment purification prior to ligation, and, in a majority of assemblies, enables selection directly in culture (i.e. without clonal isolation via plating). Products of one assembly level become sub-fragments of the next level of assembly, and assembly proceeds until the full-length product is constructed.
Figure 1.
Pairwise selection assembly. A target assembly sequence is broken down into sub-fragments (I–IV) that are synthesized with flanking tags (grey arrows). At PSA level 0, sub-fragments are inserted into one of two PSA vectors where tags activate two, divergently oriented selectable markers (A and B). Level 0 sub-fragment pairs are excised so that only one activation tag is retained for each sub-fragment. Subsequent pair ligation occurs in a second PSA vector where tags activate a second set of selectable markers (C and D), producing a PSA level 1 product. This hierarchical process is repeated, switching between two vectors with different selectable markers, until the full-length product is assembled.
In PSA, the fundamental operations performed on two sub-fragments to be joined are not dependent on the sequence being constructed, and do not change throughout the assembly process. The utility of PSA was demonstrated in the construction of a completely synthetic 91-kb right arm of chromosome IX from S. cerevisiae (nucleotide sequence and description of the synthetic 91-kb chromosome arm, termed synIXR, is described elsewhere by J.S. Dymond et al., High Throughput Biology Center, Johns Hopkins University School of Medicine, submitted for publication).
MATERIALS AND METHODS
Strategy for long DNA construction via PSA
In PSA, a target sequence is first broken down into a set of initial sub-fragments that are synthesized with small, recyclable tags on their termini. These tags activate vector-encoded antibiotic resistance in one of two standard PSA vectors containing different dual selectable marker cassettes. A hierarchical build tree is generated where the number of levels in the tree scales as the binary logarithm of the initial number of sub-fragments, and each level consists of a fixed number of sub-fragment pairs (Figure 1). If the initial number of sub-fragments (Fi) is not a power of two, then a subset of initial sub-fragments is built into one vector type (VAB) while the remaining sub-fragments are built into the second vector type (VCD) so that Fi = VAB + VCD, and
This breakdown simplifies the build tree by ensuring that after the first assembly level, the number of sub-fragments is a power of two (i.e. all sub-fragments will be members of a pair in all remaining assembly levels).
DNA synthesis and vector construction
CK and TS cassettes and all 64 sub-fragments of the 91-kb synthetic sequence, including flanking activation tags, were constructed from synthesized oligonucleotides using Codon Devices’ BioFAB™ platform. CK and TS cassettes were inserted between AatII and SapI sites of pUC19, retaining the origin of replication and ampicillin resistance marker, to create pCK and pTS, respectively. pCKBAC and pTSBAC were constructed by inserting DraI-digested fragments of pCK and pTS (containing the cassette regions and the bla gene) between HpaI and BstZ17I sites of pBeloBAC11. BsmBI and BsaI sites in the remaining pBeloBAC11 backbone were removed by site-directed mutagenesis.
Strains, growth and DNA preparation
Escherichia coli strain DH10B (F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ (ara, leu)7697 galU galK λ-rpsL nupG /pMON14272/pMON7124, Invitrogen) was used for all experiments. Cells were grown in LB media (Difco) with appropriate antibiotic combinations: pCK or pCKBAC clones were grown with 12.5 µg/ml chloramphenicol and 25 µg/ml kanamycin; pTS or pTSBAC clones were grown with 5 µg/ml tetracycline and 100 µg/ml spectinomycin. Cultures were grown at 37°C and 300 rpm for 15–30 h, and plates were grown at 37°C for 18–48 h.
