Abstract
The stable cloning of giant DNA is a necessary process in the production of recombinant/synthetic genomes. Handling DNA molecules in test tubes becomes increasingly difficult as their size increases, particularly above 100 kb. The need to prepare such large DNA molecules in a regular manner has limited giant DNA cloning to certain laboratories. Recently, we found stable plasmid DNA of up to 100 kb in Escherichia coli culture medium during the infection and propagation of lambda phage. The extracellular plasmid DNA (excpDNA) released from lysed E. coli was demonstrably stable enough to be taken up by competent Bacillus subtilis also present in the medium. ExcpDNA transfer, induced by simply mixing E. coli lysate with recipient B. subtilis, required no biochemical purification of the DNA. Here, this simple protocol was used to integrate excpDNA into a B. subtilis genome, designated the ‘BGM vector’. A slightly modified protocol for DNA cloning in BGM is presented for DNA fragments >100 kb. This technique should facilitate giant DNA cloning in the BGM vector and allow its application to other hosts that can undergo natural transformation.
INTRODUCTION
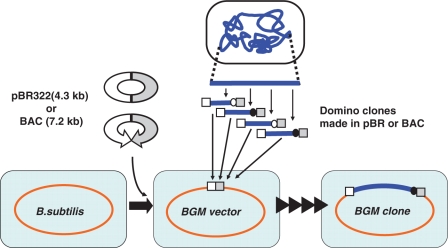
The production of recombinant DNA is the first step in all manipulations in synthetic genome biology (1). Practical methods for producing recombinant bacterial genomes of >500 kb are limited to just two techniques: the use of different microbial hosts, Bacillus subtilis (2), or the yeast Saccharomyces cerevisiae (3 4). B. subtilis, a Gram-positive bacterium, has been exploited as a platform for giant DNA cloning (5). B. subtilis derivative strains that carry integrated pBR322 sequences are collectively called ‘BGM (Bacillus genome) vectors’ (6,7). As indicated in Figure 1, the step-by-step connection of overlapping pieces of DNA allows the reconstruction of genomes in the BGM vector. Each DNA fragment, called a ‘domino clone’, must be prepared in an Escherichia coli plasmid (6–10). pBR322 derivatives with antibiotic marker for B. subtilis are used as the plasmid vectors for the dominos (9), and their sequences provide sites for homologous recombination during the integration of the fragment into the BGM vector. As shown in Figure 1, a bacterial artificial chromosome (BAC) vector, an F-plasmid-based vector that is also propagated in E. coli, is used to prepare large dominos of >100 kb (7,8,10).
Figure 1.
Design and production of recombinant genomes using the BGM vector. B. subtilis strains containing the E. coli plasmid vector pBR322 (9) or BAC (8) are designated BGM(pBR) or BGM(BAC) vectors, respectively. Those sequences preinstalled in the genomes provide cloning sites into which can be inserted DNA in pBR322- or BAC-derived vectors, respectively, prepared in E. coli. Integration of domino clone was monitored by the alternative use of the two antibiotic markers indicated by closed or open circles (9). The molecular mechanism for domino integration is shown in Figure 3.
However, the manipulation of DNA >100 kb becomes increasingly difficult in solution because of the intrinsic nature of fragile high-molecular-weight polymers. They readily shear into smaller pieces during normal DNA handling processes or with nonselective digestion by contaminating nucleases during storage (8).
