Abstract
Cyanobacteria are suitable for sustainable, solar-powered biotechnological applications. Synthetic biology connects biology with computational design and an engineering perspective, but requires efficient tools and information about the function of biological parts and systems. To enable the development of cyanobacterial Synthetic Biology, several molecular tools were developed and characterized: (i) a broad-host-range BioBrick shuttle vector, pPMQAK1, was constructed and confirmed to replicate in Escherichia coli and three different cyanobacterial strains. (ii) The fluorescent proteins Cerulean, GFPmut3B and EYFP have been demonstrated to work as reporter proteins in cyanobacteria, in spite of the strong background of photosynthetic pigments. (iii) Several promoters, like PrnpB and variants of PrbcL, and a version of the promoter Ptrc with two operators for enhanced repression, were developed and characterized in Synechocystis sp. strain PCC6803. (iv) It was shown that a system for targeted protein degradation, which is needed to enable dynamic expression studies, is working in Synechocystis sp. strain PCC6803. The pPMQAK1 shuttle vector allows the use of the growing numbers of BioBrick parts in many prokaryotes, and the other tools herein implemented facilitate the development of new parts and systems in cyanobacteria.
INTRODUCTION
Cyanobacteria are sunlight-driven cell factories that can be harnessed to sustainably produce numerous products, such as biofuels, foods, feeds and other biomaterials. They may also serve as nitrogen-fixing biofertilisers or be employed for their bioremediatory potential (1,2). Cyanobacteria may use solar energy as their energy source, CO2 from the air as their carbon source, and, some of them, atmospheric N2 as their nitrogen source. With such favorable basic characteristics, well developed molecular tools (3), and existing systems biology analyses (4), cyanobacteria are ideal candidates for a synthetic biological approach.
For some years the field of Synthetic Biology has been evolving (5–8), and it is believed to be a future technology with high impact, connecting several research fields like genetic engineering, systems biology, protein design and computational modeling. Synthetic Biology is the design and construction of new biological parts, devices and systems, as well as the redesign of existing biological systems for useful purposes (http://syntheticbiology.org). Basic principles of Synthetic Biology are:
the use of standardized and well characterized building blocks (BioBricks),
the hierarchical design of nature-inspired, artificial genetic circuits and proteins in silico, and
the use of chemical DNA-synthesis which allows production of DNA sequences that are not found in nature.
The BioBrick standard, originally proposed by Knight in 2003 (9), consists today of thousands of BioBrick parts, which are available at the ‘Registry of Standard Biological Parts’ (http://partsregistry.org). Biological parts are e.g. promoters, ribosome-binding sites, terminators, coding sequences and replication origins, all in a standardized format for easy assembly. The parts are flanked by a standard set of restriction sites and the assembly of several parts can be performed by established cloning techniques. With these features a rapid construction of newly engineered organisms is possible, and genetic constructs can easily be combined and exchanged between different research groups.
Whereas so far most work in Synthetic Biology has been done in Escherichia coli, we aim to use the principles of Synthetic Biology for the design and construction of cyanobacterial molecular tools (e.g. vectors, promoters), and to build up a repository of well characterized cyanobacterial parts. For the introduction of BioBrick parts into cyanobacteria, a BioBrick-compatible cyanobacterial shuttle vector was constructed. This vector contains the replicon of the broad-host-range plasmid RSF1010, designed for replication in different cyanobacterial, and even non-cyanobacterial, strains. The RSF1010 replicon of the IncQ group (10) is well known for allowing plasmids to replicate in a large variety of gram negative bacteria, but there are examples of successful replication in gram positive bacteria as well (11). In fact, the replicative and conjugal abilities of IncQ type plasmids make them the most wide-spread bacterial replicons known (12). This includes, for example, bacteria from the genus Pseudomonas (13), cyanobacterial genera such as Synechococcus (14,15), Prochlorococcus (16) and Agrobacterium tumefaciens (17).
