Abstract
We have achieved the ability to synthesize thousands of unique, long oligonucleotides (150mers) in fmol amounts using parallel synthesis of DNA on microarrays. The sequence accuracy of the oligonucleotides in such large-scale syntheses has been limited by the yields and side reactions of the DNA synthesis process used. While there has been significant demand for libraries of long oligos (150mer and more), the yields in conventional DNA synthesis and the associated side reactions have previously limited the availability of oligonucleotide pools to lengths <100 nt. Using novel array based depurination assays, we show that the depurination side reaction is the limiting factor for the synthesis of libraries of long oligonucleotides on Agilent Technologies’ SurePrint® DNA microarray platform. We also demonstrate how depurination can be controlled and reduced by a novel detritylation process to enable the synthesis of high quality, long (150mer) oligonucleotide libraries and we report the characterization of synthesis efficiency for such libraries. Oligonucleotide libraries prepared with this method have changed the economics and availability of several existing applications (e.g. targeted resequencing, preparation of shRNA libraries, site-directed mutagenesis), and have the potential to enable even more novel applications (e.g. high-complexity synthetic biology).
INTRODUCTION
In the past 20 years, the chemical synthesis of nucleic acids has overwhelmingly been performed using variations of the phosphoramidite chemistry on solid surfaces (1,2). The reasons for this universal adoption are both the efficiency of the chemistry and its versatility resulting in numerous adaptations to achieve specific needs. For instance, phosphoramidite based methods have been used to synthesize abundant base, backbone and sugar modifications of deoxyribo and ribonucleic acids (3–8), as well as nucleic acid analogs (9–11). While the phosphoramidite chemistry has been used to synthesize these analogs individually on a medium-to-large scale (nmol to mmol), high throughput biological studies that involve thousands of genes demanded new parallel DNA synthesis methods. For this purpose, the phosphoramidite chemistry has been adapted for in situ synthesis of DNA microarrays. Such synthesis has been achieved by spatial control of one step of the synthesis cycle, which results in thousands to hundreds of thousands of unique oligonucleotides distributed on an area of a few square centimeters. The main methods used to achieve spatial control include (i) control of the coupling step by inkjet printing (Agilent, Protogene) (12,13) or physical masks (Southern) (14), (ii) control of the 5′-hydroxyl deblock step by classical (Affymetrix) (15) and maskless (Nimblegen) (16) photolithographic deprotection of photolabile monomers or (iii) digital activation of photogenerated acids to carry out standard detritylation (Xeotron/Atactic) (17). More recently, oligonucleotides made on commercial microarrays have been cleaved from their solid surface and pooled to enable new applications such as shRNA libraries (18), gene synthesis (19,20) and site-directed mutagenesis (21). While there are other avenues to create pools of oligonucleotides of length <100mer (22,23), microarray-based synthesis of oligonucleotides is a method particularly well adapted to any application that requires minute amounts of thousands of different DNA sequences in a cost effective manner.
Important features of oligonucleotide libraries are the quantity of material recovered and the quality of the individual oligonucleotides. While the limitations on quantity can be mitigated relatively easily by PCR amplification, the sequence integrity of the nucleic acids must be maximized chemically during synthesis, and/or by post-synthesis purification of failure sequences such as by enzymatic assisted selection (24,25). In addition, the maximum length of oligonucleotides synthesized will significantly affect the versatility of the libraries. While the recent studies cited above reported the use of oligonucleotides between 40 and 97mers, we have received requests for libraries of high-quality oligonucleotides beyond that length. The rationale for such libraries of long oligonucleotides is 2-fold. First, long oligonucleotides will increase the library complexity, i.e. the number of individual bases per library, and hence increase the extent of genetic variation to be studied simultaneously. Second, long-oligonucleotide libraries will enable researchers to embed increasingly complex and numerous components, such as primers, binding probe(s), reporter probes, etc. within the same oligonucleotide strand.
However, chemical synthesis of long (≥150mer) oligonucleotides, either on controlled-pore glass (CPG) or non-porous solid surfaces as used in microarrays, present a number of challenges. As the oligonucleotide length starts to exceed 100 nucleobases, the yield of desired products becomes limited by side reactions and even modest inefficiencies within the stepwise chemical reactions have large effects. Successful synthesis of long oligonucleotides requires a stepwise coupling yield ≥99.5%. If the coupling efficiency falls below 99%, the impact on sequence integrity follows one of two scenarios. If capping is used, the low coupling efficiency will be evidenced by short, truncated sequences. If capping is not used, or if capping is unsuccessful, single base deletions will occur in the oligonucleotide and as a consequence, a large number of failure sequences lacking one or two nucleotides will be formed. To achieve high product yields, it is also imperative that the removal of the 5′-hydroxyl protecting group occurs with 100% efficiency within each cycle. If this step is not optimized, the final product mixture will be a family of oligomers with single base deletions in addition to the desired product. Moreover, for synthesis of long oligonucleotides, it is important to minimize the most prevalent side reaction—depurination (24). Depurination results in the formation of an abasic site that does not interfere with chain extension. Instead, the critical DNA damage occurs during the final nucleobase deprotection under basic conditions, which also cleaves oligonucleotide chains at abasic sites. The effect of depurination on sequence integrity is the generation of short, truncated sequences that can typically be mapped to purine nucleobases. Overall, high yield, high quality synthesis of oligonucleotides requires control of depurination combined with highly efficient coupling and 5′-hydroxyl deprotection reactions. Without high coupling yields and low depurination, the final product will require extensive purification and PCR amplification to compensate for the low yield.
The Agilent in situ microarray synthesis process has been designed for the synthesis of high-quality nucleic acids and is built on two fundamental technologies. First, inkjet printing within an anhydrous print chamber allows for the spatial control of the phosphoramidite coupling step, which results in the synthesis of thousands of unique sequences on a 2D planar surface. Second, a flowcell reactor within the Agilent microarray synthesizer is utilized for the remaining chemistry steps. Oxidation and detritylation reactions are carried out in the flowcell by immersion, or flooding, of the growing oligonucleotides with the respective oxidation and detritylation reagents. While the standard Agilent microarray consists of oligonucleotides 60 nucleobases in length, printing oligonucleotides exceeding 100 nucleobases would appear to be attainable considering the high stepwise coupling step with Agilent’s print chamber. Such yields are the result of jetting small volumes of highly concentrated reactants onto an oligonucleotide chain growing from a solid surface. The excess concentration of the phosphoramidite and tetrazole solutions is further enhanced because the quantity of DNA within each spatially controlled feature does not exceed the femtomolar range. The added benefit of a proprietary anhydrous chamber ensures that moisture is kept to a minimum, allowing the coupling reaction to proceed more efficiently. With such reproducibly high coupling yields, the traditional capping step has been eliminated from the oligonucleotide synthesis cycle. Given the successful optimization of stepwise coupling yield with the Agilent platform, the next step toward the synthesis of oligonucleotides exceeding 100 nucleobases is minimization of depurination.
When studying depurination within a flowcell reactor, one naturally refers to the vast databank of depurination on CPG (26–29). However, oligonucleotide synthesis within a CPG column differs greatly from oligonucleotide synthesis within a flowcell reactor. The most obvious difference is that the phosphoramidite coupling step has been removed to a specialized print chamber when synthesizing oligonucleotides on microarrays. The synthesis surface is also very different. While CPG is a 3D surface, that maintains a uniform surface characteristic throughout oligonucleotide chain extension, the glass substrates of the microarray process are typically flat, non-porous solid surfaces that generate a patterned array of oligonucleotides. The Agilent microarray flowcell also has a small surface area covered by a large reagent volume, typically holding 20 ml. This is the polar opposite of a 1 µmol CPG column, which has a large surface area covered by a small reagent volume (∼0.4 ml). The smaller capacity of the CPG column means that less reagent is used per chemical step and that multiple column volumes of washing solution can be used between chemistry steps. The high volume requirements result in the flowcell supplying, at best, one to two flowcell volumes of washing solution between chemistry steps.
A detailed study of fluid mechanics within the Agilent flowcell was undertaken in order to understand and control depurination within the microarray synthesis process. This article describes a new chemical strategy to optimize reagent flows so that oligonucleotide depurination is not only controlled but also minimized. This new depurination-controlled process is utilized to synthesize oligonucleotides of up to 150 nucleobases in length on microarrays. The success of the depurination-controlled process was confirmed by PCR, sequence analysis, and direct 5′-end labeling followed by PAGE.
MATERIAL AND METHODS
Materials
Titanium Taq polymerase and dNTPs were obtained from BD Biosciences Clontech, Palo Alto, CA. Restriction endonucleases (AscI, BstXI, ClaI), T4 polynucleotide kinase (PNK), T4 DNA ligase and Uracil-DNA glycosylase (UDG) were obtained from New England Biolabs Inc. Shrimp alkaline phosphatase was obtained from Promega Corp., Madison WI. Denaturing acrylamide gel solution (8%, 19:1 acrylamide: bisacrylamide, 7 M urea, 1× TBE), 10× TBE buffer and SFR agarose were obtained from Amresco Inc.; Phosphoramidites and tetrazole reagents were obtained from Sigma-Aldrich (St Louis, MO), Glen Research (Sterling, VA) and Chemgenes (Wilmington, MA). All other nucleic acid reagents were obtained from Honeywell (Muskegon, MI).
Array synthesis
Unless otherwise indicated, DNA microarrays were manufactured according to the legacy Agilent manufacturing process as described elsewhere (12,18,30,31). Overall, an automated tool designed by Agilent Technologies was used to enable the standard phosphoramidite chemistry on a silylated 6.625 × 6 in. wafer using the following major modifications. First, the solid support was a flat, non-porous surface rather than a locally curved, porous surface. Second, the coupling step was controlled in space using inkjet-printing technologies to deliver the correct amount of activator and phosphoramidite monomer to the appropriate spatial location on the solid support. Third, the oxidation and detritylation reactions were performed in dedicated flowcells whose mechanical operations are described in the next paragraph. Oxidation solution was with 0.02 M I2 in THF/pyridine/H2O and detritylation solution used 3% dichloroacetic acid (DCA) in toluene. Deprotection and cleavage of the DNA from the surface was performed as described by Cleary et al. (18). Oligonucleotides were recovered after cleavage by lyophylization in Eppendorf tubes. Each array could contain up to 22 575 features. When fewer oligonucleotides were desired, the appropriate number of locations were left blank.