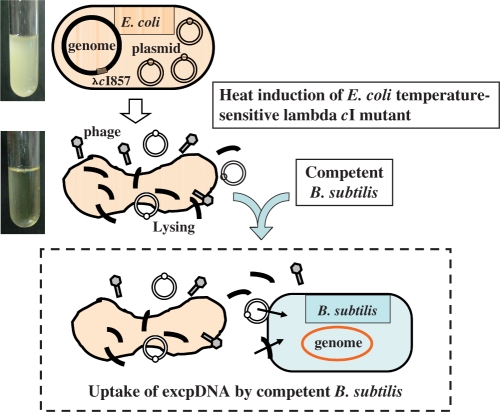
Recently, we found that plasmid DNA released from E. coli into the culture medium during lambda prophage induction was amazingly stable (11). The extracellular plasmid DNA, designated ‘excpDNA’ throughout this paper, was initially thought to be vulnerable to cellular nucleases and to be damaged by physical shearing, particularly large fragments. However, to our surprise, the excpDNA remained in the closed circular form for at least 2 h after lambda phage induction. Binary plasmids as large as 100 kb, which were also capable of replication in B. subtilis, were efficiently transferred into competent B. subtilis that was also present in the medium, as shown in Figure 2. The addition of DNase1 completely abolished the transfer of the excpDNA in the culture medium. We designated this simple protocol for the transfer of DNA from E. coli to B. subtilis the culture mix method or CMM. Its most fundamental advantage is that the plasmid DNA requires no biochemical purification. CMM was instantly applicable to the emerging BGM cloning technology. In this study, we examined whether CMM could be substituted in the current domino integration protocol, to remove the requirement for DNA purification before use.
Figure 2.
Culture mixed method (CMM) of transferring plasmids from E. coli to competent B. subtilis via extracellular plasmid DNA. (Top) E. coli lysogenized with lambda containing the cI857 mutation grow normally at 30°C. (Middle) The lysogen exposed at 37°C, causing lambda phage induction, results in a clear lysate after 2 h (11). Plasmid DNA released from the lysed E. coli into the culture medium persists as stable extracellular DNA (excpDNA) for at least 2 h. (Bottom) The excpDNA is the substrate for competent B. subtilis also present in the medium (light blue back). The integration of excpDNA into the BGM vector is described in Figure 3.
MATERIAL AND METHODS
Bacterial strains and plasmids
The E. coli strains and plasmids used are listed in Table 1. The B. subtilis strains used in this study are also listed in Table 1. Both bacteria were grown in Luria–Bertani (LB; Difco, Sparks, MD, USA) broth at 37°C unless otherwise specified. Solid medium was prepared by the addition of agar (1.5% w/v) to LB. Kanamycin (Km, 25 µg/ml) and ampicillin (Ap, 50 µg/ml) were used for E. coli selection. Neomycin (Nm, 3 µg/ml), spectinomycin (Sp, 50 µg/ml) and tetracycline (Tc, 10 µg/ml) were used for B. subtilis selection. Strains containing multiple antibiotic-resistance genes were tested using a replica plating method. The preparation and transformation of competent E. coli (12) and B. subtilis (9) cells were as previously described. The preparation of competent BGM cells specific for CMM is described in the text and Figure 4. Lambda/HindIII size markers and type II restriction enzymes were purchased from TaKaRa (Kyoto, Japan) and Toyobo (Tokyo, Japan), respectively. Molecular biology grade DNase1 was obtained from Sigma (St Louis, MO, USA).
Table 1.