For the rational design of artificial genetic circuits the behavior of each individual part has to be well characterized. Promoters, especially, play an important role in the regulation of genetic circuits. Recently, Kelly et al. (18) presented a standardized way for promoter characterization in Relative Promoter Units (RPU), where the promoter activity is measured in relation to an in vivo reference standard based on fluorescence intensities of expressed green fluorescent protein. However, in cyanobacteria a standardized promoter characterization has not yet been published. Additionally, the use of fluorescent proteins as reporters has to be investigated in cyanobacteria, because a high abundance of photosynthetic pigments could impede the fluorescence signals. Furthermore, inducible or repressible promoters are common when working with expression systems based on E. coli, but rare for cyanobacteria. As shown in this work, inducible promoters that are frequently used in E. coli, e.g. Plac and Ptet, function rather poorly, or not at all, in Synechocystis. It is known that the RNA polymerases (RNAPs) of E. coli and cyanobacteria have structural differences and show different expressional behaviors under the control of a particular promoter (19), which makes host-dependent characterization necessary. To keep the regulatory response times of artificial genetic circuits low, the lifetimes of regulatory proteins (e.g. transcription factors) have to be sufficiently short. A stable protein may accumulate inside a cell after the promoter that drives its expression is shut down, and consequently dependent circuits will not follow the desired regulation. This problem may be solved by the use of degradation tags, which cause proteases to degrade the tagged protein (20,21). In E. coli, degradation tags have been used successfully in genetic circuits (6), but to our knowledge nothing has been published about the use of degradation tags in cyanobacteria.
In this investigation, a broad-host-range BioBrick shuttle vector was constructed and tested. Furthermore, important molecular tools like fluorescent proteins, degradation tags and several constitutive and repressible/inducible promoters were developed and tested in cyanobacteria for use in Synthetic Biology approaches.
MATERIALS AND METHODS
Chemicals
BioBrick DNA parts, identified by ‘BB’, and all BioBrick plasmids, identified by ‘pSB’, were obtained from the Registry of Standard Biological Parts (http://partsregistry.org). The GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich) was used for all plasmid preparations. For purifications of DNA from agarose gels or PCR reactions the Illustra DNA and gel band purification kit (GE Healthcare) was used. For all preparative PCR reactions the Phusion High Fidelity DNA Polymerase (Finnzymes Oy) was used, and native Taq polymerase (Fermentas) was used for colony PCR. Colony PCR for BioBrick plasmids was performed with the standard primers VF2 (BBa_G00100) and VR (BBa_G00101). All restriction enzymes were obtained from Fermentas. The Quick Ligation Kit (New England Biolabs) was used for all ligations. All oligonucleotides for PCR amplifications were purchased from Thermo Fisher Scientific GmbH (Germany) and are listed in Table 1.
Table 1.
Oligonucleotides used in this study
| Primer | Sequence 5′→3′ | Purpose of primer |
|---|---|---|
| pAWG02-MunI-f | TTACAATTGAGTTCTTTTACCCTCAGCCG | Forward primer for RSF1010 origin |