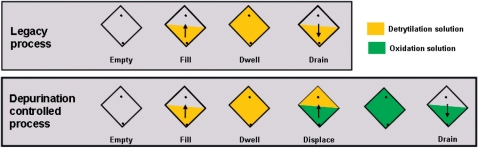
Oxidation and detritylation reactions were carried out by flood steps in a flowcell as depicted in Scheme 1. The Agilent flowcell is constructed such that a glass substrate carrying the microarrays forms one wall of the reactor chamber. The substrate is brought to bear upon a seal embedded in the perimeter of the fixed reactor cell thus forming a high aspect ratio sealed chamber, where the aspect ratio is defined as the ratio of the planar flowcell width L to the lateral gap height h. Typically, the planar width of the substrate is 15–20 cm while the gap is 0.5–1 mm. Active liquid reagents, wash solvents and gases are introduced into the flowcell through two ports. One port is located at the bottom corner of the cell and one at the top. A series of solenoid valves control the inflow and outflow of reagents to these two ports. The flowcell is mounted such that the walls of the flowcell are vertical so that gravity assists draining. During a typical synthesis cycle, the reagents are first introduced into the flowcell from the bottom port until the flowcell is filled (fill time). The reagents are then left in the flowcell without mixing for 30 s (oxidation) and 60 s (detritylation). We define these periods (when the flow within the flowcell is stationary) as the dwell time. Finally, the reagents are drained from the bottom port (drain time), followed by washes using two flowcell volumes of acetonitrile (ACN). This is typically 30–50 ml and is dependent upon the particular flowcell geometry. In the depurination controlled detritylation process, the detritylation solution is driven out of the flowcell through the top port of the flowcell using an inflow of oxidation solution through the bottom port. The plumbing connected to the outlet port of the flowcell is carefully constructed to avoid excessive restriction to the exiting liquid.
Scheme 1.
Schematic of the detritylation process in the flowcell. Schematic description of the detritylation process used in the legacy process (top) and depurination controlled process (bottom). The diamond symbol represents the wafer surface when mounted in the flowcell, and the two black dots represent the bottom and top reagent ports allowing flow in and out of the flowcell. In the legacy process, starting from an empty flowcell, the detritylation solution is filled from the bottom port, resides without mixing (dwell), then drained through the bottom port using gravity. In the depurination controlled process, the detritylation solution is displaced after dwelling through the top port using oxidation solution introduced through the bottom port. The oxidation solution is then drained through the bottom port. Management of the stratification of the detritylation and oxidation solution is critical to the control the depurination side reaction and the reproducibility of the detritylation process.
Fidelity analysis by radio labeling of individual oligonucleotides
For initial fidelity analysis, three sequences were synthesized on separate microarray slides. The sequences were 3′-CCTATGTGACTGGTCGATGCTACTA-5′, 3′-AACAGTATGAAGAGTACCAACCTATGTGACTGGTCGATGCTACTA-5′ and 3′-TTTTTTTTTTAACAGTATGAAGAGTACCAAGTGTGCCTATGTGACTGGTCGATGCTACTA-5′. All features of each individual microarray slide were used for each synthesis. After cleavage of oligonucleotides from the microarray surface by treatment with NH4OH, 32P labeling of all the reaction products was performed with a mixture of 1 µl of 10× buffer (supplied with the enzyme), 1 µl of gamma-ATP (3000–6000 Ci/mmol, Amersham, Piscataway, NJ), 1 µl kinase (T4 polynucletoide kinase 10 U/µl, NEB #M0201S) and doubly distilled water in a 10 µl reaction volume. The mixture was incubated at 37°C for 30 min and then quenched with 10 µl of loading buffer (90% deionized formamide, 1× TBE, bromophenol blue, xylene cyanol). The labeled oligonucleotides were analyzed by gel electrophoresis on denaturing polyacrylamide gels (18% acrylamide, 29:1 acrylamide:bisacrylamide, 8 M urea, 1× TBE) using 2000 V for 2 h. Data acquisition was performed using a Molecular Dynamics Inc. Storm Scanner and the data analysis performed using the ImageQuant software.
Depurination metrics
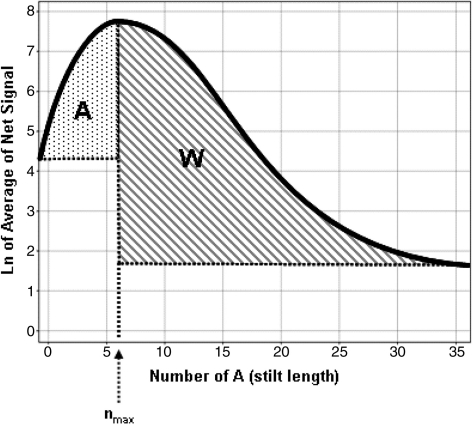
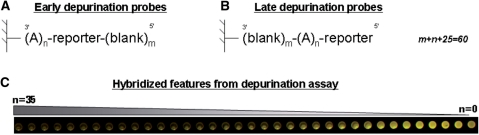
The oligonucleotide sequence set used to calculate the depurination metrics were 3′-(A)n-reporter-(blank)m-5′ for the early probes and 3′-(blank)m-(A)n-reporter-5′ for the late probes, where n + m = 35, where ‘blank’ means that no coupling was performed in that synthesis cycle and where the reporter sequence was 3′-CCTATGTGACTGGTCGATGCTACTA-5′. Each probe was repeated 12 times at random positions within the array. The hybridization was performed using 15 pM of Cy3 and Cy5 5′ labeled target having a sequence complementary to the reporter deoxyoligonucleotide. Hybridization and wash conditions were as described previously (12). Following scanning on an Agilent Technologies scanner (Part number G2565BA), the hybridization signals for every feature were obtained using the Agilent Technologies Feature Extraction software v8.1. The text files output were then loaded and combined in a Microsoft Access database (Richmond, WA). The depurination metrics calculation was performed for each channel (Cy3 and Cy5) using an internally developed code written in C++ as follows. The natural log of the signals from each feature, as obtained by the subtraction of the background signal from the raw signal, were smoothed as a function of the depurination stilt length, n or number of A nucleotides, using Lowess to obtain a depurination profile (Scheme 2). The depurination stilt length that corresponds to the maximum observed signal, nmax, was then calculated and the depurination metrics obtained by integrating the areas shown as A and W on Scheme 2.
Scheme 2.
Schematic description of the depurination metrics calculation. The depurination metrics are derived from the signal intensities of the depurination probes described in the ‘Materials and Methods’ section and in Figure 2A and B. The natural log of their average net signal as a function of the depurination stilt length (number of A nucleotides in the stilt), is smoothed using Lowess and plotted as represented. The profiles were experimentally observed to monotonically increase starting from a null number of A, and then monotonically decrease after reaching a maximum. The depurination metrics A and W are calculated by integrating the shaded areas on the plot. In some cases, the extent of depurination and/or detritylation would be such that the signal profile would be monotonically increasing or decreasing with increasing number of A nucleobases in the stilts. In those cases, one of the depurination metrics would be null (depurination metric A = 0 when nmax = 0; and depurination metric W = 0 when nmax = 35).
Flow visualization
Characteristics of the bulk flow were studied using straightforward flow visualization. Fortunately, variation in the liquid color allowed us to examine details of the miscible liquid front between the oxidation solution laden with I2 and the transparent detritylation solution. Detritylation solution displacement events were recorded on a simple digital video camera. Ambient lighting was usually adequate to provide bulk flow characteristics. Details of the miscible front often required high-intensity lighting. The lights were situated >1 m from the flowcell to avoid heating the liquid and a Canon Optura PI large field camera was used to record the flow characterization.
Details of liquid behavior on the substrate were visualized using high-magnification lenses mounted on CCD cameras. The camera was a Sony XC-75 CCD video camera module equipped with a Navitar 12× PN 1-50486 lens and a 2× Adapter PN 1-6030 adapter. The substrate and interfacial flow were back-lit using an LED shining through the substrate and liquid layer. The CCD camera assembly was situated such that the LED light was focused directly into the lens that had been mounted on the opposite side of the substrate. The LED was pulsed at 1–10 kHz depending on the amount of light required for each level of magnification. Lighting the flow this way provided adequate illumination with minimal thermal effects.
Most of the flowcell characteristics derive from the very large aspect ratio. As such the critical flowcell dimension for a given substrate size is the gap height, h, between the substrate and the reactor wall. The gap width, fluid material properties and liquid speed set a basic non-dimensional parameter: the Reynolds numbers (Re). Here, Re = ρuh/2µ, where ρ is the liquid density, u is average speed and µ is dynamic viscosity. The Re directly influences primary flow qualities during the filling and draining process. For the present case, h restricts fluid motions to small Re 0(100) for typical fluids used in phosphoramidite chemistry. This Re is far below the transitional Re ∼ 3000 where this family of flows—pressure driven flows in a slot or Poiseuille flows—transition to turbulence. As such, in this geometry, these fluid motions are necessarily laminar. These types of flows are often referred to as Hele Shaw flows (32).
Near the inlet to the flowcell, the flow is not laminar. As it enters the flowcell, the flow is best described as a normally directed jet. As such the flow is laden with vortical eddies. Once formed, these eddies persist for a short distance before dissipating which is typical of this type of flow (33). Typically these structures dissipate within 20 gap widths of the inlet. Our observations agree qualitatively with this assessment? We also note that flow in such a device is not symmetric between the top and bottom port in this respect. The flow is fully developed and laminarized by the time it reaches the exit port. The top port therefore, acts very much like an ideal sink since there is no source of vorticity being ejected into the flow.