For high-throughput processing of inputs for PSA rounds 1–3, a Qiagen BioRobot 8000 automated workstation using a QIAPrep 96 Turbo BioRobot kit was used for DNA preparation, and DNA quantification was performed using a SpectraMax M5 plate reader (Molecular Devices). Custom software was written to normalize DNA concentration to 50 ng/µl using the Qiagen BioRobot workstation and a Savant ISS110 SpeedVac concentrator. The Qiagen BioRobot 8000 transfers 500 ng (10–50 µl) from the initial DNA prep plate to a normalization plate that is placed in the SpeedVac concentrator; DNA is then resuspended in 10 µl dH2O. For the smaller number of inputs to PSA rounds 4–6, a QIAprep Spin Miniprep kit (Qiagen) or a QIAGEN Plasmid Maxiprep kit (Qiagen) were used for high-copy and low-copy backbones, respectively, and a Nanodrop ND-1000 Spectrophotometer was used for DNA normalization.
RecA-mediated blocking and PSA
All enzymes and buffer solutions were from New England Biolabs (NEB). PSA sub-fragments undergo a four-step process prior to ligation that includes (i) RecA polymerization of blocking oligonucleotides, (ii) D-loop formation, (iii) methylation and (iv) digestion. Blocking oligonucleotides (Supplementary Table S1) are incubated with RecA at a nucleotides:RecA ratio of 3:1 in a 15 µl polymerization reaction that includes 3 µg RecA, 0.27 µM blocking oligo mix (2 pmol each oligo) and 0.67 mM ATPγS (Sigma) in NEB buffer 2 (50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM DTT). The polymerization reaction is incubated at 37°C for 20 min. After RecA polymerization, 9 µl of normalized plasmid DNA (450 ng) and 1 µl of 10 × NEB buffer 2 are added to the reaction, and incubated an additional 30 min at 37°C for D-loop formation. A 5 µl methyltransferase mix of 0.5 units M.SssI and 3200 pmol S-adenosylmethionine (SAM) in NEB buffer 2 is then added directly to the reaction, and incubated at 37°C for an additional 3–4 h. The 30 µl reaction is then heated to 60°C for 25 min to inactivate the RecA protein, and either 1 unit of BsmBI (L sub-fragments), or 2 units of BtgZI (R sub-fragments) are added directly to the reaction. Digestion proceeds at 55°C for 50 min, and the enzymes are then heat-inactivated at 80°C for 25 min. In the three problematic cases where a sub-fragment junction matched a vector overhang, alkaline phosphatase (5 units) was added directly to the digest of the sub-fragment with directional, matching ends (L sub-fragment if TGAG junction, R sub-fragment if TTCA junction). In these cases, alkaline phosphatase was also used in the linearization of the destination vector to remove the 5′ phosphate from the vector overhang that matched the sub-fragment junction.
Pair ligations include equal volumes of each heat-inactivated sub-fragment reaction (3–6 µl), 200 units T4 DNA ligase and 40 ng of linearized PSA vector in T4 buffer (50 mM Tris–HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT). When necessary, a QIAquick Gel Extraction kit (Qiagen) was used to purify digested sub-fragments prior to ligation. Ligations were incubated at room temperature for 0.5–3 h, or at 16°C for 12–16 h. Chemically competent DH10B cells (Invitrogen) were transformed with assemblies for PSA levels 1–4, and electrocompetent DH10B cells (Invitrogen) were transformed via electroporation with assemblies for PSA levels 5–6 using a BTX ECM630 (BTX Harvard Apparatus) and 1 mm gap cuvettes (BTX). Both transformation procedures followed the manufacturer's instructions. For PSA levels 5 and 6, a simple de-salting step (19) was added prior to both ligation and electroporation.
Assembly verification
Prepped PSA product was analyzed by restriction digestion and agarose gel electrophoresis. At PSA level 3, all constructs were sequenced prior to further assembly using an ABI 3730 DNA Analyzer and BigDye Terminator reagent. The final, 91-kb construct was analyzed by field inversion gel electrophoresis (FIGE) with a BioRad FIGE MAPPER running program 3 (switch time ramp 0.1–2 s, linear shape, forward voltage 180 V, reverse voltage 120 V, 16 h run time). Two full-length clones selected either in culture or clonally on an LB agar plate were sequenced. A ∼300 bp region of the full-length product was highly repetitive and not covered by high quality sequencing reads (see J.S. Dymond et al., submitted for publication).