Bacterial strains and plasmids
| Bacterial strains and plasmids | Relevant genotypesa | Antibiotic selectionb | Reference or source |
|---|---|---|---|
| E. coli | |||
| LE392 | F−supE44 supF58 lacY1 or del(lacIZY)6 trpR55 galK2 galT22 metB1 hsdR514(rK- mK+) | (16) | |
| MIC128 | lysogenic LE392 by λgt11 | (11) | |
| MEC5754c | pSHINE2121 | Ap | This study |
| MEC5768c | pGETS1021 | Km | This study |
| MEC5769c | pGETS1023 | Km | This study |
| MEC5770c | pGETS1036 | Km | This study |
| B. subtilis | |||
| RM125 | leuB8 arg-15 ΔSPβ hsdR hsdM | (17) | |
| BEST310 | RM125 plus proB::pBR[BAC, cI-spc] Pr-neo | Sp | (8) |
| BEST6606 | RM125 plus proB::pBR[BAC, cI-spc] Pr-neo, leuB::cat | Sp, Cm | This study |
| BEST9279 | RM125 plus proB::pBR[BAC, cI-spc] Pr-neo, leuB::tet | Sp, Tc | This study |
| Plasmids | |||
| pSHINE2121 | Derived from pBR322. bla, cat, bsr, PS10-GFPuv | (18) | |
| pGETS1021 | Derived from pGETS118. km, tetL, mtDNA(101 kb), oriS, repA | (8) | |
| pGETS1023 | Derived from pGETS118. km, tetL, mtDNA(100 kb), oriS, repA | (8) | |
| pGETS1036 | Derived from pGETS118. km, tetL, mtDNA(80 kb), oriS, repA | (8) | |
aAntibiotic-resistance genes indicated are as follows: bla, β-lactamase gene; cat, chloramphenicol acetyltransferase gene; bsr, blasticidin-S-resistance gene; km, kanamycin-resistance gene; tetL, tetracycline-resistance determinant gene (for B. subtilis). oriS functions in the replication of E. coli. repA functions in the replication of B. subtilis; binary plasmids (except pSHINE2121) replicate in both E. coli and B. subtilis.
bAp, ampicillin resistance; Km, kanamycin resistance; Sp, spectinomycin resistance; Cm, chloramphenicol resistance; Tc, tetracycline resistance.
cDerived from MIC128.
Figure 4.
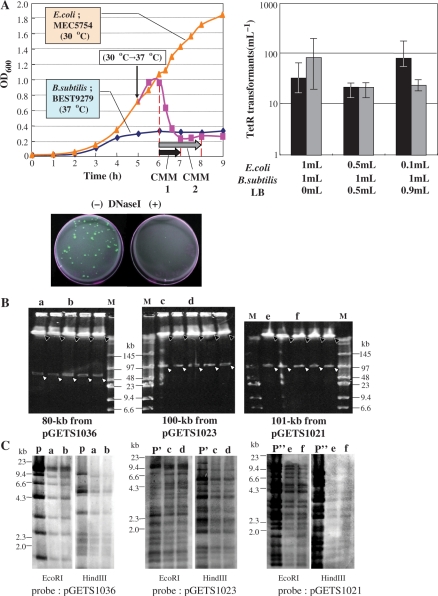
Integration of DNA in the BGM(BAC) vector. (A) Filled triangles (orange) with solid line show the complete growth curve of MEC5754 (pSHINE2121) in LB medium at 30°C. Filled squares (pink) with solid line show growth after the temperature was increased to 37°C. Solid diamonds (blue) with solid line show the growth curve for B. subtilis BEST9279, BGM(pBR), which developed competency at 5–6 h. The different culture mix periods, CMM1 and CMM2, are shown by black and gray horizontal arrows, respectively. The frequencies (colonies/ml) of colonies resistant to Cm and Tc, measured for CMM1 and CMM2 with three different mix ratios, are shown. All BGM colonies formed on LB plates supplemented with Cm and Tc were fluorescent under UV irradiation because they had acquired the GFPuv gene. The BGM colonies were completely suppressed because excpDNA is sensitive to the addition of DNase1. (B) The genomes of strains obtained through spontaneous integration acquired two I-PpoI sites, as shown in Figure 3B. Five representative strains, each of which generated 80-kb, 100-kb or 101-kb fragments are indicated by open arrowheads. Solid arrowheads indicate the BGM. Size markers are shown in lane M. Running conditions for CHEF: 3 V cm–1, 18-s pulse time, 16-h running time, at 14°C. (C) Southern hybridization of EcoRI and HindIII digests of the genomes of the two representative strains examined in Figure 4B. Lane P, P′ and P′′′ include original BAC plasmid digested with the indicated enzyme. Whole plasmid was used as the probe for each.