Oligonucleotide designs
For comparative studies of oligonucleotide synthesis via standard phosphoramidite chemistry (CPG) and synthesis on surfaces (chips), a set of 48 oligonucleotides with lengths between 84 and 99 nt were designed and prepared by each method. CPG oligonucleotides were synthesized and purified by reverse-phase chromatography at Biosearch Technologies Inc. Equal molar amounts of each oligonucleotide were pooled for multiplex PCR experiments. A corresponding set of oligonucleotides was synthesized on microarrays using the depurination controlled oligonucleotide synthesis process described previously. All oligonucleotides were released from the microarray surface by treatment with NH4OH as previously described and used as template for PCR without additional purification. A linker made of five dT was added at the 3′ end of oligonucleotides synthesized on microarrays. An additional set of 578 oligonucleotides with a maximal length of 120 bases were included in the microarray synthesis in order to test the length, composition and dT-linker effect. A total of 626 different oligonucleotides varying in lengths and compositions were also synthesized on another chip. All sequences were uniformly distributed among the 22 575 chip features. Specific primer pairs were designed to amplify each individual sequence or multiple sequences by PCR. Unique restriction sites were incorporated in each sequence for subsequent identification.
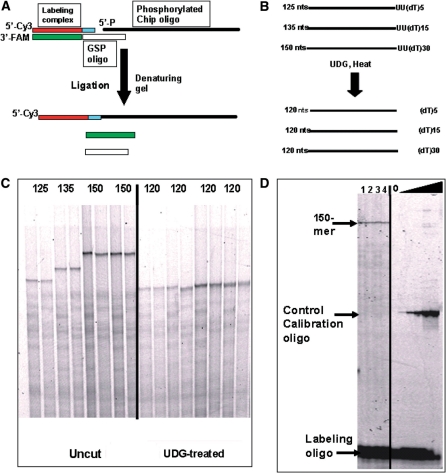
For the synthesis of extremely long oligonucleotides on microarrays, two sequences of 118 nt were chosen, and oligonucleotides with sizes 125, 135 and 150 were synthesized by adding a linker of 5, 15 and 30 deoxythymidine nucleotides at the 3′-end of sequences (Figure 9B). Two dU-residues were inserted between the sequence and poly(dT) linker and used for cleaving the poly(dT) linker from the sequence of interest by uracil-DNA glycosylase treatment. Separate chips were used to synthesize each oligonucleotide.
Figure 9.
Synthesis of 125, 135 and 150mer oligonucleotides on microarray surfaces and analysis of full-length products. (A) Scheme for the 5′-end labeling of oligonucleotides by ligation. Chip synthesized oligonucleotides were phosphorylated, annealed to gene-specific oligonucleotide, and ligated to the labeling duplex. As a result, only full length oligonucleotides are ligated to Cy-3 labeled oligonucleotides. (B) Oligonucleotide design. (C) Left panel: denaturing gel analysis of 5′-end labeled 125, 135 and 150mer oligonucleotides. Right panel: the same products after UDG treatment. (D) Estimation of yield for full-length product.
PCR analyses
PCRs were performed with Titanium Taq polymerase (BD Biosciences) according to manufacturer’s instructions. PCR parameters were as follows: 3 min at 95°C, two cycles of 5 s at 95°C, 90 s at 68°C and 33 cycles of 5 s at 95°C, 15 s at 65°C, 30 s at 72°C, and final elongation step of 3 min at the same temperature. PCR products were analyzed by gel electrophoreses on 4% agarose gel in TBE buffer containing 93 mM Tris–borate (pH 8.3), 2 mM EDTA, visualized by Cyber-Green I staining according to manufacturer’s recommendation (Molecular Probes).
Fluorescein dye labeling of PCR product by ligation
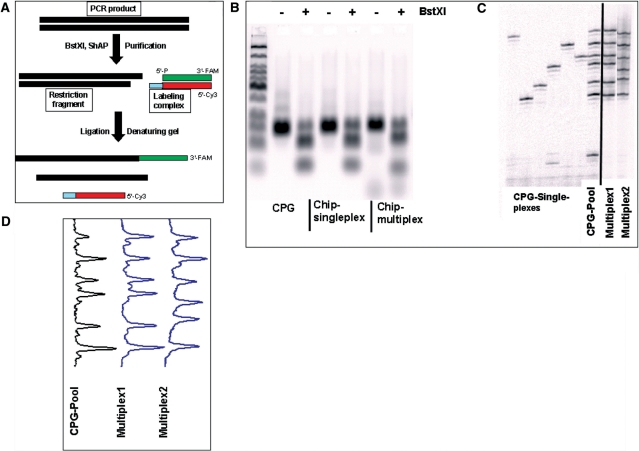
Two complementary oligonucleotides of 20 and 24 nt labeled at the 3′-end with fluorescein and at 5′-end by Cy3, respectively, were mixed together to form a double-stranded DNA complex with a 4-nt 5′-recessed terminus (Figure 8A). A non-palindromic sequence at the 5′-recessed termini was used to prevent self-ligation.
Figure 8.
Oligonucleotides synthesized on microarray surfaces are uniformly represented. (A) Scheme of the labeling PCR product by ligation. Approximately 100 mol of individually CPG-synthesized oligonucleotides, normalized pool of the CPG-synthesized oligonucleotides, or 1 µl of 1000-fold diluted Chip oligo pool were amplified by 35 cycles of PCR and digested with BstXI restriction endonuclease. Aliquots of PCR amplified DNA and digestion reaction fragments were analyzed by agarose gel electrophoresis (B). Digested PCR products were labeled by ligation and analyzed by denaturating gel electrophoresis (C). Fragment profiles were generated for each multiplex PCR analysis (D).
PCR products were cleaved at a BstXI site followed by dephosphorylation so that only the 3′ overhanging termini generated by BstXI cleavage were available for ligation with the 5′-recessed termini of the labeling duplex. Next, 2 pmol of purified digested PCR products, 10 pmol of pre-annealed labeling duplex, and 200 units of T4 DNA ligase were incubated at 16°C in 20 µl of T4 ligase reaction buffer containing 50 mM Tris–HCl, pH 7.5 at 25°C, 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol, 25 µg/ml BSA. After 1 h, the reaction was terminated by heating for 15 min at 65°C and by addition of EDTA to 10 mM. The products of the reaction were analyzed by gel electrophoresis on denaturing polyacrylamide gels (8% acrylamide, 19:1 acrylamide:bisacrylamide, 7 M urea, 93 mM Tris–borate, 2 mM EDTA, pH 8.3). Electrophoresis was carried out in 1× TBE (pH 8.3) at 2000 V for 2 h. The fluorescent image was produced on Typhoon 9410 scanner and analyzed using ImageQuant 5.2 software (Molecular Dynamics Inc.)
Cy5 dye labeling of long oligonucleotides by ligation
Labeling of the 5′-end of a fully synthesized oligonucleotide was performed by using the same labeling duplex as described above. A gene specific (GSP) oligonucleotide was designed to anneal at the 5′-end of the synthesized oligonucleotide, so that in the resulting duplex only the 4-nt 5′ recessed termini of the analyzed oligonucleotide was available for ligation with the 3′ overhanging termini of labeling duplex (Figure 9A). First, the long oligonucleotide was released from microarrays and desalted on a Micro Bio Spin P30 columns (Bio-Rad Laboratories). Then, the 5′-end of the oligonucleotide was phosphorylated using T4 polynucleotide kinase in T4 ligase reaction buffer. In the labeling reaction, the labeling oligonucleotides and gene-specific oligonucleotides were added to concentrations of 1 µM each. The reaction mixture was heated at 80°C for 2 min, and oligonucleotides were annealed at 45°C for 45 min. T4 DNA ligase was added to 20 U/µl, and the ligation proceeded at 16°C for 1 h. Reactions were terminated by heating at 65°C for 15 min.
To cleave oligonucleotides at the dU base, uracil-DNA glycosylase was added to the ligation mixture to 0.1 U/µl. After 30 min of incubation at 37°C, the generated AP-sites were cleaved by heating the mixture at 95°C for 10 min. The products of the ligation and cleavage reactions were separated on 8% denaturing polyacrylamide gel and analyzed.