| pAWG02-KpnI-r | GGTAGGTACCATGGTATTACCAATTAGCAGG | Reverse primer for RSF1010 origin |
| pSB03-KpnI-f | CAGGGTACCGCTCTGCCAGTGTTACAAC | Forward primer for BioBrick cloning site and antibiotic cassettes |
| pSB03-MunI-r | GATCAATTGATTACCGCCTTTGAGTGAGC | Reverse primer for BioBrick cloning site and antibiotic cassettes |
| BB-Ptrc-2O-f1 | GGACGAATTGTGAGCGCTCACAATTCGAACG GTTCTGGCAAATATTC | Forward primer for Ptrc |
| BB-Ptrc-2O-f2 | TTTGAATTCGCGGCCGCTTCTAGAGCGAATT GTGAGCGCTC | Forward primer for Ptrc |
| BB-Ptrc-2O-r1 | AGCCTGCAGCGGCCGCTACTAGTATGTGTGA AATTGTTATCC | Reverse primer for Ptrc |
| BBC-lacI-f1 | GCCGAATTCGCGGCCGCTTCTAGATGGTGAA TGTGAAACCAGTAACG | Forward primer for LacI |
| BBC-lacI-r1 | TAACTGCAGCGGCCGCTACTAGTATTATTAC TGCCCGCTTTCCAGTC | Reverse primer for LacI |
| FP_noCO2 | TGGAATTCGCGGCCGCATCTAGAGCAGTCAA TGGAGAGCATTGCCAT | Forward primer for PrbcL1A, PrbcL1B, PrbcL1C |
| FP_CO2noEcoRI | TGGAATTCGCGGCCGCATCTAGAGTCACCAT TTGGACAAAACATCAGCAATTC | Forward primer for PrbcL2A, PrbcL2B, PrbcL2C |
| RP_TSP | AGCCTGCAGCGGCCGCTACTAGTAGCGAAAT TTGGCAATATAAAGCCTAC | Reverse primer for PrbcL1C, PrbcL2C |
| RP_noBBRBS | AGCCTGCAGCGGCCGCTACTAGTAAAACATT GAATAGCCTAGCTTTCTCC | Reverse primer for PrbcL1B, PrbcL2B |
| RP_BBRBS | AGCCTGCAGCGGCCGCTACTAGTATTTCTCCT CTTTAAACATTGAATA | Reverse primer for PrbcL1A, PrbcL2A |
| Fexpro1124 | TGGAATTCGCGGCCGCATCTAGAGTGGTGCA AAACCTTTCGC | Forward primer for Ptrc1O |
| Rsppro1124 | AGCCTGCAGCGGCCGCTACTAGTATGTGTGAA ATTGTTATCCGCTC | Reverse primer for Ptrc1O |
| Fxba | CATCTAGAGATGGTGAGCAAGGGCGAG | Forward primer for Cerulean |
| Rbcu | TACTAGTATTATTACTTGTACAGCTCGTCCATGC | Reverse primer for Cerulean |
| Rceru | GTCGTGCTGCTTCATGTGGT | Confirmation primer for Cerulean |
| REco31I | AATGATACCGCGAGACCCAC | Reverse primer for protease tag |
| FBsp14cfp | AGCTGTACAAGAGGCCTGCT | Forward primer for protease tag |
| VF2 (BBa_G00100) | TGCCACCTGACGTCTAAGAA | Forward primer for sequencing/amplifying BioBrick parts |
| VR (BBa_G00101) | ATTACCGCCTTTGAGTGAGC | Reverse primer for sequencing/amplifying BioBrick parts |
| lacI_flip_r1 | AACGCGGGAAAAAGTGG | Confirmation primer for LacI construct |
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich.
Organisms and growth conditions
Escherichia coli strains DH5α, DB3.1 (both Invitrogen) and HB101 (22), were grown with LB medium at 37°C rotary shaking at 225 rpm, or on agar plates, supplemented with relevant antibiotics depending on the plasmid: pAWG1.1, pRL623 and pSB1AC3 were grown with 35 µg/ml chloramphenicol, pSB1AK3 and pPMQAK1 were grown with 50 µg/ml kanamycin, pSB1A2 and pRL443 were grown with 100 µg/ml ampicillin. When both pRL623 and pPMQAK1 were grown simultaneously in HB101 cells the media were supplemented with 30 µg/ml chloramphenicol and 25 µg/ml kanamycin. For promoter characterization, E. coli was grown in M9 minimal medium (23), supplemented with 0.4% (w/v) glucose.
All cyanobacterial strains were grown in 50 ml E-flask batch cultures at 30°C with continuous shaking in continuous illumination of 50 µmol photons m−2s−1, or on agar plates. Synechocystis sp. strain PCC 6803 (Synechocystis) was grown in BG11 medium (24), supplemented with 50 µg/ml kanamycin when containing pPMQAK1 constructs. Nostoc sp. strain PCC 7120 (Nostoc 7120) and Nostoc punctiforme strain ATCC 29133 (N. punctiforme) were grown under nitrogen fixing conditions in BG110 minimal medium (25), supplemented with 25 µg/ml neomycin when containing pPMQAK1 constructs.