RESULTS
PSA component design
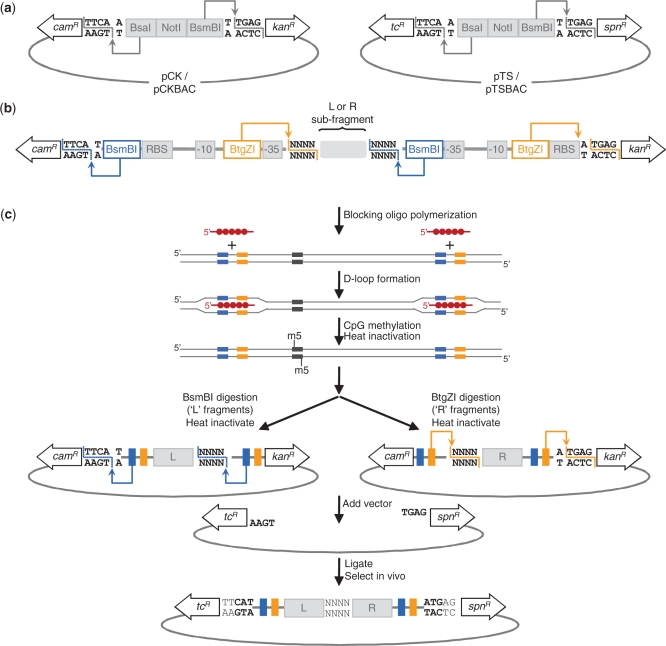
Two cassettes containing divergently-oriented, non-functional antibiotic resistance markers were synthesized. Chloramphenicol and kanamycin resistance markers were incorporated into a ‘CK’ cassette, while tetracycline and spectinomycin resistance markers were incorporated into a ‘TS’ cassette (Supplementary Table S1). Both cassettes were designed so that the incorporated antibiotic resistance markers lack promoter elements, are separated by a 20-base intergenic region containing abutting BsaI–NotI–BsmBI restriction sites, and have modified TTG start codons (Figure 2a). Digestion with BsaI and BsmBI excises the intergenic DNA, including the initial thymine base of the modified TTG start codons, leaving four-base, 5′ overhangs. In both cassettes, a BsaI cut produces a 5′-TTCA overhang, while a BsmBI cut produces a 5′-CTCA overhang (Figure 2a). Importantly, the first amino acid downstream of the start codon in the chloramphenicol resistance marker was mutated from glutamic acid to asparagine (GAG > AAC) so that the four-base overhang produced at the chloramphenicol end is identical to the overhang produced at the tetracycline end. Identical overhangs for each cassette are necessary for seamless transition from one cassette to the other as PSA progresses from one assembly level to the next.
Figure 2.
PSA components and assembly scheme. (a) Two PSA cassettes contain non-functional chloramphenicol (camR) and kanamycin (kanR), or tetracycline (tcR) and spectinomycin (spnR) resistance markers that are separated by abutting BsaI, NotI and BsmBI sites. Cassettes are inserted in high-copy and single-copy backbones to create pCK and pTS or pCKBAC and pTSBAC, respectively. The arrows and dotted lines show restriction sites for the type IIS BsaI and BsmBI enzymes that cut outside of their recognition sequences. (b) Flanking activation tags are composed of E. coli promoters embedded with BsmBI and BtgZI restriction sites which, when digested, retain either the L (BsmBI digest) or R (BtgZI digest) tag for activation of vector-encoded resistance markers (arrows and dotted lines indicate type IIS restriction sites). (c) Illustration of the steps involved in a single PSA reaction. Universal blocking oligos (red lines), polymerized with RecA (red circles), form D-loops at complementary sites within activation tags, blocking CpG methylation. After heat inactivation of RecA, L and R sub-fragments are digested with BsmBI (blue arrows) or BtgZI (orange arrows) so that one activation tag is retained per sub-fragment. Overhang compatibility ensures directional ligation with pTS vector (linearized with BsaI and BsmBI), and activation of tcR and spnR markers. Reconstituted start codons are shown in bold, and growth in tetracycline and spectinomycin media selects for a correctly paired product. Use of recyclable activation tags ensures that the product of a PSA reaction can be used as either an L or R sub-fragment in a subsequent PSA reaction.