RESULTS
Evaluation of the major synthetic failure modes that affect sequence integrity for oligonucleotides synthesized on microarrays
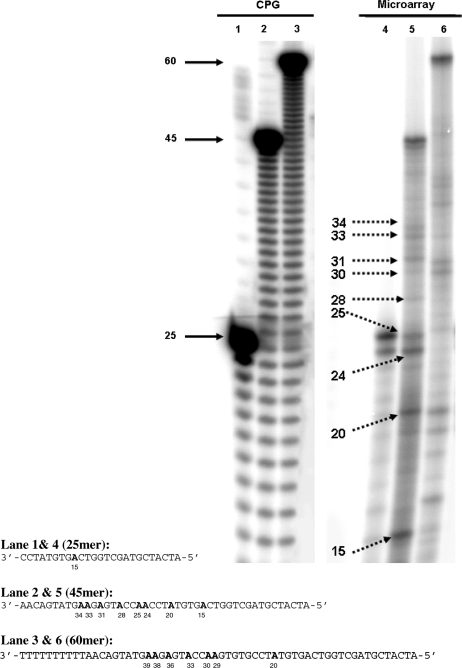
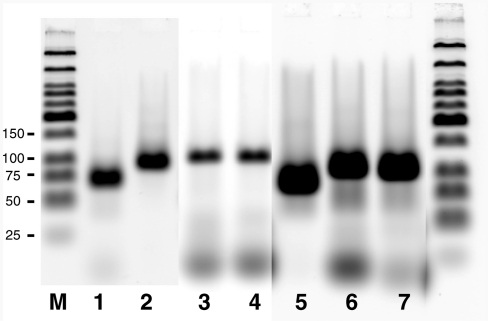
Initial attempts to quantify the sequence integrity of oligonucleotides synthesized on flat solid surfaces were carried out by preparing oligonucleotides on a microarray, removing them from the surface through chemical cleavage, and then measuring the distribution of different lengths by a separation technique. In order to facilitate analysis, the same oligonucleotide was originally synthesized on all available features of a single microarray surface so that no confounding effects would be introduced by the presence of multiple sequences. Results obtained using denaturing gel electrophoresis as a separation technique (Figure 1) document the observed failure modes when the initial microarray manufacturing process was used. For the synthesis of heteropolymers of length 25, 45 and 60, the observed profile of failed sequences is characterized by an n − 1 band (where n is length of the intended oligonucleotide) and by discrete bands that align with positions of the adenine (A) nucleobases on the oligonucleotide sequences. The n − 1 band could be due the coupling not being 100% effective. However, since no appreciable levels of n − 2 and n − 3 product were obtained without capping, it is more likely that the n − 1 band is due to the failure of the initial coupling (34). Finally the discrete bands at A positions, which are characteristic of short truncated sequences, are due to depurination leading to the formation of abasic sites that are cleaved during the final deprotection. It should be noted that in order to observe these failure sequences, the photographic film had to be significantly over-exposed, which prevents an accurate quantification with respect to the full length sequence. Nevertheless, it is apparent that no significant amount of n − m bands (m > 1) are present, which would have been due to the cumulative effect of incomplete coupling steps (capping was not used in this experiment). Overall, the main limitation to the synthesis of long oligonucleotides using this initial microarray manufacturing process appears to be the depurination side reaction since, no evidence of significant coupling failure is observed even in the absence of capping. This observation is in general agreement with previous observations using CPG as the solid support (34).
Figure 1.
Identification of synthesis integrity using the legacy process. Oligonucleotides of length 25, 45 and 60mer were obtained from synthesis on CPG or from synthesis on microarray surfaces using the legacy process. The oligonucleotides were labeled with 32P at their 5′-end and analyzed using denaturing gel electrophoresis. Profiles from the oligonucleotides synthesized on microarray surfaces show the presence of full length product (full arrow), n − 1 product32 in the 25 and 45mer samples (lanes 4 and 5), and discrete bands (indicated by dotted arrows). The sizes of the discrete bands match the A nucleobase positions indicated in bold in the sequences and the pattern is characteristic of depurination side reactions during synthesis. The offset between the actual migration length on the gel (indicated by numbers below the sequences) and the position of the A nucleobases relative to the 5′ end of the sequence (−2 bases) is attributed to the chemical functionality of the 3′-end of the chain cleaved by depurination.
Hybridization-based depurination assay and depurination metric
In order to understand the origin of the higher than expected level of depurination during oligonucleotide synthesis on microarrays, and to assist in the development of a process where depurination was controlled and/or eliminated, we sought to develop a fast and robust depurination assay. The intent was to eliminate the labor intensive and slow process of physically removing the oligonucleotides from the surface, labeling them and analyzing the size distribution by a size separation method. Taking advantage of the ability to synthesize in parallel oligonucleotides of diverse compositions on a microarray, we investigated the use of a hybridization-based depurination assay. This assay relies on the relative sensitivity of different depurination probes to the detritylation condition used during synthesis. The depurination probes evaluated (described in Figure 2A and B for a 60mer microarray) are composed of a depurination sensitive stilt of variable length (An, n = 10–35) and a common 25mer reporter sequence. It was expected that, as the number of A nucleobases increase in the stilt, the probability of depurinating at least one A nucleobase would increase. After further cleavage of the abasic site during the final, basic deprotection step, the amount of reporter probes present on the array surface would be reduced. Therefore, following hybridization with a labeled oligonucleotide complementary to the reporter sequence, the observed fluorescent signal for this probe would be correlated to the extent of depurination. This prediction is experimentally confirmed as shown in Figure 2C where a typical series of depurination probes have decreasing signal intensities with increasing number of A nucleobases in the stilt.
Figure 2.
Hybridization based depurination assay. A hybridization based depurination assay was used to measure the extent of the depurination side reaction during synthesis of microarrays using the legacy process. The depurination probes are composed of a depurination sensitive stilt of variable length (An, n = 10–35) and a common 25mer reporter sequence. The length of each depurination probe is n + 25 and their synthesis was staggered during the 60-nt synthesis cycles used to synthesize the 60mer microarray. Early depurination probes (A) are such that their synthesis is initiated as early as possible, i.e. m blank synthesis cycles are performed after the probe synthesis so that m + n + 25 = 60. Late depurination probes (B) are such that their synthesis is initiated as late as possible, i.e. m blank synthesis cycles are performed prior to the probe synthesis so that m + n + 25 = 60. All probes are synthesized simultaneously and hybridized to a labeled oligonucleotide complementary to the reported probe. The early depurination probe hybridization signals show a decrease in signal intensity with increasing number of n due to a loss of reporter probe (C). A similar profile is observed for late depurination probes.
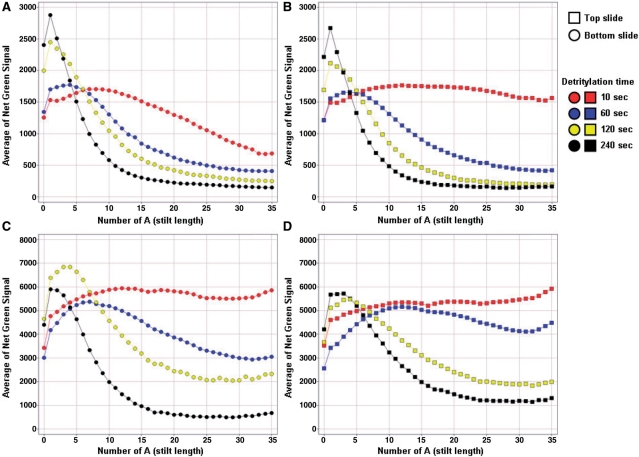
A more detailed and quantitative analysis of the measured fluorescent intensity signals as a function of the number of A nucleobase in the stilt is presented in Figure 3, where each data point is an integration of the signal intensity obtained for sequences described in Figure 2A and B. It is apparent from the plot at 60 s detritylation time in Figure 3A–D that the signal profile as a function of the number of A nucleobase is a composite of two distinct effects. First, at low number of A nucleobases, the signals are increasing with increasing number of adenines. This is consistent with a stilt effect resulting in more favorable thermodynamic parameters, especially entropy, when probes are placed further away from the surface by the addition of stilts. Second, after a peak in signal intensity, the signals are decreasing with increasing number of adenines in the depurination sensitive stilt. This is consistent with the incrementally increasing probability of depurination in incrementally longer A stretches which generates a significantly larger portion of the reporter sequence being cleaved from the array. The data for different detritylation times, as shown in Figure 3A–D (10, 60, 120 and 240 s) have expected signal profiles where the severity of signal loss at higher A stilt length is correlated with the longer detritylation times. Figure 3A–D also shows that the depurination probes synthesized as early as possible have a higher level depurination than probes synthesized as late as possible as indicated by the lower overall signal intensities (1500 versus 8000) and the more rapid decrease in signal intensities, especially at low detritylation time (compare 10 s detritylation time in Figure 3A versus C). This is fully expected since the stilts of the early probes shown in Figure 2A will experience a higher cumulative number of detritylation steps than the late probes shown in Figure 2B, thus resulting in more extensive depurination and larger signal loss. Similar experiments were performed where the concentration of acid in the detritylation solution was varied at a constant detritylation time. The observed profiles were consistent with lower acid concentrations resulting in less depurination (data not shown). When the acid concentration and detritylation times were kept constant, and the contact angle of the wafer surface was varied, lower amount of depurination was observed on surfaces of higher surface energy, i.e. lower contact angle (data not shown). Overall, the most salient observation is that, while the trends of the signal profiles are consistent with the theoretical expectations, a significant difference is observed between the slides from the same wafer made at the top of the flowcell (top slide) versus those made at the bottom (bottom slide) (Figure 3A versus B and C versus D). Therefore, the flowcell process is not only characterized by a higher than usual extent of depurination compared to CPG, but also by a depurination gradient resulting in a variable oligonucleotide quality over the area of the wafer.
Figure 3.
Signal profiles of depurination probes and the effect of slide location on the wafer and detritylation time. Representative signal profiles of the depurination probes manufactured with the legacy process are plotted as a function of the number n of A nucleobases in the depurination stilt for early (A and B) and late (C and D) depurination probes (early and late depurination probes were described in Figure 2A and B). The shape of the depurination intensity profiles are characterized by a decrease in signal after the signal reaches a maximum. This loss in signal is consistent with the loss of the reported probe following depurination-induced cleavage in the A nucleobase stilt. The signal decrease is correlated with the detritylation time, with higher detritylation times resulting in steeper declines past the maximum signal. Additionally, the profiles for slides synthesized close to the bottom port, also called bottom slide (A and C) have significantly higher signal loss than the profiles of slides synthesized close to the top port, also called top slide (B and D).