DNA isolation and manipulation
Plasmid DNA was isolated from E. coli with the alkali–sodium dodecyl sulfate method (12). The plasmid DNA was isolated from B. subtilis similarly but with an increased lysozyme concentration (10 mg/ml) in solution I. B. subtilis genomic DNA was extracted with the liquid isolation method (13) and used for Southern analysis. Intact unsheared B. subtilis genomic DNA was prepared in agarose gel plugs for contour-clamped homogeneous electric field (CHEF) gel electrophoresis, as described elsewhere (13). I-PpoI endonuclease was purchased from Promega (Madison, WI, USA).
Electrophoresis and Southern hybridization
CHEF gel electrophoresis was conducted in agarose gels (1.0% w/v) in TBE solution [50 mM Tris–borate (pH 8.0), 1.0 mM ethylenediaminetetraacetic acid (EDTA)], with running conditions as described in the legend to Figure 4B. Agarose gels (1.0% w/v) in TAE solution [50 mM Tris–acetate (pH 8.00, 1.0 mM EDTA] were used for conventional gel electrophoresis at room temperature.
After electrophoresis, the gels were stained with ethidium bromide solution and visualized under ultraviolet (UV) light. A nonradioactive labeling nucleotide, digoxigenin-11-dUTP, was used for Southern hybridization. The probes were prepared with the random primer included in the DIG-High Prime kit (Roche, Mannheim, Germany). Labeled bands were detected with the DIG Nucleic Acid Detection Kit (Roche), and visualized with 5-bromo-4-chloro-30-indolylphosphate and nitroblue tetrazolium salt (Sigma–Aldrich, St Louis, MO, USA).
Small domino integration with the standard CMM protocol
pSHINE2121 (Figure 3) can replicate in E. coli only and must be integrated into the pBR322 sequence of the BGM vector for chloramphenicol (Cm) selection. The donor E. coli MEC5754 was obtained by the transformation of MIC128 cells (lambda lysogen) (11) with the pSHINE2121 plasmid, with selection at 30°C on LB plates supplemented with Ap. A culture in LB medium containing Ap was incubated at 30°C for 17 h and was diluted 1:200 (v/v) in 20 ml of prewarmed LB supplemented with the same antibiotic in a 100-ml flask. The medium was shaken at 120 r.p.m. for 5 h at 30°C, after which the temperature was increased to 37°C to induce temperate λgt11(cI857). As described in Figure 4A, 1 ml of lysed E. coli medium, incubated for 1 h at the increased temperature, was mixed with the same volume of competent B. subtilis (BGM vector BEST9279), which had been simultaneously prepared from a starting culture in TFI medium [1.4% K2HPO4, 0.6% KH2PO4, 0.2% (NH4)2SO4, 0.1% Na-citrate, 0.5% glucose, 0.02% MgSO4.7H2O, 0.05 mg/ml Trp, 0.05 mg/ml Arg, 0.05 mg/ml Leu, 0.05 mg/ml Thr] at 37°C. The mixed culture (2 ml) was continuously shaken for 1 or 2 h at 37°C. An aliquot (200 µl) of this culture was spread onto LB plates supplemented with Cm and Tc. All experiments were conducted in triplicate. Freshly prepared DNase1 was added, if necessary, to the mixture at a final concentration of 3.4 µg/ml. The growth curve measured at 120 r.p.m. and 37°C is shown in Figure 4A.
Figure 3.