Construction of broad-host-range BioBrick shuttle vector pPMQAK1
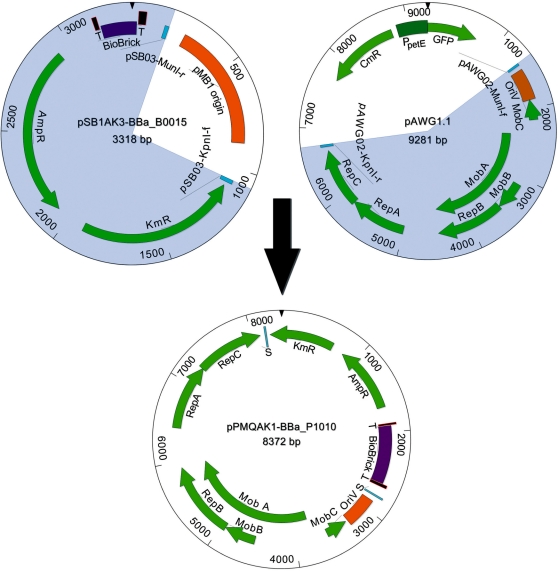
The plasmid DNA fragment necessary for self-replication corresponding to the 5382-bp sequence in plasmid RSF1010 (10) defined by primer sequences pAWG02-MunI-f and pAWG02-KpnI-r (Table 1) was PCR amplified using the RSF1010 derivative pAWG1.1 as template (Figure 1). The 2434-bp DNA fragment containing the BioBrick cloning site and cassettes encoding resistance for ampicillin and kanamycin was PCR amplified with primers pSB03-KpnI-f and pSB03-MunI-r (Table 1) using the plasmid pSB1AK3 carrying the part BBa_B0015, a double transcriptional terminator, as template (Figure 1). The pAWG1.1 and the pSB1AK3-BBa_B0015 PCR products carry MunI and KpnI restriction sites introduced by the primers. Using these restriction sites, the PCR products were cloned together to form the plasmid pPMQAK1 containing BioBrick part BBa_B0015.
Figure 1.
Construction of the broad-host-range BioBrick shuttle vector pPMQAK1. The shaded 3318-bp DNA fragment of pSB1AK3-BBa_B0015, which includes the BioBrick cloning site containing part BBa_B0015 flanked by terminators and cassettes conferring resistance against ampicillin and kanamycin, was amplified with PCR using primers pSB03-KpnI-f and pSB03-MunI-r (Table 1). The shaded 9281-bp DNA fragment of pAWG1.1, which includes the replicon derived from RSF1010, was amplified with PCR using primers pAWG02-MunI-f and pAWG02-KpnI-r (Table 1). Intermediate plasmid pPMQAK1-BBa_B0015 (data not shown) was formed by digesting the two DNA fragments with KpnI and MunI and joining the resulting fragments. Finally, BioBrick part BBa_B0015 was exchanged with part BBa_P1010 to produce the pPMQAK1-BBa_P1010 plasmid. Abbreviations: BioBrick (BioBrick cloning site), T (double transcriptional terminator BBa_B0015), AmpR (ampicillin resistance cassette), KmR (kanamycin resistance cassette), CmR (chloramphenicol resistance cassette), RepA, RepB and RepC (replication proteins A, B and C), MobA, MobB and MobC (mobilization proteins A, B and C), OriV (vegetative origin of replication), GFP (green fluorescent protein), PpetE (petE promoter) and S (ligation site of the two DNA fragments).
Finally, the BBa_B0015 part was exchanged with part BBa_P1010, encoding the DNA gyrase toxin ccdB (26), from plasmid pSB1AK3 using the EcoRI and PstI sites of the BioBrick cloning site, to produce the shuttle vector pPMQAK1 carrying the part BBa_P1010 (Figure 1). The sequence of the new shuttle vector was verified by sequencing (Macrogen, Republic of Korea) and submitted to GenBank (GU933126).
Construction of DNA parts compatible with the BioBrick standard
DNA parts compatible with the BioBrick standard (9) were, unless otherwise stated, obtained with PCR using forward primers containing the BioBrick prefix (5′ gaattcgcggccgcttctagag 3′) and reverse primers containing the BioBrick suffix (5′ tactagtagcggccgctgcag 3′) flanked by a few extra bases to allow for efficient restriction enzyme digestion (Table 1). PCR products were cloned into pSB1A2 usually using the restriction enzyme sites EcoRI and PstI of the BioBrick cloning site.
The coding sequence for the fluorescent protein Cerulean was obtained using the Addgene plasmid 15214 (Addgene) as template and primers Fxba and Rbcu. Since the primers only contained truncated versions of the BioBrick prefix and suffix (Table 1), XbaI and SpeI were used to clone the PCR product and primer Rceru was used together with VF2 for colony PCR to confirm the correct insertion of Cerulean in the plasmid.
For obtaining lacI, the gene encoding the lac repressor, genomic DNA from E. coli strain JM107 (27) was used as template with primers BBC-lacI-f1 and BBC-lacI-r1. Primer BBC-lacI-f1 changes the guanine in the native GTG start codon to an adenine, producing an ATG start codon. Primer BBC-lacI-r1 changes the native TGA stop codon to two TAA stop codons. For complete sequence of the mutated lacI see Supplementary Data.