While the signal profiles as function of stilt length in Figure 3 correlate well with controlled variations in process parameters (i.e. detritylation time, acid concentration, surface energy), the results are still mostly qualitative. To address this concern, we developed quantitative depurination metrics from the experimentally observed profiles. The computation consists in first smoothing out the signal profiles of the log of the signals, and then integrating specific areas of the smoothed curve. As shown in Scheme 2, the area W is limited in the x-axis by the stilt length with the maximum signal and stilt length 35, and is limited in the y-axis by the maximum and minimum signal. As the depurination increases and the rate of signal loss increased, the defined area will increase resulting in a larger depurination metric W value. As depurination decreases and the profile become flatter, the depurination metric value decreases. At the extreme, when the depurination is so low that the signal is monotonically increasing as a function of stilt length, the stilt length with the maximum signal will be 35 and the depurination metric value will be equal to 0. While we did not attempt to correlate the values of this depurination metric with a stepwise depurination yield, we observed that, as long as the target concentration is held constant across hybridization events, this intuitive metric is very robust and stable. The depurination metric A calculated by integrating the area shown in Scheme 2 was expected to be complementary to W since it would be zero at high depurination level and increase as depurination decreases. In other words, it would increase the dynamic range of the depurination quantification and be sensitive with chemical conditions where depurination was so low that the depurination metric W would be close to zero. Also, it was expected that the A depurination metric may indicate conditions in which the detritylation reaction itself may become incomplete. However, in practice, this metric was not used since the tested conditions were in the depurination range where it was mostly insensitive. Finally, for each depurination metric, we calculated their values for both the ‘early’ and ‘late’ versions of the depurination probes. Overall, we obtained four depurination metrics per slide (A and W, early and late) and eight per wafer (top and bottom slide). In the remaining part of this article, the quantitative depurination metric early W will be used to compare the extent of relative depurination in different chemical processes and to develop an optimized chemical process where depurination is significantly reduced. The studies have led to the synthesis of oligonucleotides of previously unachievable length.
Delivery and control of reagents in a thin gap flowcell
To understand the source of higher than expected depurination levels, we carefully examined details of the flowcell process. Casual observation of reagent flow can mistakenly leave a simplistic impression of the filling and draining process (Scheme 1). In the legacy process, the flowcell is simply filled with reagent through the bottom port for a duration tf. The reagent dwells within the flowcell for a time td to allow completion of the reaction. After td, the liquid is drained from the flowcell taking time te which approximately equals the fill time. Typical values of te and tf are 8–10 s.
Careful visual observation of the draining process reveals several more subtle and important variables that led us to classify the draining flow into three regimes. First, we define stage 1 as the first 8–10 s after the detritylation solution begins to drain from the flowcell. During this time we observe the simple motion of the bulk or centerline interface. This is the most easily observed motion of the interface. At first glance, this process seems to leave the flowcell interior completely evacuated. However, sustained observation shows that a film is actually left on both walls of the flowcell. This film gradually becomes visible as large-scale features form under the influence of gravity and surface tension. In this first stage, motion of the liquid–gas interface in proximity to a wall becomes complex. The necessity of material adherence to the solid wall—the no-slip condition—must be satisfied by zero fluid velocity at the wall. Also, the dynamics of the triple phase line that forms and starts to migrate across the surface complicates observations. The non-dimensional capillary number defined by Ca = µu/γ gives a convenient means to characterize a flow where the viscous dissipation of energy competes with surface energy. For our flows, front speeds µ range from 1 to 10 cm/s. Values of viscosity taken from the literature are ∼0.552 mPa s and surface tension γ is 28.4 mN/m. This yields Ca in the range of 2.8 × 10−4 for the slowest front speeds to 2.8 × 10−3 for the fastest.
The literature contains many insightful examinations of film formation in thin gap flows (35). A salient feature of this process is that the thickness of the film increases with increasing Ca. In simple terms, for a given set of material properties, the film thickness increases with increasing front speed. In addition, Halpern and Gaver (36) numerically modeled this process over a wide range of Ca. The authors fit their data to Equation (1) correctly predicting a thicker film thickness with increasing Ca:
| (1) |
where β is the fraction of channel width occupied by the finger of gas penetrating through the liquid. Our lowest Ca number lies just outside of their predictions but the high end of our range lies within their data. Their model predicts a film thickness no greater than 0.015 h corresponding to film thicknesses of 15 µm.
We also define an effective reaction time based on the accelerated kinetic rate due to increased concentration. The effective time is taken to be the equivalent time needed to drive the reaction to the same state holding concentration and temperature fixed and only varying time. For this first stage of fluid motion we denote this time as τ1. This effective time differs from the real exposure time due to increased concentration that arises from even small amounts of evaporation in the very thin resident film.
Second, we define stage 2 beginning 10–100 s after draining begins. This corresponds to the times where the film drains and sufficiently thins to the point that it begins to rupture, exposing the underlying substrate. As the substrate-scale film continues to drain it bursts at discrete but unpredictable locations. The film begins to contract away from these nucleation sites. The three phase intersection of the gas, liquid and substrate is termed the dynamic contact line. This contraction of the dynamic contact line is essentially independent of gravity as it moves just as quickly upward as downward at a particular nucleation site. This flow is purely driven by surface tension–substrate interaction. Its progression is retarded at a meso-scale by large-scale patterned regions of regularly patterned DNA features. Our cursory observations showed no preference to the sites where the film bursts. The dynamic contact line begins to recede in all directions from the disruption. As the dynamic contact line recedes, its motion is retarded by the surface energy of the substrate.
To go further we must introduce a characteristic of oligonucleotide chains grown on a 2D planar solid support. While the solid support is uniformly hydrophobic at the beginning of the microarray synthesis, the synthesis of oligonucleotide features affects its surface energy. Indeed, we observed that oligonucleotide features gain substantial surface energy with increasing oligonucleotide length. Generally, these sites or features consisting of protected oligonucleotide acquire enough surface energy to become spontaneously wetting to high surface tension organic solvents such as propylene carbonate after 10–20 synthesis cycles. This creates a regularly patterned surface of disparate surface energies. At the feature scale (µm scale), higher surface energy features are surrounded with lower energy surface. At a microarray scale (cm scale) the aggregate features forming the microarrays creates an average surface energy higher than the surrounding un-patterned regions. As a result the draining fluid tends to aggregate on the microarray while liquid in the surrounding regions tends to sheet off these low surface energy regions easily after the film has broken. We also now define our second effective time τ2 which reflects the increased effective reaction time due to increased reagent exposure from increased average surface energy of the microarray surface.
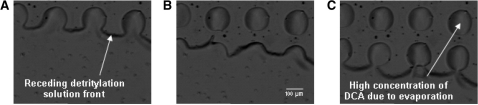
In the third and final stage, the meso-scale film resident upon the microarray continues to recede and evaporate but leaves a sessile droplet of detritylation solution on each of the high surface energy DNA features. This process begins to occur in 10–100 s after the meso-scale film begins to form on the microarray. Some portions of the film that begins to migrate downward will coalesce into large millimeter-scale droplets on the microarray. Careful inspection of the dynamic contact line at the DNA feature scale shows the receding dynamic contact line leaving sessile droplets on each of the feature locations. An example of this is shown in Figure 4. We have not attempted to measure the dependence of droplet size on oligo length although it is reasonable to expect one. Video of the patterned microarray shows that the sessile drops formed on the patterned features persist indefinitely. Patterned droplets on the features also persist indefinitely if the microarray is exposed to an open atmosphere and the detritylation solution is flooded over the surface. Patterning was also compared between arrays that patterned during the legacy process as well as pre-synthesized arrays where the surface had been pre-washed with an organic solvent before exposure to detritylation solution. We undertook this experiment to ensure that the detritylation patterning was not dependent on a previous flowcell step. Since DCA is much less volatile than toluene we surmise in this final state that the droplets pinned to the features are essentially pure DCA. This would of course substantially alter the reaction kinetics and damage the molecule.
Figure 4.
Visualization of the flow of detritylation solution on the microarray surface. Visualization of the flow of the detritylation solution film on the microarray surface following draining of the bulk detritylation solution from the flowcell. Frames A, B and C were taken at 300 ms intervals and show that detritylation solution is trapped on the feature after the receding front has passed. This phenomenon is due to the difference between the hydrophilic nature of the features (growing oligonucleotide chain) and the hydrophobic nature of the substrate surface. The formation of droplets on every feature is followed by rapid evaporation of the toluene solvent carrier leading to a high DCA concentration and significant depurination side reactions.
Removing the film and controlling depurination
Identifying the likely source of depurination as the resident film of detritylation solution adhering to the surface led to experiments where we attempted to inhibit film formation while not harming the DNA.
Our first attempts at removing the detritylation solution was to introduce less dense acetonitrile through the top port of the flowcell in an attempt to displace the denser detritylation solution as it drains through the bottom port of the flowcell. However, this sets up an inherently first in last out (FILO) process for the active reagents and introduces exposure differences on the order of substrate length/front speed. Although the process produced a noticeable reduction in observed extent of depurination (data not shown), it is not well suited for a dimensionally scaled process on large planar substrate and was abandoned.
We created the most effective depurination controlled process by displacing the resident detritylation solution with the oxidizing solution used in the standard phosphoramidite chemistry. The oxidation solution provides two key features. First, the chemical composition contains an aggressive quenching agent in the form of pyridine. Secondly, the density is slightly greater than the detritylation solution. This allowed the introduction of the reagent through the bottom port thus creating a first in first out (FIFO) process. Here the first reagent elements introduced into the flowcell becomes the first to be displaced.
We found that great care needed to be exercised when constructing this flow. The purging process relies on gravity stabilization at the density interface to ensure the interface remains level. If the oxidizer is introduced too quickly, the inertia of the displacing liquid dominates the stratification effect and the fluid channels along the centerline of the flowcell towards the exit port. A sense of the density stratification strength is given by the Atwood number where
The extremes of A range of −1 for unstable cases with a heavier fluid above a lighter to 1 which is the most stable case such as the surface of a pool. For our displacing flow, A ∼ 0.02, which closely corresponds to a homogenous fluid where fluid inertia dominates. While the gravity is still able to maintain stratification, it requires that the miscible interface moves rather slowly in order for the interface to remain close to level which avoids stagnant zones on the substrate during the displacement.