CK and TS cassettes were inserted in a pUC19 backbone creating the high-copy pCK and pTS vectors, respectively (Figure 2a). To enable PSA-based construction of DNA fragments larger than those carried efficiently by high-copy vectors, a second set of single-copy PSA vectors were created by inserting the CK and TS cassettes into a BAC-based vector, creating pCKBAC and pTSBAC. Both sets of vectors were verified as not conferring resistance to the working concentrations of chloramphenicol and kanamycin (12.5 and 25 µg/ml, respectively), or tetracycline and spectinomycin (5 and 100 µg/ml, respectively).
Short activation tags located at the termini of each sub-fragment to be assembled via PSA contain divergently-oriented promoters designed to activate otherwise non-functional markers in pCK or pTS vector cassettes. These ∼65 bp tags differ in sequence content (to avoid inverted repeats), and are designed to promote efficient expression in E. coli based on the architecture of σ70 promoters (20). Restriction sites for two type IIS enzymes, BsmBI and BtgZI, are embedded within each promoter so that overhangs generated by digestion at sites proximal to the antibiotic resistance markers are compatible with overhangs generated by linearization of pCK or pTS, while digestion at sites distal to the markers produces compatible overhangs between adjacent sub-fragments (Figure 2b).
The pairing of two pCK sub-fragments in a pTS vector is illustrated in Figure 2c. The upstream, or ‘left’ pCK sub-fragment (L) is digested with BsmBI to retain the upstream activation tag, and the downstream, or ‘right’ pCK sub-fragment (R) is digested with BtgZI to retain the downstream activation tag. Overhangs created at the pCK cassette-tag junctions are compatible with linearized pTS. Sub-fragments are synthesized with 4 bp overlaps to ensure that the overhangs created at the junction between sub-fragments are compatible. After ligation and transformation, a correctly paired product will activate tetracycline and spectinomycin antibiotic resistance markers, enabling in vivo selection. The paired product in pTS now has flanking activation tags and becomes an L or R sub-fragment for downstream assembly in a pCK vector. The digestion–ligation–selection process is repeated until the desired DNA sequence has been constructed.
Because the activation tags are recycled, and the embedded BsmBI and BtgZI sites are used to excise L and R sub-fragments throughout the assembly process, it is important that these sites are made to be unique. Each non-palindromic six-base recognition sequence would be expected to be present at a frequency of 1/2048 bp, normally necessitating modification of the internal restriction sites prior to assembly, or requiring the use of alternate rare cutters. To avoid this issue, a RecA-assisted cleavage technique (21,22) is used to block restriction at sites located within the sequence being assembled (Figure 2c). Two sets of universal blocking oligonucleotides (Supplementary Table S2), designed with complementarity to the activation tag regions, form a RecA-mediated structure that blocks DNA methylation by CpG methyltransferase. The BsmBI and BtgZI enzymes chosen for use in PSA are sensitive to CpG methylation (23), enabling selective cleavage at the blocked sites within the activation tags upon removal of the RecA complex (Figure 2c). This process allows the use of a single set of enzymes to effectively excise full-length sub-fragments throughout the assembly process, regardless of whether or not the sites are present in the sequence being assembled.
