Abstract
GeneDesign is a set of web applications that provides public access to a nucleotide manipulation pipeline for synthetic biology. The server is public and freely accessible, and the source is available for download under the New BSD License. Since GeneDesign was published and made publicly available 3 years ago, we have made its code base more efficient, added several algorithms and modules, updated the restriction enzyme library, added batch processing capabilities, and added several command line modules, all of which we briefly describe here.
INTRODUCTION
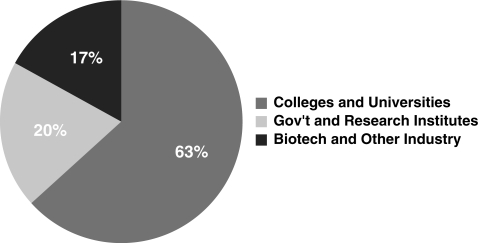
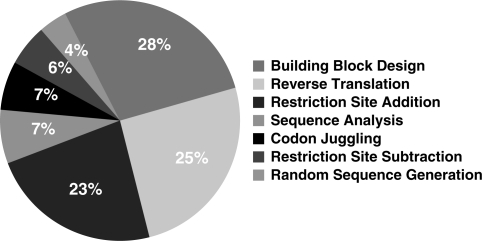
GeneDesign was originally developed as an in-house project to automate the design of oligonucleotides for the construction of individual synthetic genes (1). Gene synthesis is becoming more practical as synthesis costs decrease, and many excellent computational tools have been introduced to aid in the design of synthetic constructs, the most recent of which are compared in Table 1. In 6 months from June 2009 to December 2009, GeneDesign was accessed over 2000 times from cities all over the globe (Figure 1). College and university networks account for 63% of GeneDesign access (Figure 2). The most popular modules are Building Block Design, Reverse Translation and Restriction Site Addition, the three of which account for three quarters of traffic (Figure 3). Since our original publication we have adapted GeneDesign for the construction of entire chromosomes (2). This genome-scale project has necessitated new modules, a new approach to assembly of multikilobase genes from oligonucleotides, and significant improvement to the efficiency of the underlying code.
Table 1.
Synthetic design tools
| Program name | Availability | Open source? | Manipulations offered | Batch processing? |
|---|---|---|---|---|
| GeneDesign (1) | Free webserver, source code downloadable scripts (PC/Mac/Linux/Unix) | Yes | Codon, restriction enzyme, reverse translation, large gene assembly, general sequence, assembly oligonucleotides | Yes |
| GenoCAD (7) | Free webserver | No | Construct assembly, semantic analysis | No |
| TmPrime (8) | Free webserver | No | Codon, large gene assembly, assembly oligonucleotides | No |
| OPTIMIZER (9) | Free webserver | No | Codon, restriction enzyme, reverse translation | No |
| Gene Designer (10) | Free executable (PC/Mac) | No | Codon, restriction enzyme, reverse translation, general sequence, sequencing oligonucleotides | No |
| Gene2Oligo (11) | Free webserver | No | Assembly oligonucleotides | No |
| DNAWorks (12) | Free webserver | No | Codon, restriction enzyme, reverse translation, assembly oligonucleotides | Yes |
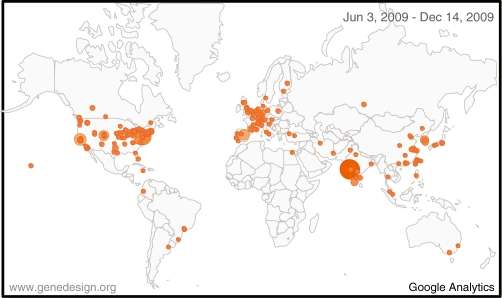
Figure 1.
Visits to GeneDesign in a 6-month period have come from 368 cities in 47 countries, as measured by Google Analytics. The diameter of the circle corresponds to number of visits.
Figure 2.
Distribution of visiting network types to GeneDesign in a 6-month period, as measured by Google Analytics.
Figure 3.
Distribution of access to GeneDesign modules in a 6-month period, as measured by Google Analytics.