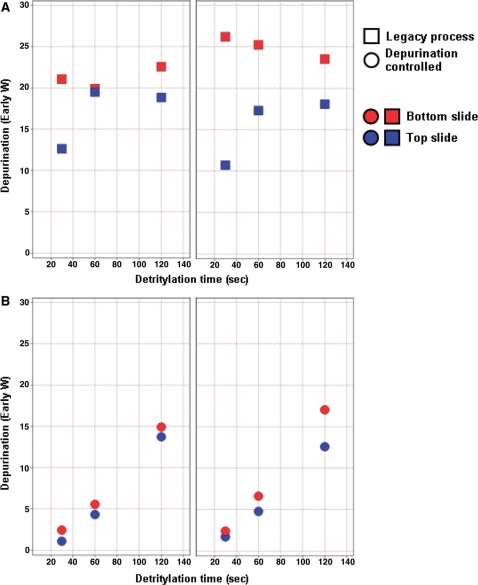
The impact of this novel depurination controlled chemical process on the efficiency of the oligonucleotide synthesis, and especially the control of the side reactions during the detritylation step, was quantified using the depurination metrics described above. Figure 5 shows the effect of varying detritylation dwell time on the extent of depurination for both processes as measured with the depurination metrics ‘early W’. It is apparent that, as expected, the depurination values on the legacy process are high, with a significant gradient between the top and bottom part of the flowcell. In addition, the correlation between depurination values and detritylation time is not strong, suggesting that simple detritylation time estimate is not the major parameter driving the formation of depurination side products. We also note that the difference in depurination metrics for the top and bottom is counterintuitive when analyzed in this context of a simple fill and drain process. This difference arises from the complex local film characteristics close to the entrance and exit ports. We have not attempted to understand the local flow complexities that create these differences but rather sought only to eliminate them using the detritylation solution displacement process.
Figure 5.
Depurination metric as function of detritylation process and detritylation time. The detritylation metric W was measured as a function of detritylation time for slides manufactured using the legacy (A) and depurination controlled (B) process. The data points are colored by slide type (red are bottom slides, blue are top slides) and the two panels in A and B represent two repeats of the same experiment. It is apparent that the depurination controlled process better controls the detritylation reaction, producing depurination metrics that are low, correlated with detritylation time and more reproducible spatially (top versus bottom slide) and in time (from experiment to experiment).
On the contrary, using the novel displacement process, the depurination levels as well as the difference between top and bottom slide are significantly decreased. More importantly, the extent of depurination is now tightly correlated with the detritylation time. These results confirm that elimination of the detritylation solution film, following draining of the flowcell, positively influences control over the extent of depurination side reaction and spatial uniformity of the oligonucleotide synthesis. There is a much weaker but still present counterintuitive depurination effect where the top slide suffers greater depurination than the bottom. We ascribe this much smaller effect to entrance and exit effects of the flow.
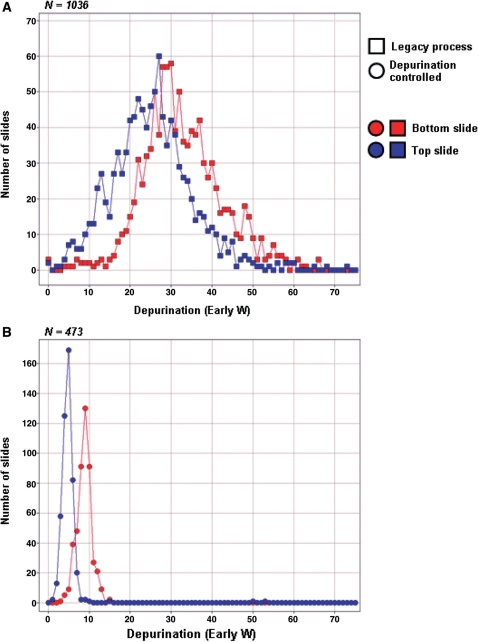
In order to test robustness, stability and reproducibility of the new chemical process, a comparison of the depurination was performed on a large number of slides (N = 3018) made by the legacy process (FILO delivery of detritylation reagent) and the depurination-controlled process (FIFO delivery of detritylation reagent by reagent displacement). For this evaluation, the comparison was made between slides manufactured by the two processes over 20 weeks and over unavoidable variations in process parameters, such as reagent consumables (i.e. reagent lots), substrate lots, and normal temperature and humidity variations within the manufacturing facility. Figure 6 shows the distribution of the depurination values experimentally observed. It is very apparent that both the average and standard deviation of the depurination values observed on the depurination controlled process have been significantly decreased compared to the legacy process. The results obtained in Figure 6B for the depurination process are representative of the reproducibility routinely observed in the manufacturing of microarrays and oligonucleotide libraries within Agilent Technologies’ manufacturing facilities. Such control over relative variations and absolute extent of depurination enable the synthesis of oligonucleotides beyond the usual limit of ∼100mers. The next section describes the quantification of the quality and sequence integrity of oligonucleotides up to 150mers in length.
Figure 6.
Reproducibility of the depurination controlled process. The reproducibility of the depurination controlled process was compared to the reproducibility of the legacy process by evaluating the distribution of the depurination values experimentally observed over a large number of experiments. The data was gathered over a period of 20 weeks and consisted of 1036 wafers synthesized with the legacy process (A) and 473 wafers synthesized with the depurination controlled process (B). The average and standard deviation of the depurination values observed from the depurination controlled process have been significantly decreased, enabing the synthesis of high quality, long (100–150mer) oligonucleotitdes.
Characterization of synthesis efficiency of long oligonucleotides
Uniformity of oligonucleotide synthesis on microarrays
For most biological applications, uniform representation of each sequence within a complex oligonucleotide library is extremely important. Constructing such a library from individual sequences can be expensive and labor intensive. An inexpensive and convenient alternative is the massively parallel synthesis of multiple sequences on a microarray. However, in this approach there is only limited control over the relative sequence representation in the resulting oligonucleotide libraries because post-synthesis purification and/or normalization of individual sequences is not possible. Therefore, it is important that in such microarray-based approaches the amount of each oligonucleotide produced within a feature not vary as a function of length or nucleotide composition. To examine the uniformity of microarray-based synthesis, 626 sequences varying in size from 68 to 120 nt and GC content from 40 to 70% were designed and synthesized on a microarray. Nineteen features randomly distributed on the microarray surface were used for synthesis of each sequence. PCRs were performed with specific primers to amplify each synthesized sequence in singleplex or multiplex reactions. PCR products were detected in all reactions, and no differences in amplification performance were observed in either singleplex or multiplex reactions. Figure 7 presents gel analysis for representative PCRs. The amplification of correct sequences was confirmed by subsequent restriction digestion of PCR product (data not shown). Two sequences A3 and B3 were cloned and their identities were confirmed by sequencing. The synthesis of a dT6 linker at either the 5′- or 3′-end of the oligonucleotide had no effect on PCR performance.
Figure 7.
PCR analysis of oligonucleotides synthesized on microarray surfaces. Six hundred and twenty-six oligonucleotides were released in 100 µl of TE buffer and diluted 1/1000. An aliquot (1 µl) of the diluted pool was used for PCR with a specific set of PCR primers. PCR products with correct lengths were produced after amplification with primers corresponding to A3 oligonucleotide of 78 nt (lane 1) and B3 oligonucleotide of 104 nt (lane 2). Synthesis of a dT6 linker at either the 5′ (lane 3) or 3′-end (lane 4) of oligonucleotides 114 nucleobase in length had no effect on amplification. PCR products were also detected after amplification with corresponding primers which were designed to simultaneously amplify six (lanes 5 and 6), or 18 (lane 7) sequences. The lengths of oligonucleotides vary from 80 to 99 nt; 104 to 120 nt; and 106 to 120 nt, respectively. M-Low Molecular Weight DNA Ladder DNA (New England Biolabs Inc.) was used as size marker.
The results of PCR analysis indicate that there is no significant difference in the yields among the different sequences synthesized on a microarray. To examine this point further, an additional experiment was performed to determine the synthesis uniformity. Several sets of sequences were synthesized both individually and on microarrays that had identical flanking sequences for simultaneous multiplex PCR amplification. Forty-eight oligonucleotides were synthesized individually on CPG. Each such sequence had identical counterparts among the 626 microarray sequences and consisted of several sets of either 6 or 12 oligonucleotides having identical flanking primer binding sites, so that PCRs having multiplex levels of 6 or 12 were possible. Individually synthesized oligonucleotides were purified, quantified, and mixed to form uniform pools of either 6 or 12 sequences. Multiplex PCRs were performed to amplify (i) the individually prepared oligonucleotides, (ii) a uniform pool made up of the same oligonucleotides, or (iii) the pool of 626 oligonucleotides synthesized on microarrays. The PCR products were digested at a BstXI site and labeled by ligation as described in ‘Materials and methods’ section. The products of the labeling reaction generated a ladder of fragments after electrophoresis, where each fragment corresponds to one product generated from one sequence (shown in Figure 8). All tested multiplex PCR pools generate patterns specific to the synthesized sequences, as shown in Figure 8C where six sequences were amplified either individually from CPG-produced material (‘CPG singleplexes’), or from pooled CPG-produced material (‘CPG-pool’), or from microarray-produced material (‘Multiplex1’ and ‘Multiplex2’). The sequences synthesized on microarrays show no difference in the pattern or intensity of the bands relative to the multiplex PCR products generated from uniform pools of individually prepared oligonucleotides. These results demonstrate that oligonucleotides synthesized with the depurination controlled chemistry are uniformly represented after release from microarrays. The level of uniformity observed with oligonucleotides synthesized on microarrays is at least as good if not better than that achieved by manual normalization of individually synthesized probes.