Assembly of a synthetic 91-kb S. cerevisiae chromosome arm via PSA
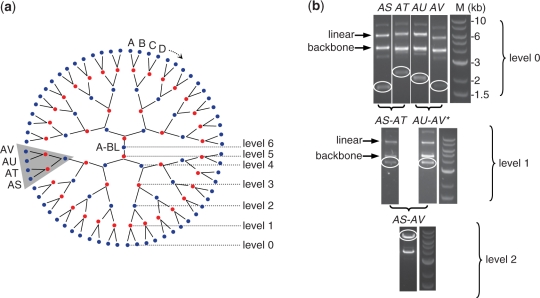
A designed 91010 bp right arm of S. cerevisiae chromosome IX termed synIXR (J.S. Dymond et al., submitted for publication) was broken down into 64 sub-fragments averaging ∼1500 bp. Sub-fragments were constructed with flanking activation tags and were sequence-verified in pCK vectors. A hierarchical build tree (Figure 3a) illustrates the 63 pairing reactions spanning six PSA levels necessary for assembly of the full-length product. All level 0 L and R sub-fragments in pCK vectors were blocked and methylated (regardless of whether or not an internal site was present), digested and mixed with linearized pTS in a ligation (see ‘Materials and Methods’ section). The reaction mix differed for L and R sub-fragments only in terms of the blocking oligos and restriction enzymes used. Importantly, the restriction enzymes were heat-inactivated prior to mixing and ligation to avoid unwanted cross-digestion of the protected tag regions. Agarose gel electrophoresis of the blocked and digested pCK sub-fragments indicates a mixture of plasmid forms, with double-cut vector (where the sub-fragment is excised from the backbone) as well as single-cut vector (where one of the two target sites is not cut) (Figure 3b, Supplementary Figures S1 and S3).
Figure 3.
Hierarchical PSA tree and assembly of 91-kb synthetic chromosome arm. (a) 64 sub-fragments are assembled to make a single 91-kb product over six PSA levels. Transition from one PSA level to the next involves transition between CK backbones (blue circles) and TS backbones (red circles). Sub-fragments are labeled alphabetically (A through BL), and successive products retain the outermost label (e.g. A–B + C–D = A–D), ensuring that all intermediates have unique identifiers, and order information is preserved. (b) Assembly of the AS through AV portion of the build tree. PSA level 0 and level 1 images represent results from blocking/digestion of sub-fragments prior to assembly (asterisk for AU–AV indicates that an internal BtgZI site was blocked); PSA level 2 image is a BsmBI digest screen of a correctly assembled product selected in culture. The same DNA molecular weight marker (M) is used in each image.
After transformation and recovery, cells were both plated on agar plates and diluted 1:50 in liquid media with 5 µg/ml tetracycline and 100 µg/ml spectinomycin. After ∼16 h of growth, plasmid DNA was extracted from non-plated cultures and screened via digestion to determine the extent of selection of the correct product in culture. Of the 32 products formed at PSA level 1, 69% (22 products) showed a correct band pattern, indicating selection of a homogeneous population of correctly assembled pairs. These constructs were used as inputs to PSA level 2, and were not clonally isolated by picking colonies on plates (e.g. see Figure 3b). The restriction digest screen of the remaining 10 products showed a mixture of correct and incorrect bands, indicating that a heterogeneous population had been selected. In these cases, four colonies were picked from plates for growth, DNA preparation and screening via digestion. Seven of the 10 remaining products were obtained by clonal selection for a total PSA yield of 29/32 or ∼91% (Table 1). The remaining 3 pairings were repeated with size-purified fragments and correct products were obtained either by selection in culture (2 products) or by clonal selection on plates (1 product). A second, independent pairing of all 64 sub-fragments showed that 71% (23 products) were successfully selected in culture as indicated by restriction digest (Supplementary Figure S2).
Table 1.
PSA details and yield for assembly of 91-kb synthetic chromosome arm
| PSA level | Number of products | Average product base pairs | Average (max.) sites blocked per sub-fragment | PSA vector | Fraction (number) of products selected |
Total PSA yield (%)c | |
|---|---|---|---|---|---|---|---|
| Culturea | Clonal/plateb | ||||||
| 0 | 64 | 1422 | 0.3 (2) | pCK | – | – | – |
| 1 | 32 | 2844 | 0.8 (3) | pTS | 0.69 (22) | 0.22 (7) | 91 |
| 2 | 16 | 5688 | 1.4 (4) | pCK | 0.31 (5) | 0.56 (9) | 88 |
| 3 | 8 | 11 376 | 3.0 (6) | pTS | 0.75 (6) | 0 | 75 |
| 4 | 4 | 22 752 | 5.5 (11) | pCK | 0.5 (2) | 0.5 (2) | 100 |
| 5 | 2 | 45 504 | 14.5 (19) | pTSBAC | 0 | 1.0 (2) | 100 |
| 6 | 1 | 91 010 | – | pCKBAC | 1.0 (1) | – | 100 |
aProducts selected in culture required a single digest screen.