NEW MODULES
Generate relative synonymous usage values
The relative synonymous usage (RSCU) value for a codon is the ratio of how often that codon is seen over how often it would be expected to be seen, given a completely random distribution (3). GeneDesign is equipped with a canonical set of RSCU values from early studies in codon distribution on a set of genes with observed high rates of expression (4). However, at the time the study was performed, there were only 809 gene sequences available from six model organisms. Although we have enjoyed experimental success with the sequences designed using the old RSCU data set, we wanted to be able to determine the RSCU values for genes in much more targeted subsets; for example, in designing a yeast histone gene, we may wish to use RSCU values derived from many yeast histone genes as a reference set rather than values derived from the whole yeast genome. This module takes as input a single gene or a list of genes in FASTA format and returns a table of RSCU values, as well as a table of the most often used codons for each residue, which may be directly used in the Reverse Translation module. This module is now available as a command line script.
Codon bias graphing
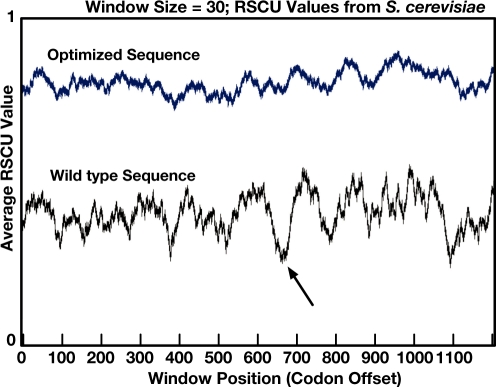
It is sometimes helpful to visualize the change in RSCU values across a gene. Significant deviations from the average RSCU value in a sub-sequence may indicate a nucleotide requirement (such as an effect on local translation rate, which might affect folding) that would be disrupted by manipulation. In Figure 4, we show the wild type and optimized sequences for the integrase and reverse transcriptase open reading frame from the Saccharomyces cerevisiae Ty1 transposon. The wild-type sequence has a significant dip in RSCU averages that is completely disrupted by optimization; this valley corresponds to the nucleotides at the boundary between the two genes, an area which has been shown to have an impact on the expression of reverse transcriptase (5). The new Codon Bias Graphing module accepts FASTA input and generates a graph of the average RSCU value across the length of each gene.
Figure 4.
Output from the codon bias graphing module visualizing the average RSCU value across the wild-type and optimized sequences for the integrase/reverse transcriptase polyprotein from yeast Ty1. The boundary between the two protein coding regions can be seen in the wild-type curve as a deep valley at offset 600; the optimized curve does not have this valley.
Building block design
The original release of GeneDesign employed a very simple model of gene assembly. Multikilobase coding regions were designed to be synthesized as ∼500 bp segments, or building blocks, that overlapped at restriction enzyme recognition sites. Building blocks were assembled by restriction digestion and ligation. This works well for genes on the small side of the multikilobase scale; otherwise, practical restriction enzymes sites are used up rather quickly. We currently enjoy success in synthesizing ∼750 bp building blocks and have updated GeneDesign's defaults to reflect this. We have also moved away from the restriction enzyme model for the assembly of large genes (and even small chromosomes) and have therefore added a module that designs oligos and primers for the assembly of building blocks based on a uracil excision reaction (USER) protocol (6). This module carves up chunks of 10 kb or longer into building blocks of any desired size, and defines endpoints at sequences conforming to the consensus ANxT, where x is an odd integer and where the chosen sequence is shared between adjacent building block ends. Picking an odd integer ensures that the overhang will be non-palindromic and hence will assemble in only one orientation. The default lets x be any odd number between 5 and 11 (generating USER overhangs of 7, 9, 11 or 13 base pairs). We empirically determined that with yeast DNA, a mixture of x values results in the nearly constant building block lengths that are desirable for production synthesis. GeneDesign also ensures that every building block except the adjacent ends meant to assemble together, will have incompatible overhangs, facilitating assembly in a single defined order and orientation. A third option for building block synthesis is to simply have GeneDesign assign overlaps of a constant length. This can be used to assemble building blocks by overlap extension PCR or exonuclease-based methods. This module is now available on the command line, where it offers the ability to design building blocks and oligos from multiple sequences at once.
UPDATED MODULES
All codes have been revised for efficiency, consistency and compatibility. In addition, all modules that output gene sequences now offer FASTA output.
Reverse Translation
The Reverse Translate module takes a protein sequence and replaces each residue with a user-determined codon. Typically, users select one of GeneDesign's codon sets, but they may also define their own. This module now offers Bacillus subtilis and Drosophila melanogaster RSCU data sets. It will now accept a set of protein sequences in the FASTA format. This module is now available as a command line script, where it offers the ability to design sequences reverse translated for more than one organism at a time.