Syntheses of long oligonucleotides on microarrays and direct analysis
PCR analysis provides valuable information about oligonucleotide quality, but direct sequence analyses of full-length oligonucleotides provides a more complete assessment. To achieve this goal, we synthesized only one oligonucleotide per microarray. Two sequences of 118 bases were used and a special design was applied (Figure 9B). Three sets of microarrays were used to synthesize each sequence. Additional poly(dT) linkers of 5, 15 or 30 bases were synthesized at the 3′-end to give lengths of 125, 135 or 150 bases, respectively. The products of the microarray synthesis were released from the surface, labeled by ligation, and analyzed by denaturing polyacrylamide gel. The ligation scheme used (Figure 9A) selectively labels only oligonucleotides with a complete sequence at the 5′-end. Major products corresponding to the designed sequences were detected in all experiments (Figure 9C, left panel). No difference in yields of 125 or 150mer oligonucleotides for both sequences was observed in the labeling experiments. Taken together, these results indicate that the sequences with different nucleotide compositions are synthesized with similar yield and the efficiency of the coupling reaction does not change with an increasing number of cycles.
To estimate the yield of full-length 150mer oligonucleotides, we compared 150mer oligonucleotides synthesized on microarrays with labeled control oligonucleotides of known concentration (Figure 9D). The results reveal that ∼1 pmol of full-length product is synthesized on one microarray. The full-length product consists of ∼4–5% of total crude oligonucleotides as measured by OliGeen reagent. This data agrees with PCR titration experiments (data not shown).
In some applications, the removal of undesired sequences from the 3′-ends of oligonucleotides after synthesis is required for direct usage. To examine the possibility of such applications, two dU-residues were synthesized between the sequence of interest and the poly(dT) linker. The treatment of labeled oligonucleotides with uracil-DNA glycosylase and subsequent cleavage of the generated apurinic site by heating to 95°C resulted in the predicted mobility change of the labeled material (Figure 9C). This result shows that dU residue can be incorporated during oligonucleotide synthesis and effectively cleaved if desired to generate a functional 3′-hydroxyl group.
DISCUSSION
We have shown that the principal limitation to the synthesis of long oligonucleotides on a flat solid support is depurination when using inkjet technologies to spatially control the coupling step. Typically, the extent of depurination of a DNA synthesis process has been measured by physical separation of synthesis product, followed by quantifying the distribution of oligonucleotide lengths. Physical separation methods usually include denaturing gel electrophoresis, high performance liquid chromatography or capillary electrophoresis, and detection methods are generally based on radioactive labeling or UV absorbance. We initially used these methods to demonstrate the relative importance of controlling depurination on our platform compared to yield improvements in other DNA synthesis steps. However, it quickly became apparent that these physical separation methods were not suited for the development of chemical processes useful for array-based DNA synthesis. The main reason was that only one sequence per array could be synthesized and analyzed using these procedures. Such pathways for analysis do not take full advantage of the massively parallel synthesis capability of microarray technology, where the ability to simultaneously synthesize tens of thousands of unique oligonucleotides is available. Instead a hybridization-based depurination assay was developed which enabled quick and reliable on-microarray quantification of the depurination side reaction. This assay relies on the use of probes that each consist of a common reporter segment and a segment with differing sensitivity to depurination. The probes amplified the rather small probability of depurination to a level that could be measured by simple microarray hybridization. Since this assay needs only a small number of probes, it is especially efficient and has been incorporated with other quality control assays as a measurement of reliability and stability of the array manufacturing process. The assay helped the rapid and effective characterization of the spatial dependence of chemical processes, such as depurination, between the top and bottom part of the flowcell. The assay was also used to quickly compare and evaluate the performance of the multiple chemical processes that were tested for removal of the detritylation solution film. Finally, the concept of hybridization-based assays to measure synthesis efficiency is useful for development of all microarray platforms.
To determine the best scheme for improving the oligonucleotide manufacturing process, we investigated the detritylation process as a source of unacceptably high levels of depurination side reactions. Examination of depurination metrics from many experiments showed little or no correlation between the values of the metrics and detritylation exposure times to acid, which was contrary to the expected response dose correlation. Instead, it became apparent that the legacy microarray synthesis was subject to an uncontrolled phenomenon during the detritylation process step, which directed our attention to an examination of the fluid mechanics and mass transfer within the flowcell.
Experimental results showed that the principle cause of excessive depurination was accelerated reaction kinetics induced by increased DCA concentration. This occurs when the toluene carrier solvent evaporates from the film of detritylation solution that is deposited on the substrate after the flowcell is drained. What remains is a concentrated solution of dichloroacetic acid. A naïve view of the draining fluid process assumes the dynamic contact line keeps pace with the centerline interface and leaves a dry substrate. However, some of the more interesting and fundamentally important aspects of the drain step in the legacy process are due to the physics of the dynamic contact line. Careful observation confirms that the dynamic contact line does not keep pace with the fluid centerline. Instead, a film is deposited over the entire substrate. The presence of this film is a well known consequence of draining a liquid between two parallel plates separated by a thin gap (35). Although we were not able to directly measure the film thickness, numerical models and measurements in the literature suggest a film thickness of about 10 µm. The film bursts onto the substrate in an unpredictable way during drainage. This creates an expanding dynamic contact line that retracts from these nucleation sites. Subsequent passage of the dynamic contact line over the patterned microarray surface leads to feature scale patterning that leaves a sessile droplet of concentrated DCA on each feature.
We found that the detritylation reaction could be effectively and reproducibly terminated by using oxidizer solution to displace the detritylation solution from the flowcell. This process of detritylation displacement by oxidation contributes to a controlled chemical process in three ways. First, this method prohibits formation of uncontrolled films or droplets of highly concentrated DCA. Second, this process provides a rapid quenching agent to terminate the reaction. Finally, by taking advantage of the lower density of the detritylation solution, we were able to convert the FILO reagent control of the legacy process into a more effective FIFO reagent control of the displacement process. Thus, the displacement process has been optimized to adequately control the chemical dosage on the surface within a useful range. We define dosage as the summed kinetic effects of time, concentration and temperature on both the completion of the intended reaction (detritylation) and the extent of the side reaction (depurination). With the detritylation displacement by oxidation process, the expected chemical dose correlates with detritylation reaction time and DCA concentration (data not shown). Unfortunately, the impact of temperature on the process could not be measured because it is passively controlled in the facility used for this investigation; hence temperature is a constant for the purpose of experimentation. However, it is expected to also be primary variable. Another aspect of this process is the significant improvement observed in the extent of depurination between the top and bottom slide of the flowcell. Finally, the robustness and reproducibility of the process over large numbers of syntheses was demonstrated by the tight distribution of the depurination metric.
From a manufacturing and process control perspective, the depurination controlled process presented above has several additional advantages. First, the displacement process is scalable to larger substrates. Indeed small substrates are less prone to excessive detritylation. This is likely due to the speed at which reagents can be changed within the flowcell. As the substrate grows, the timescales between reagent changes become long enough for deleterious effects to become apparent. Second, the quenching process holds an advantage over simple displacement with an inert solvent such as toluene because it actively quenches the reaction rather than relying upon simple dilution. The restrictive geometry of the flowcell and low Reynolds numbers of the reagent flows suppresses mixing processes and hence does not favor simple dilution.
Prior to settling on this strategy to control depurination, we carefully reviewed the existing literature. The replacement of DCA with Lewis acids had shown to be an effective alternative (37). However, reports show that the detritylation reaction proceeds at significantly lower rates, which is less desirable in a large scale manufacturing process. Krotz and Cole have documented the use of an aqueous buffer to effectively prevent depurination (38). While this method was appealing, it would not be amenable to a high throughput commercial process because of the long reaction times required to complete the detritylation reaction. Another effective method consists of modifying the C-6 protection of purines to decrease the pKa of the N-7 position (39,40). Although effective, this method utilizes base protecting groups, such as amidinines that are not compatible with fast deprotection strategies, which decreases the throughput and complicates the industrial process. Alternatively, exocyclic amine protecting groups could be omitted, but this approach results in the need for additional washes and reactions to eliminate branching adducts (41). Finally, the two-step chemistry proposed by Sierzchala et al. (42) definitively resolves the issue of depurination by eliminating the use of acid deprotection of the 5′ or 3′ ends of the growing chain. This chemistry is in an early stage of industrial scale development and holds great promise. Meanwhile, the depurination controlled process presented here is an alternative that is well suited for industrial scale applications.
It is interesting to put the results obtained using the depurination controlled process described here in perspective with the synthesis integrity of sequences synthesized on CPG. The mass transfer mechanics for DNA synthesis on a planar solid support are much different than the classical CPG bead. For example, the detritylation process is a flood step performed within a high aspect ratio flowcell. The active reagent is delivered in the bulk reagent that fills the flowcell but must ultimately diffuse to the globally 2D surface from the bulk laminar flow. Hence, over a large substrate we must be aware of concentration depletion in the bulk. Typically reagents are run in massive excess and the depletion is rapidly replenished. This is in contrast to the reagent delivery in porous media such as CPG where the active reagent must ultimately diffuse into a small pore with a size on the order of hundreds of Angstroms. This implies that the detritylation and washing processes must rely on the solvent and reactant being diffused into the pore as there is no chance of convection. This mass transfer effect is compounded by the fact that acid binding to oligonucleotides significantly impacts the efficiency of detritylation as the DNA mass increases (29). Therefore, higher acid concentrations and/or longer detritylation times are required on CPG to achieve acceptable detritylation yields. On microarrays, we speculate that acid binding is not impacting the detritylation efficiency since the boundary layer is more easily replenished and since the loading of oligonucleotide is significantly lower (on the order of pmol instead of µmol). Overall, as long as the detritylation solution can be removed without draining, a lower detritylation dose is necessary on flat solid support, which results in lower depurination.