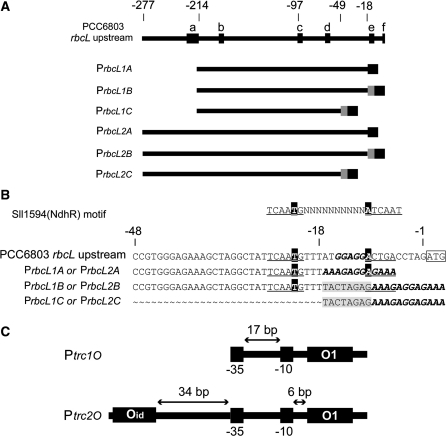
Different variants of the promoter for the large subunit of Rubisco, PrbcL, were constructed by PCR using genomic DNA from Synechocystis as template. The construction details of the rbcL promoter variants that follow are illustrated in Figure 2A and B. The rbcL2A promoter, PrbcL2A, was composed by the sequence from –277 to −18 bp upstream the rbcL start codon, making it the longest version of the rbcL promoters, with the RBS added directly to the 3′-end using PCR. PrbcL2B was composed of the same sequence as PrbcL2A, but the RBS was added using normal BioBrick assembly, creating an 8-bp scar sequence between the promoter and the RBS. PrbcL2C was composed of a shorter sequence of the promoter, stretching from –277 to –49 relative to the rbcL start codon, and the RBS was added as for PrbcL2B, producing a scar sequence. An AT-rich element was observed between –250 and –215 bp upstream of the rbcL start codon. To determine the effect of this element on the rbcL promoter activity, it was excluded from the promoters PrbcL1A, PrbcL1B and PrbcL1C. These three promoters were designed as the PrbcL2A, PrbcL2B and PrbcL2C promoters, respectively, except that they all start at −214 bp upstream the rbcL start codon instead of at −277 bp. PrbcL1A was produced with primers FP_noCO2 and RP_BBRBS, PrbcL1B with FP_noCO2 and RP_noBBRBS, PrbcL1C with FP_noCO2 and RP_TSP, PrbcL2A with FP_CO2noEcoRI and RP_BBRBS, PrbcL2B with FP_CO2noEcoRI and RP_noBBRBS, and PrbcL2C with FP_CO2noEcoRI and RP_TSP (Table 1). In the primer FP_CO2noEcoRI an EcoRI restriction site in the native upstream region of rbcL was removed by changing the sequence to CAATTC. For all sequences of the rbcL promoter variants see Supplementary Data.
Figure 2.
Construction of the rbcL promoter variants and the structure of Ptrc1O and Ptrc2O. (A) The upstream region (–277 to –1, relative to the ATG) of the rbcL gene in Synechocystis sp. strain PCC 6803. a: predicted NtcA-binding site (57), b: putative –10 element (57), c: putative –35 element (56), d: –10 element (56), e: putative ribosome-binding site (RBS) (56), f: start codon ATG of the rbcL gene. The variants ‘1’ (PrbcL1A, PrbcL1B, PrbcL1C) lack the predicted NtcA-binding site and the AT-rich region upstream (–277 to –215, relative to the ATG of the rbcL gene), whereas in the variants ‘2’ (PrbcL2A, PrbcL2B, PrbcL2C) it is present. In the variants ‘A’ (PrbcL1A, PrbcL2A) the BioBrick RBS (black box), replacing the native RBS, was introduced on the primer, whereas in the variants ‘B’ (PrbcL1B, PrbcL2B) the BioBrick RBS was attached by standard assembly, resulting in an additional 8-bp scar (TACTAGAG, gray box). The variants ‘C’ (PrbcL1C, PrbcL2C) lack part of the 3′-end (–48 to –18, relative to the ATG of the rbcL gene), and the RBS (black box) is attached by standard assembly, resulting in an additional 8-bp scar (TACTAGAG, grey box) as in the variants ‘B’. (B) Alignment of the rbcL promoter variants ‘A’, ‘B’ and ‘C’ at the 3′-end. The first 48 bp of the rbcL gene upstream of the ATG and the consensus Sll1594 (NdhR) DNA-binding motif TCAATG(N10)ATCAAT are shown as references. The consensus Sll1594 (NdhR) DNA-binding motif is underlined, the consensus LysR-type motif T(N11)A in black boxes, the putative native RBS and the BioBrick RBS in bold/italic, the 8-bp scar in gray boxes, and the start codon ATG boxed. (C) The structure of Ptrc1O and Ptrc2O. In Ptrc1O, the –35 (TTGACA) and –10 (TATAAT) elements are divided by a 17-bp spacer. The 21 bp O1 lac operator is located 6-bp downstream of the –10 element. In Ptrc2O, the 20 bp ideal Oid lac operator is located 34-bp upstream of the –35 element. The distance between the middle of the two operators is 88 bp.