bFor products that were not selected directly in culture, four cfu were grown and screened via digestion.
cPercent of products at each PSA level obtained from one round of culture/clonal screening.
This process was repeated for levels 2–4 in the PSA build tree, switching between pTS and pCK vectors and the associated dual antibiotic media. Yields for selection in culture for PSA levels 2–4 were 31% (5/16), 75% (6/8) and 50% (2/4), respectively (Table 1). The lower yield for selection in culture at PSA level 2 may have been due to the use of a an incorrectly prepared, low-concentration antibiotic, and a second, independent pairing of the 32 sub-fragments with the correct antibiotic preparation showed that 63% (10 products) were successfully selected in culture as indicated by restriction digest (Supplementary Figure S4). As with level 1, four individual colonies were selected for growth and screening via digestion for products that were not obtained by selection in culture. The combined yield at PSA levels 1–4 of the correct product selected either in culture, or by picking four plated colonies, was ∼88% (53/60) with an average of 2.2 screening assays for each of the 53 correct products. The success of selection of the correctly assembled product was not dependent on the need for restriction site blocking (due to the presence of internal restriction site(s) in one or both of the sub-fragments). For PSA levels 1 and 2, 55% of the 29 products that required blocking were selected in culture, and 38% were selected on plates. A similar breakdown was observed for the 19 products that did not have an internal restriction site where 58% were selected in culture and 26% were selected on plates. After PSA level 2, all pairs had at least one sub-fragment that required blocking.
There were seven cases, distributed over the first three PSA levels, where the correct-sized product was not obtained either by initial selection in culture, or subsequent screening of plated colonies. In these cases, the correct product was obtained by repeating the PSA process using sub-fragments that had been size-purified via agarose gel electrophoresis prior to ligation. One notable failure mode was the propagation of a single sub-fragment and its corresponding tag in the destination vector, despite the lack of the second activation tag. Of the seven cases where a correct-sized product was not initially obtained, there were three specific cases where ligation of a single sub-fragment was favored due to an exact match between the 4 bp sub-fragment junction and one of the two constant vector overhangs (Figure 2a). Although a single sub-fragment contains only one activation tag, it was observed that the ‘non-tag’ junction end could activate the downstream non-functional marker, as indicated by resistance to the associated antibiotic. There was a single case where a sub-fragment junction matched one of the two vector overhangs, but did not result in propagation of a single sub-fragment. In this case, it is notable that the sub-fragment provides a stop codon in-frame with the associated ‘promoter-less’ antibiotic resistance marker 11 codons upstream from the start codon.
To determine whether errors were incorporated either in the assembly process, or through replication in the E. coli host, the eight sub-fragments at PSA level 3 were sequenced before progressing to PSA level 4. Four errors were detected in three of the sub-fragments. The errors occurred only at sub-fragment junctions, and consisted of either deletions of one to five bases, or, in one case, a three-base insertion. Notably, three of these errors occurred in pair junctions that matched a constant vector overhang (as described above) where additional steps for size-purification were required. Mutations were ‘corrected’ by stepping back in the build tree and re-assembling from sub-fragments that were verified as error-free. In addition to size-verifying all subsequent assembly products, the three ligation junctions between vector and sub-fragments were sequence-verified to ensure junction fidelity.