Finally, the synthesis and characterization of the sequence integrity of libraries of 150mer oligonucleotides based on this novel depurination controlled process represents a new milestone in the long and continuous development of DNA synthesis methodologies. Our results show that it is possible to obtain high-quality oligonucleotide libraries up to 150mer in length using the phosphoramidite chemistry without capping and purification. In conclusion, this new process is characterized by two advantages compared to other strategies for the creation of oligonucleotide libraries. First the synthesis of high-quality long oligonucleotides of length up to 150mer is now possible on the Agilent microarray platform while current methods rarely generate oligomers in excess of 100 nt. Second, the Agilent microarray platform permits the synthesis of libraries of oligonucleotides in which the stoichiometry of each oligo is tightly controlled and tunable by varying the relative number of features synthesized. This dramatically reduces the need to pool individually synthesized oligonucleotides or to develop an appropriate normalizing scheme. We believe that microarray-based synthesis of high-quality, long-oligonucleotide libraries, such as presented herein, will become critical components of novel life science assays and tools, such as those aimed at advancing the understanding of biology and/or at the diagnostic, prevention and cure of human diseases. Already, libraries of long-oligonucleotides made using this improved method have been used for SNP analysis by Targeted Resequencing (43–46), Methylation analysis (47–49), RNA-editing discovery (50), RNA allelotyping analysis (51), Histone characterization (52), high complexity shRNA silencing analysis (53–55) and high throughput mutagenesis (56). Undoubtedly, they will also prove amenable to large-scale synthetic biology applications due not only to the significantly reduced cost per base of the oligonucleotide starting material, but also to the potential improvements in assembly costs compared to methods using oligonucleotides of shorter lengths.
FUNDING
Funding for open access charge: Agilent Technologies.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Dr Xiaohua Huang for technical discussions on purification; Dr Diane Ilslee for radio labeling and gel analysis of oligonucleotides; Mika Illouz for preparing a code prototype to calculate the depurination metrics; Dr Mel Kronick, Dr Doug Amorese and Dr Jeff Sampson for discussions and encouragement; Dr Joel Myerson for technical discussions; the R&D writer engineering team for technical assistance, in particular Stan Woods and Yat-Kwong Ip; Dr Dianne Rees for review of the manuscript; and Dr Steve Macevicz for coordinating the manuscript preparation process.
REFERENCES
- 1.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 2.Caruthers MH. Gene synthesis machines – DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 3.Beaucage SL, Iyer RP. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 1992;48:2223–2311. [Google Scholar]
- 4.Beigelman L, Matulic-Adamic J, Karpeisky A, Haeberli P, Sweedler D. Base-modified phosphoramidite analogs of pyrimidine ribonucleosides for RNA structure-activity studies. Methods Enzymol. 2000;317:39–65. doi: 10.1016/s0076-6879(00)17005-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Dudgeon N, Shen L, Wang JH. Chemical modification of gene silencing oligonucleotides for drug discovery and development. Drug Discov. Today. 2005;10:587–593. doi: 10.1016/S1359-6446(05)03426-4. [DOI] [PubMed] [Google Scholar]
- 6.Pankiewicz KW. Fluorinated nucleosides. Carbohydrate Res. 2000;327:87–105. doi: 10.1016/s0008-6215(00)00089-6. [DOI] [PubMed] [Google Scholar]
- 7.Lesnikowski ZJ, Shi J, Schinazi RF. Nucleic acids and nucleosides containing carboranes. J. Organometallic Chem. 1999;581:156–169. [Google Scholar]
- 8.Foldesi A, Trifonova A, Kundu MK, Chattopadhyaya J. The synthesis of deuterionucleosides. Nucleosides Nucleotides Nucleic Acids. 2000;19:1615–1656. doi: 10.1080/15257770008045450. [DOI] [PubMed] [Google Scholar]
- 9.Leumann CJ. DNA Analogues: from supramolecular principles to biological properties. Bioorg. Med. Chem. 2002;10:841–854. doi: 10.1016/s0968-0896(01)00348-0. [DOI] [PubMed] [Google Scholar]
- 10.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 11.De Mesmaeker A, Altmann K-H, Waldner A, Wendeborn S. Backbone modifications in oligonucleotides and peptide nucleic acid systems. Curr. Opin. Struct. Biol. 1995;5:343–355. doi: 10.1016/0959-440x(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 12.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 13.Butler JH, Cronin M, Anderson KM, Biddison GM, Chatelain F, Cummer M, Davi DJ, Fisher L, Frauendorf AW, Frueh FW, et al. In situ synthesis of oligonucleotide arrays by using surface tension. J. Am. Chem. Soc. 2001;123:8887–8894. doi: 10.1021/ja003758r. [DOI] [PubMed] [Google Scholar]
- 14.Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992;13:1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- 15.Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SPA. Light-generated oligonucleotide arrays for rapid dna-sequence analysis. Proc. Natl Acad. Sci. USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh-Gasson S, Green RD, Yue YJ, Nelson C, Blattner F, Sussman MR, Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 17.Gao XL, LeProust E, Zhang H, Srivannavit O, Gulari E, Yu PL, Nishiguchi C, Xiang Q, Zhou XC. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001;29:4744–4750. doi: 10.1093/nar/29.22.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleary MA, Kilian K, Wang YQ, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nature Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 19.Richmond KE, Li MH, Rodesch MJ, Patel M, Lowe AM, Kim C, Chu LL, Venkataramaian N, Flickinger SF, Kaysen J, et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian JD, Gong H, Sheng NJ, Zhou XC, Gulari E, Gao XL, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 21.Saboulard D, Dugas V, Jaber M, Broutin J, Souteyrand E, Sylvestre J, Delcourt M. High-throughput site-directed mutagenesis using oligonucleotides synthesized on DNA chips. BioTechniques. 2005;39:363–368. doi: 10.2144/05393ST04. [DOI] [PubMed] [Google Scholar]
- 22.Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc. Natl Acad. Sci. USA. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl Acad. Sci. USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr PA, Park JS, Lee YJ, Yu T, Zhang SG, Jacobson JM. Protein-mediated error correction for de novo dna synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binkowski BF, Richmond KE, Kaysen J, Sussman MR, Belshaw PJ. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res. 2005;33:e55. doi: 10.1093/nar/gni053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efcavitch JW, Heiner C. Depurination as a yield decreasing mechanism in oligodeoxynucleotide synthesis. Nucleosides Nucleotides. 1985;4:267–267. [Google Scholar]
- 27.Adams SP, Kavka KS, Wykes EJ, Holder SB, Galluppi GR. Hindered dialkylamino nucleoside phosphite reagents in the synthesis of two DNA 51-mers. J. Am. Chem. Soc. 1983;105:661–663. [Google Scholar]
- 28.Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3053–3058. doi: 10.1093/nar/24.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul CH, Royappa AT. Acid binding and detritylation during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3048–3052. doi: 10.1093/nar/24.15.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchard AP, Kaiser RJ, Hood LE. High-density oligonucleotide arrays. Biosens. Bioelectron. 1996;11:687–690. [Google Scholar]
- 31.Lefkowitz SM, Fulcrand G, Dellinger DJ, Hotz CZ. US Patent; 2002. Functionalization of substrate surfaces with silane mixtures. 6,444,268. [Google Scholar]
- 32.Lajeunesse E, Martin J, Rakotomalala N, Salin D, Yortsos YC. Miscible displacement in a Hele-Shaw cell at high rates. J. Fluid Mech. 1999;398:299–319. [Google Scholar]
- 33.Drazin PG, Reid WH. Hydrodynamic Stability. Cambridge: Cambridge University Press; 1991. p. 452. [Google Scholar]
- 34.Temsamani J, Kubert M, Agrawal S. sequence identity of the n-1 product of a synthetic oligonucleotide. Nucleic Acids Res. 1995;23:1841–1844. doi: 10.1093/nar/23.11.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saffman PG, Taylor GI. the penetration of a fluid into a porous medium or hele-shaw cell containing a more viscous liquid. Proc. Roy. Soc. Lond. A. 1958;245:312–329. [Google Scholar]
- 36.Halpern D, Gaver DP. Boundary element analysis of the time-dependent motion of a semi-infinite bubble in a channel. J. Comput. Phys. 1994;115:366–375. [Google Scholar]
- 37.Matteucci MD, Caruthers MH. The use of zinc bromide for removal of dimethoxytrityl ethers from deoxynucleosides. Tetrahedron Lett. 1980;21:3243–3246. [Google Scholar]
- 38.Krotz AH, Cole DL, Ravikumar VT. Synthesis of antisense oligonucleotides with minimum depurination. Nucleosides Nucleotides Nucleic Acids. 2003;22:129–134. doi: 10.1081/NCN-120019499. [DOI] [PubMed] [Google Scholar]
- 39.McBride LJ, Kierzek R, Beaucage SL, Caruthers MH. Amidine protecting groups for oligonucleotide synthesis. J. Am. Chem. Soc. 1986;108:2040–2048. [Google Scholar]
- 40.Froehler BC, Matteucci MD. Dialkylformamidines: depurination resistant N6-protecting groups for deoxiadenosine. Nucleic Acids Res. 1983;11:8031–8036. doi: 10.1093/nar/11.22.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gryaznov SM, Letsinger RL. Synthesis of oligonucleotides via monomers with unprotected bases. J. Am. Chem. Soc. 1991;113:5876–5877. [Google Scholar]
- 42.Sierzchala AB, Dellinger DJ, Betley JR, Wyrzykiewicz TK, Yamada CM, Caruthers MH. Solid-phase oligodeoxynucleotide synthesis: A two-step cycle using peroxy anion deprotection. J. Am. Chem. Soc. 2003;125:13427–13441. doi: 10.1021/ja030376n. [DOI] [PubMed] [Google Scholar]
- 43.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nature Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 44.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nature Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tewhey R, Nakano M, Wang X, Pabón-Peña C, Novak B, Giuffre A, Lin E, Happe S, Roberts DN, LeProust EM, et al. Enrichment of sequencing targets from the human genome by solution hybridization. Genome Biol. 2009;10:R116. doi: 10.1186/gb-2009-10-10-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li JB, Gao Y, Aach J, Zhang K, Kryukov GV, Xie B, Ahlford A, Yoon JK, Rosenbaum AM, Zaranek AW, et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 2009;19:1606–1615. doi: 10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 51.Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nature Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 54.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl Acad. Sci. USA. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nature Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]