The promoters Ptrc1O, the trc promoter which contains one lac operator (28), and Ptrc2O, a modified version of Ptrc1O with two lac operators (this work), were both produced using the plasmid pTrc99A as template. Ptrc1O was obtained with primers Fexpro1124 and Rsppro1124. The Ptrc2O promoter was constructed in two consecutive PCR steps; first primers BB-Ptrc-2O-f1 and BB-Ptrc-2O-r1 were used to add on the second lac operator, leading to the same inter-operator distance as for a previously constructed LacI repressible promoter (29). Then the first PCR product was used with primers BB-Ptrc-2O-f2 and BB-Ptrc-2O-r1 to add on the BioBrick cloning site (Figure 2C). For sequences of Ptrc1O and Ptrc2O, see Supplementary Data.
The rnpB promoter, PrnpB, from the Ribonuclease P gene of Synechocystis (CyanoBase, http://genome.kazusa.or.jp/cyanobase, Synechocystis sp. strain PCC6803, chromosome: 152 958–153 165) was synthesized with BioBrick prefix and suffix by Epoch BioLabs, Inc.
All parts are summarized in Table 2.
Table 2.
BioBrick parts and plasmids
| BioBrick part | Description | Source |
|---|---|---|
| Cerulean | Coding sequence of the Cerulean fluorescent protein, obtained by PCR | This work. Template: plasmid Addgene 15214 (Addgene) |
| LacI | Variant of the lac repressor with the start codon GTG changed to ATG, and the stop codon TGA changed to TAATAA. Obtained by PCR | This work. Template: Genomic DNA from E. coli strain JM107 (27) |
| PrbcL1A, B and C | Different versions of the promoter for the large subunit of Rubisco, where an AT-rich upstream region has been excluded. Obtained by PCR | This work. Template: Synechocystis genomic DNA |
| PrbcL2A, B and C | Different versions of the promoter for the large subunit of Rubisco where an AT-rich upstream region has been included. Obtained by PCR | This work. Template: Synechocystis genomic DNA |
| Ptrc1O | The trc promoter (28), which contains one lac operator. LacI repressible, obtained by PCR | This work. Template: plasmid pTrc99A |
| Ptrc2O | A modified version of Ptrc1O with two lac operators. LacI repressible, obtained by PCR | This work. Template: plasmid pTrc99A |
| PrnpB | Promoter of the rnaseP gene. Synthesized DNA | This work. Template: Synechocystis genomic DNA |
| ccdB | DNA gyrase toxin ccdB | BBa_P1010 (Registry) |
| Double terminator | Double transcriptional terminator | BBa_B0015 (Registry) |
| Ribosome-binding site | Ribosome-binding site | BBa_B0034 (Registry) |
| Plac | Wild-type promoter of the E. coli lacYZA operon. LacI repressible | BBa_R0010 (Registry) |
| Ptet | TetR repressible promoter | BBa_R0040 (Registry) |
| Pr | Based on the pR promoter, repressible by the cI repressor | BBa_R0051 (Registry) |
| Promoterless GFPmut3B reporter construct | Contains the ribosome-binding site, the GFPmut3B gene (BBa_E0040) and the double terminator | BBa_I13504 (Registry) |
| Promoterless EYFP reporter constructs with or without protease degradation tags | Contains the ribosome-binding site, the EYFP gene without (BBa_E0030) or with protease degradation tags from the ssrA system, and the double terminator | BBa_E0430, BBa_E0436, BBa_E0434 and BBa_E0432 (Registry) |
| pPMQAK1 | Broad-host-range shuttle vector constructed from the RSF1010 replicon of pAWG1.1, and the BioBrick cloning site and the two antibiotic resistance cassettes for ampicillin and kanamycin from pSB1AK3 | This work |
| pSB1A2 | High copy number plasmid (E. coli) with BioBrick cloning site and ampicillin resistance | pSB1A2 (Registry) |
| pSB1AC3 | High copy number plasmid (E. coli) with BioBrick cloning site and ampicillin and chloramphenicol resistance | pSB1AC3 (Registry) |
| pSB1AK3 | High copy number plasmid (E. coli) with BioBrick cloning site and ampicillin and kanamycin resistance | pSB1AK3 (Registry) |
Registry = Registry of Standard Biological Parts.