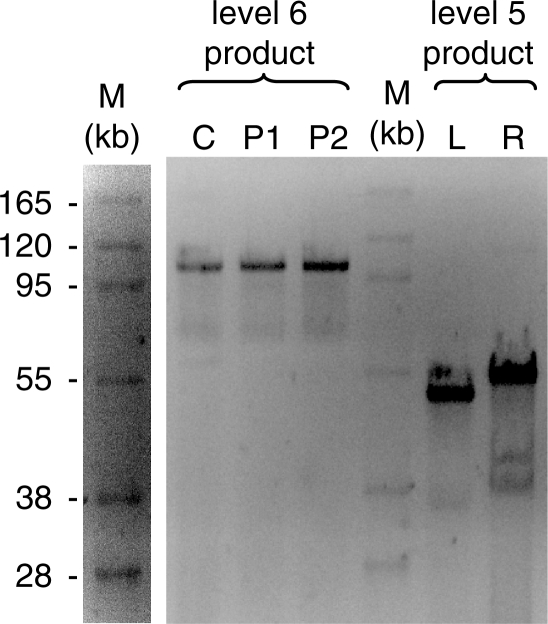
The transition was made from high-copy to single-copy PSA vectors at level 5 in the build tree due to the large size (>45 kb) of the PSA level 5 product. As anticipated, the transition from pCK to pTSBAC was seamless, and the correct product was selected based on resistance to tetracycline and spectinomycin. At this level of the assembly process, electroporation was used to increase the yield of a transformed product. Size-verified level 5 sub-fragments were combined in the final assembly step to obtain the full-length 91-kb construct (Figure 4). In the final assembly step, L and R sub-fragments contained 10 internal BsmBI sites and 19 internal BtgZI sites, respectively. Size-verification (Figure 4) and sequence-verification were both used to validate that the full-length construct was correctly assembled. Despite verifying all sub-fragments at PSA levels 0 and 3, a single C > T transition mutation was detected in the full-length product. This mutation was not present in the fully sequenced sub-fragment covering that region at PSA level 3, and is not located at a junction between two sub-fragments, possibly indicating that the mutation was generated in vivo. Importantly, the full-length 91-kb construct was selected in culture, indicating that the key features of PSA, including reaction standardization, sequence-independence and stringent selection, are scalable over a sub-fragment range of ∼1.5–45 kb.
Figure 4.
Full-length synthetic chromosome arm. FIGE with ∼45-kb L and R sub-fragments in pTSBAC, and the full-length synthetic chromosome arm in pCKBAC selected in culture (C) or on plates (P1, P2). DNA molecular weight marker (M) is a supercoiled BAC ladder (Epicentre) enhanced to show contrast.
DISCUSSION
We have demonstrated the utility of the PSA method by constructing a completely synthetic 91-kb fragment of DNA. This fragment, termed synIXR, is a completely synthetic version of the right arm of the S. cerevisiae chromosome IX (J.S. Dymond et al., submitted for publication). Yield of correctly paired sub-fragments, selected either directly in culture, or on plates, was 89% (56 out of 63 products). The remaining seven products were constructed using the fundamental PSA process, but required additional processing for sub-fragment purification prior to ligation (and in two cases, this purification was sufficient to enable selection in culture). A major failure mode involved cases where the four-base junction between sub-fragments matched one of the two destination vector overhangs exactly. Propagation of the single sub-fragment with vector-compatible ends may be due to spurious transcription of the ‘promoter-less’ resistance marker from a non-canonical promoter located within the insert DNA (20). These cases are easily avoided in future assemblies by ensuring that the 4 bp sub-fragment junctions do not include TTCA or TGAG sequences. Further constraints, such as the elimination of palindromic overhangs, can be placed on the junctions to minimize the titration of sub-fragments into non-productive complexes which may decrease the yield of a correctly ligated product. In addition, future improvement to increase selectivity in culture (e.g. by modifying the activation tags or selection stringency) may facilitate more rapid construction by further reducing or eliminating the need for clonal isolation.