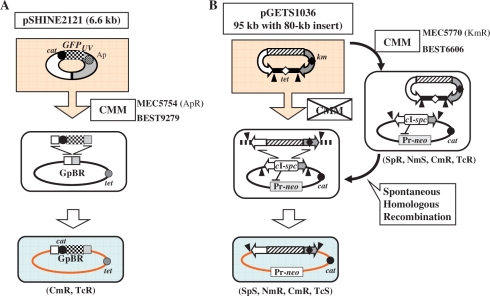
Integration of excpDNA into the genome. (A) Transferred excpDNA must be integrated in the genome to be selected with Cm (Bottom). The recipient BGM(pBR) vector BEST9279, derived from BEST310, contains the pBR322 sequence (4.3 kb) in the proB gene and tet (TcR) marker in the leuB gene (Table 1). The integration of pSHINE2121, including the GFPuv gene, occurred by homologous recombination, as depicted (Middle). (B) BEST6066, a BGM(BAC), includes the BAC sequence and a neomycin-resistance gene (Pr-neo) inserted in the proB gene and between the NotI sites of yvfC and yveP, respectively (8). The BAC-based binary plasmid pGETS1036 is composed of three parts: BAC (two curved arrows, one with the kan marker for E. coli, shown as the shaded box); an 80-kb Arabidopsis thaliana mitochondrial DNA insert and a replicon for B. subtilis (the tet-marked horizontal bar) (8). The two closed triangles indicate the sites for I-PpoI. The two-step integration is shown, in which the first step of plasmid transfer was performed with CMM, as shown in Figure 4A. The markers that are altered are indicated below the cell. cI-spc indicates a cassette in which two genes are combined: cI857 and the spc gene conferring resistance to Sp (8). The spontaneous homologous recombination that occurs in the second step is described in the text.
RESULTS
Integration of domino DNA in pBR322 plasmid
pBR322-based plasmids are unable to replicate in B. subtilis and undergo direct integration into the BGM vector by the molecular mechanism shown in Figure 3A. B. subtilis that lacked the genomic pBR322 sequence, such as RM125 in Table 1, produced no transformants. The pSHINE2121 plasmid transferred by the CMM protocol from lysed E. coli to competent B. subtilis, BEST9279, should produce transformants resistant to Cm only when the plasmid integrates by homologous recombination between the shared pBR322 sequences. As shown in Figure 4A, all the Cm-resistant (CmR) colonies exhibited clearly visible green fluorescence under UV light of 365 nm. The number of CmR transformants did not vary significantly with different mixing ratios. This is consistent with our previous observations and suggests that the DNA for the competent B. subtilis cells was saturated (11). The complete suppression of the transfer when DNase1 (3.4 µg/ml) was added supports the presence of pSHINE2121 as excpDNA in the CMM. This result indicates that the integration of the domino DNA into the BGM vector can be conducted with the same standard method that is used for plasmid delivery into B. subtilis.
Modified protocol used to integrate BAC-based dominos
The integration of dominos larger than pSHINE2121 could be based on BAC clones. For this, a combination of BAC dominos and a BGM vector containing BAC sequences instead of pBR322 sequences is required, as shown in Figure 3B. As BACs do not contain an antibiotic selection marker for B. subtilis, we constructed a BGM vector suitable for BAC integration (8). Several E. coli–B. subtilis binary BACs of around 100 kb were constructed using the BGM vector (8). We have previously reported that one of these BACs, pGETS1036 (95 kb), was successfully transferred by CMM and established as a plasmid in B. subtilis (11). Therefore, the two-step integration protocol described in Figure 3B was tested using binary BACs and the BGM(BAC) vector BEST6606. In the first step, the binary BAC pGETS1036 (95 kb) was transferred from E. coli (MEC5770) to BEST6606 with the standard CMM, and established as a plasmid. Among the seven colonies selected with the plasmid-associated Tc-resistance marker (tet), five representatives were analyzed for their plasmid structure. Two I-PpoI recognition sequences were inserted, one at each end of the BAC vector region, as shown in Figure 3B. Digestion with the enzyme generated two fragments, an 80-kb insert and the vector region for B. subtilis (including the tet gene) (data not shown). BEST6606 contains regions homologous to the BAC part of plasmid pGETS1036. As pGETS1036 replicates and is maintained as a single copy per cell (8), homologous recombination causes the integration of the long insert when it replaces the pre-existing cI repressor gene, as indicated in Figure 3B. Pr-neo plays a crucial role in the selection of the integration event. The Pr promoter regulates the expression of the neo gene. The expressed CI857 protein binds to the Pr promoter and represses the expression of the neo gene of BEST6066. The loss of the cI gene with its replacement by the BAC insert derepresses the Pr promoter activity, resulting in the full expression of the neo gene, conferring Nm resistance on the strain. Integration and replacement occurred spontaneously at a low but specific frequency during growth (Tsuge,K. and Itaya,M., unpublished data). pGETS1036/BEST6606 grown in LB medium without antibiotics produced colonies (several hundreds/ml) on LB plates supplemented with Nm at 5 µg/ml. Five randomly chosen representative colonies were examined for other markers. They were all sensitive to 10 µg/ml Tc and 50 µg/ml Sp, consistent with the loss of both the replicon for B. subtilis and the genomic cI857 gene, as shown in Figure 3B. The presence of the complete 80-kb insert was confirmed by I-PpoI digestion of the genomes as shown in Figure 4B. Southern blot analysis of EcoRI and HindIII digests of two strains verified that the 80-kb DNA insert came from pGETS1036, based on its structure (Figure 4C). These findings demonstrate that no structural alteration of the insert occurred during the two-step integration process.