Assembly constructs made of BioBrick Standard Biological Parts
Constructs of parts conforming to the BioBrick standard were produced using standard cloning techniques with the BioBrick restriction enzymes EcoRI, XbaI, SpeI and PstI, as described in the original BioBrick proposition (9), or using the three antibiotic assembly technique (3A assembly) (30). The 3A assembly makes use of three BioBrick plasmids with different antibiotic resistance cassettes and the negative selection marker ccdB (BBa_P1010), a DNA gyrase toxin (26). The technique allows for iterative directional assembly of two parts into a third recipient vector without insert or vector gel purification steps, effectively speeding up the cloning process. A strain resistant to the ccdB toxin, such as DB3.1, is required to propagate a ccdB expressing plasmid.
To produce GFPmut3B (BBa_E0040) reporter constructs for testing promoter strengths, the promoters Plac (BBa_R0010), Ptet (BBa_R0040), PR (BBa_R0051), Ptrc1O, Ptrc2O, PrbcL1B, PrbcL1C, PrbcL2B, PrbcL2C and PrnpB were assembled with a promoterless GFPmut3B reporter construct (BBa_I13504 carried by pSB1A2) into pSB1AC3. PrbcL1A and PrbcL2A, which already contain the BioBrick RBS (BBa_B0034), were assembled with a GFPmut3B reporter construct, which lacks the RBS (BBa_I13401 carried by pSB1A2), into pSB1AC3.
To characterize the LacI repressible promoters Ptrc1O and Ptrc2O two cassettes for lacI expression, consisting of a double terminator (BBa_B0015), a promoter driving lacI transcription (either PrnpB or PrbcL2A), a ribosome-binding site (BBa_B0034), the BioBrick standardized lacI gene (this work) and a double terminator (BBa_B0015), were assembled into pSB1AK3. For avoiding read-through transcription affecting the promoter characterization the lacI cassettes were set in opposite direction next to the Ptrc1O or Ptrc2O GFPmut3B reporter constructs in pSB1AC3. This was done by opening each pSB1AC3 reporter plasmid with XbaI, extracting the lacI cassette with XbaI and SpeI from pSB1AK3, inserting it into the open XbaI site of the pSB1AC3 reporter plasmid, and identifying clones containing the reversely inserted cassettes with PCR using primers VF2 and lacI_flip_r1 (Table 1). The first double terminator in the lacI cassettes was inserted to spatially separate the promoter of the lacI cassette from the promoters to be tested, in order to reduce possible crosstalk.
To test the function of different fluorescent proteins, Ptrc1O was assembled with a ribosome-binding site (BBa_B0034), the Cerulean fluorescent protein and a double terminator (BBa_B0015), or with a promoterless enhanced yellow fluorescent protein (EYFP) reporter construct (BBa_E0430), into pSB1AC3. Furthermore, to characterize the effect of protease degradation tags that cause degradation by the ClpXP and ClpAP cytoplasmic proteases (20,21), Ptrc1O was assembled with three different versions of promoterless EYFP reporter constructs differing in the C-terminal ssrA protease degradation tags of EYFP: EYFP-ASV (BBa_E0436), EYFP-AAV (BBa_E0434) and EYFP-LVA (BBa_E0432), into pSB1AC3.
Finally, all reporter constructs were transferred from the pSB1AC3 plasmid to the pPMQAK1-BBa_P1010 broad-host-range shuttle vector using the EcoRI and PstI of the BioBrick cloning site.
Since all reporter constructs contain the same ribosome binding site and the same double terminator, reporter constructs will henceforth be referred to as ‘promoter name’—‘fluorescent protein name’, unless otherwise stated.
All constructed assemblies are summarized in Table 3.
Table 3.