Importantly, the sequence-independent PSA method enabled assembly of the entire 91-kb synIXR fragment without introducing sequence modifications to accommodate the construction method. Two common type IIS enzymes were used together with a single methyltransferase to excise sub-fragments with up to 19 internal sites. This advantage over conventional ligation-based assembly methods becomes substantial when designing and constructing pathway or chromosome-sized DNA molecules where the tolerance for sequence modification is unknown. In addition, the high selectivity of the PSA method minimizes the need for extensive handling and purification of such large fragments, limiting the potential for DNA damage. The method was effective for pairing sub-fragments ranging from ∼1.5 to ∼45 kb and may be limited only by the ability to propagate an assembled product in the chosen host. For example, Gibson et al. found that in vitro assemblies greater than ∼150 kb were not recoverable in E. coli, and moved to yeast as a more suitable host for assembly of a 583-kb synthetic chromosome (16).
PSA will be a useful tool in the assembly of next generation synthetic gene networks (24), where rationally designed libraries of components and pathways are rapidly constructed and screened for function. For example, combinatorial assembly of pathways and pathway variants from native or engineered ‘modules’ consisting of promoters, genes and other regulatory elements is feasible using pools of variant ‘L’ and ‘R’ sub-fragments rather than individual DNA molecules. The key features of PSA, including hierarchical assembly, reaction standardization and stringent selection for a correct product make this process amenable to large-scale, automated assembly of long-length DNA for a variety of synthetic biology applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Codon Devices, Inc. and the High Throughput Biology Center at The Johns Hopkins University School of Medicine, Baltimore, MD, USA. Funding for open access charge: Codon Devices, the company that sponsored and funded the work no longer exists; funds are not available for publication charges.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tim Ho, Sara Haserlat, Margot Schomp, Eric Devroe and Senthil Ramu for sub-fragment assembly and verification; Anna Huang, Gina Prophete and Caitlin Vestal for sequencing support; Jean Courtemanche, Mario Alfano and Lee Kamentsky for automation support.
REFERENCES
- 1.Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R. Engineering life: building a fab for biology. Sci. Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 2.Mandecki W, Bolling TJ. FokI method of gene synthesis. Gene. 1988;68:101–107. doi: 10.1016/0378-1119(88)90603-8. [DOI] [PubMed] [Google Scholar]
- 3.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 4.Smith HO, Hutchison CA, III, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 6.Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 8.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yount B, Curtis KM, Baric RS. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32 kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebatchouk D, Daraselia N, Narita JO. NOMAD: a versatile strategy for in vitro DNA manipulation applied to promoter analysis and vector design. Proc. Natl Acad. Sci. USA. 1996;93:10891–10896. doi: 10.1073/pnas.93.20.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight TF. Idempotent vector design for standard assembly of biobricks. MIT Synthetic Biology Working Group Technical Reports. 2003 doi:1721.1/21168. [Google Scholar]
- 13.Reisinger SJ, Patel KG, Santi DV. Total synthesis of multi-kilobase DNA sequences from oligonucleotides. Nat. Protoc. 2006;1:2596–2603. doi: 10.1038/nprot.2006.426. [DOI] [PubMed] [Google Scholar]
- 14.Menzella HG, Reisinger SJ, Welch M, Kealey JT, Kennedy J, Reid R, Tran CQ, Santi DV. Redesign, synthesis and functional expression of the 6-deoxyerythronolide B polyketide synthase gene cluster. J. Ind. Microbiol. Biotechnol. 2006;33:22–28. doi: 10.1007/s10295-005-0038-3. [DOI] [PubMed] [Google Scholar]
- 15.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 16.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 17.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., III One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 19.Atrazhev AM, Elliott JF. Simplified desalting of ligation reactions immediately prior to electroporation into E. coli. Biotechniques. 1996;21:1024. doi: 10.2144/96216bm12. [DOI] [PubMed] [Google Scholar]
- 20.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrin LJ, Camerini-Otero RD. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 22.Koob M, Burkiewicz A, Kur J, Szybalski W. RecA-AC: single-site cleavage of plasmids and chromosomes at any predetermined restriction site. Nucleic Acids Res. 1992;20:5831–5836. doi: 10.1093/nar/20.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat. Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.