The same protocol was applied to two other binary BAC plasmids (8), pGETS1023 (100-kb insert) and pGETS1021 (101-kb insert). The first step of CMM yielded eight and seven Tc-resistant (TcR) colonies, respectively. Five representatives were shown by I-PpoI digestion to contain the same plasmid, replicating as a single copy in BEST6066 (data not shown). They produced similar numbers of Nm-resistant (NmR) derivatives in the second step. Five NmR representatives of each showed the integration of the 100-kb or 101-kb DNA insert, as verified by I-PpoI digestion (Figure 4B). Southern analysis of EcoRI and HindIII digests of both transformants verified that the two-step integration was completed without structural alteration of the inserts (Figure 4C).
DISCUSSION
The design and preparation of domino clones constitute a prerequisite for giant DNA cloning in the BGM vector (5,9). The size of the domino is a particularly critical factor: the larger the individual domino size, the fewer dominos are required. Another important factor not mentioned previously is the possible automation of the domino integration step. The size of the DNA fragments that can be incorporated into competent B. subtilis cells in a single transformation has been estimated to be >200 kb (10). Unlike yeast, in which about 1 Mb of DNA can be effectively transformed (3,4,14), agarose inhibits the transformation of B. subtilis. Thus, giant DNA prepared in agarose plugs and extracted by agarase digestion is difficult to use in BGM cloning. Instead, a genetic cross between two BGMs that contain two overlapping dominos has been shown to produce a 355-kb DNA in one step (10). The BGM vector can be used to reconstruct DNA fragments as large as 3600 kb in total (2) and probably even larger after many genetic crosses. Therefore, the convenient and efficient delivery of a giant domino DNA to the BGM vector is crucial.
Dominos are currently prepared in E. coli, which is a universal host that acts not only as a DNA reservoir but also as a workhorse in various DNA manipulations. E. coli normally contains engineered DNA in a plasmid form. The immediate use of excpDNA in E. coli lysis medium as a DNA reservoir for the transformation of B. subtilis offers a new way to circumvent the biochemical preparation of DNA from E. coli in the size range above 100 kb.
Recent cutting-edge technology applied to the de novo chemical synthesis of DNA has promoted the phenomenal synthesis of designed DNA sequences from scratch (4). Although chemically synthesized DNA has not yet been tested in BGM cloning, contiguous dominos could be prepared in appropriate E. coli plasmid vectors by bottom-up assembly (15). As fewer and simpler steps are desirable in the BGM cloning system, CMM, which can handle many samples at once, may be suitable for a potential multiplexed, automated process. Several points remain to be investigated, including the effectiveness of the simultaneous incorporation for more than two donor E. coli lysates. The establishment of one-step integration requires the preparation of BAC dominos carrying a B. subtilis marker but lacking a replicon for B. subtilis. This must be examined. The BAC vector presently persists as a low-copy-number inclusion in E. coli. Medium- or high-copy-number vectors, such as fosmids or cosmids, may be more efficient. However, they reduce the size of the DNA that can be stably cloned. Furthermore, methods to lyse E. coli may not be limited to the presently used induction of a lysogenic lambda by temperature shift.