Codon Juggling
The Codon Juggling module takes a protein coding nucleotide sequence and offers several synonymous, algorithmic variations on it. This module now has a new algorithm, ‘least different RSCU’, which seeks to replace as many codons as possible while minimizing disruption of the original average RSCU value for the sequence. This module is now available as a command line script, where it offers the ability to design sequences using multiple algorithms and for more than one organism at a time.
Restriction Site Subtraction
The Restriction Site Subtraction module takes a protein coding nucleotide sequence and allows the user to specify which restriction sites will be removed without modifying the encoded protein sequence. The Subtraction module has been modified to use the ‘least different RSCU’ algorithm when replacing codons in order to minimize the impact of each edit, and to change as few codons as possible in every edit.
Restriction Enzyme Filter
The restriction enzyme database used by GeneDesign has been updated to include a more current set of commercially available enzymes and their prices (15). The enzyme choosing module has been updated to add overhang palindromy, heat inactivation, star activity, optimal incubation temperature, incubation buffer and methylation sensitivity as filter criteria.
COMMAND LINE MODULES
We have modified the most popular modules to use a command line interface, allowing high-throughput design of synthetic genes and enabling GeneDesign to be embedded in other software applications and synthesis pipelines. Currently, the Reverse Translation, Codon Juggling and Building Block Design modules are implemented as scripts executable on a POSIX command line that use the FASTA format for input and output. There is evidence that codon optimization, in some cases, is more involved than using the most highly expressed codons. The command line modules allow users to either use GeneDesign’s built-in codon definitions or to define their own codon tables for special cases. For example, recent work indicates that Escherichia coli protein expression is optimized by using codons that are charged during amino acid starvation rather than overrepresented in highly expressed proteins (16).
AVAILABILITY
An instance of GeneDesign is freely available at http://www.genedesign.org and the source is now available under the New BSD License from a github source control server at http://github.com/GeneDesign.
IMPLEMENTATION AND DEPENDENCIES
The GeneDesign libraries are written entirely in Perl; the online interface is HTML and JavaScript. GeneDesign uses the gd graphics library freely available from Boutell.com to render graphs.
FUTURE DEVELOPMENT
We will continue to expand the command line interface to the GeneDesign libraries. Planned modules will select unique PCR primers that distinguish between original and recoded sequences, suggest sequencing primers, and identify hairpins in designed sequences and oligonucleotides. We regularly solicit users for suggestions for new modules and for improving the web interface and command line version.
FUNDING
Department of Energy (grant number DE-FG02097ER25308 to S.M.R.); Microsoft Research (to J.S.B.); National Science Foundation (grant numbers MCB0718846 to J.D.B. and J.S.B., MCB-0546446 to J.S.B.). Funding for open access charge: Department of Energy (grant number DE-FG02097ER25308 to S.M.R.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
Many thanks to Jessica Dymond for insightful comments and enthusiastic beta testing, and to John Kloss for a suffix tree implementation.
REFERENCES
- 1.Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 2006;16:550–556. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dymond JS, Scheifele LZ, Richardson SM, Lee P, Chandrasegaran S, Bader JS, Boeke JD. Teaching synthetic biology, bioinformatics and engineering to undergraduates: the interdisciplinary build-a-genome course. Genetics. 2009;181:13–21. doi: 10.1534/genetics.108.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm M, Boutabout M, Wilhelm FX. Expression of an active form of recombinant ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the c-terminal region of the ty1 integrase linked to the reverse transcriptase-rnase h domain exhibits polymerase and rnase h activities. Biochem. J. 2000;348:337–342. [PMC free article] [PubMed] [Google Scholar]
- 6.Bitinaite J, Rubino M, Varma KH, Schildkraut I, Vaisvila R, Vaiskunaite R. User friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czar M, Cai Y, Peccoud J. Writing DNA with genocadtm. Nucleic Acids Res. 2009;37:W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bode M, Khor S, Ye H, Li M-H, Ying JY. TmPrime: fast, flexible oligonucleotide design software for gene synthesis. Nucleic Acids Res. 2009;37:W214–21. doi: 10.1093/nar/gkp461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–31. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285–292. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouillard JM, Lee W, Truan G, Gao X, Zhou X, Gulari E. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–80. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for pcr-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Marchisio MA, Stelling J. Computational design tools for synthetic biology. Curr. Opin. Biotechnol. 2009;20:479–485. doi: 10.1016/j.copbio.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Roberts R, Vincze T, Posfai J, Macelis D. Rebase- a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2009 doi: 10.1093/nar/gku1046. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]