Our results indicate that a circular BAC plasmid of >100 kb no longer requires careful purification when it is to be transferred into other cells of a different species or genus. In fact, a similar protocol using excpDNA is under investigation, with application to various competent recipient cells.
ACKNOWLEDGEMENTS
We thank Ms S. Segawa, Ms R. Kuniyasu and Mr K. Komai for their technical assistance. Funding for open access charge: Keio University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 2.Itaya M, Tsuge K, Koizumi M, Fujita K. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl Acad. Sci. USA. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lartigue C, Vashee S, Algire MA, Chuang RY, Bender GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 4.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 5.Itaya M. Recombinant genomes. In: Fu P, Panke S, editors. Systems Biology and Synthetic Biology. Hoboken, NJ: John Wiley & Sons, Inc.; 2009. pp. 155–192. [Google Scholar]
- 6.Itaya M, Nagata T, Shiroishi T, Fujita K, Tsuge K. Efficient cloning and engineering of giant DNAs in a novel Bacillus subtilis genome vector. J. Biochem. 2000;128:869–875. doi: 10.1093/oxfordjournals.jbchem.a022825. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko S, Tsuge K, Takeuchi T, Itaya M. Conversion of submegasized DNA to desired structures using a novel Bacillus subtilis genome vector. Nucleic Acids Res. 2003;31:e112. doi: 10.1093/nar/gng114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko S, Akioka M, Tsuge K, Itaya M. DNA shuttling between plasmid vectors and a genome vector: systematic conversion and preservation of DNA libraries using the Bacillus subtilis genome (BGM) vector. J. Mol. Biol. 2005;349:1036–1044. doi: 10.1016/j.jmb.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 9.Itaya M, Fujita K, Kuroki A, Tsuge K. Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat. Methods. 2008;5:41–43. doi: 10.1038/nmeth1143. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko S, Takeuchi T, Itaya M. Genetic connection of two contiguous bacterial artificial chromosomes using homologous recombination in Bacillus subtilis genome vector. J. Biotech. 2009;139:211–213. doi: 10.1016/j.jbiotec.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko S, Itaya M. Designed horizontal transfer of stable giant DNA released from Escherichia coli. J. Biochem. doi: 10.1093/jb/mvq012. doi:10.1093/jb/mvq012 [9 February 2010, Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EC, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Itaya M, Tanaka T. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene directed mutagenesis method. J. Mol. Biol. 1991;220:631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- 14.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 15.Tsuge K, Matsui K, Itaya M. Production of the non-ribosomal peptide plipastatin in Bacillus subtilis regulated by three relevant gene blocks assembled in a single movable DNA segment. J. Biotech. 2007;129:592–603. doi: 10.1016/j.jbiotec.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Itaya M, Crouch RJ. Correlation of activity with phenotypes of Escherichia coli partial function mutant of rnh, the gene encoding RNase H. Mol. Gen. Genet. 1991;227:433–437. doi: 10.1007/BF00273934. [DOI] [PubMed] [Google Scholar]
- 17.Itaya M, Tanaka T. Predicted and unsuspected alterations of the genome structure of genetically defined Bacillus subtilis 168 strains. Biosci. Biotechnol. Biochem. 1997;61:56–64. [Google Scholar]
- 18.Ohashi Y, Ohshima H, Tsuge K, Itaya M. Far different levels of gene expression provided by an oriented cloning system in Bacillus subtilis and Escherichia coli. FEMS Microbiol. Lett. 2003;221:125–130. doi: 10.1016/S0378-1097(03)00171-X. [DOI] [PubMed] [Google Scholar]