Constructed assemblies
| Construct name | Description |
|---|---|
| Plac-GFP | GFPmut3B reporter construct for characterization of the promoter Plac |
| Ptet-GFP | GFPmut3B reporter construct for characterization of the promoter Ptet |
| PR-GFP | GFPmut3B reporter construct for characterization of the promoter PR |
| Ptrc1O-Cerulean | Construct for characterization of Cerulean fluorescence |
| Ptrc1O-GFP | GFPmut3B reporter construct for characterization of the promoter Ptrc1O, also used for characterization of GFPmut3B fluorescence and determination of pPMQAK1 replicative ability |
| Ptrc1O-EYFP | Construct for characterization of EYFP fluorescence, and as negative control for characterization of protease degradation tags |
| Ptrc1O-EYFP-ASV | Construct for characterization of protease degradation tagss |
| Ptrc1O-EYFP-AAV | Construct for characterization of protease degradation tags |
| Ptrc1O-EYFP-LVA | Construct for characterization of protease degradation tags |
| Ptrc2O-GFP | GFPmut3B reporter construct for characterization of the promoter Ptrc2O |
| PrbcL1A-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL1A |
| PrbcL1B-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL1B |
| PrbcL1C-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL1C |
| PrbcL2A-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL2A |
| PrbcL2B-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL2B |
| PrbcL2C-GFP | GFPmut3B reporter construct for characterization of the promoter PrbcL2C |
| PrnpB-GFP | GFPmut3B reporter construct for characterization of the promoter PrnpB |
| PrnpB-LacI | Cassette for expression of the lac repressor using the promoter PrnpB (low expression) |
| PrbcL2A-LacI | Cassette for expression of the lac repressor using the promoter PrbcL2A (high expression) |
All constructs were moved from the plasmid pSB1AC3 to the pPMQAK1 shuttle vector for subsequent transfer to cyanobacteria.
Triparental mating—conjugal transfer of plasmid DNA
Triparental mating, based on a previously described procedure (31), was used to transfer the shuttle vector pPMQAK1, containing different constructs, to the cyanobacterial cells. Escherichia coli HB101 cells with the conjugal plasmid pRL443 and, for Synechocystis and N. punctiforme, E. coli DH5α cells with the cargo plasmid pPMQAK1, or, for Nostoc 7120, E. coli HB101 cells containing both the helper plasmid pRL623 and the cargo plasmid pPMQAK1, were grown over-night until stationary phase. The pRL623 plasmid contains the M.AvaI methylase that will methylate AvaI, II and III target sites, protecting pPMQAK1 from the native AvaI, II and III restriction enzymes of Nostoc 7120 which would otherwise lower the conjugation efficiency (32). Before conjugation, the cultures of the filamentous strains Nostoc 7120 and N. punctiforme were fragmented by sonication. For this, the collected culture was resuspended with 3 ml BG110 in 10-ml glass tubes, and then sonicated on ice for 30 s with 1-s pulses with an effect of about 7 W using a Sonics Vibracell instrument. This was repeated four times for each culture with about 1 min rest at room temperature in between. After sonication visual inspection by light microscopy showed that the majority of the cells were unicellular, with very few bicellular fragments and some cell debris. For recovery the sonicated cells were resuspended in 25 ml BG11 and incubated at 30°C under low-light conditions overnight. Triparental mating was then performed as described (31).
Determination of pPMQAK1 replicative ability in cyanobacteria using a GFPmut3B reporter
Wild-type cultures, or cultures carrying the pPMQAK1-Ptrc1O-GFP reporter construct, of Synechocystis, Nostoc 7120 and N. punctiforme, were imaged using a Leica TCS SP5 confocal microscope and a HPX PL Fluotar 40.0 × 0.75 dry objective. GFPmut3B was excited with 488-nm laser light and the emission detected between 500 and 540 nm. To visualize the cells, independently of GFPmut3B fluorescence but in the same focal plane, the phycobilisomes and photosystem II complexes were excited with 543-nm laser light and the red auto-fluorescence emission was detected between 650 and 690 nm as described previously (33). For producing non-confocal reference images of the cells, an additional channel with a differential interference contrast (DIC) set-up was used.
Measurement of spectra of fluorescent proteins
The excitation and emission spectra of the fluorescent proteins expressed in Synechocystis were measured using a Horiba Fluorolog-3 fluorometer (Jobin Yvon). Because of the wavelength-dependent transmission of excitation and emission monochromators and the wavelength-dependent sensitivity of the detector R928P PMT, all spectra were corrected using the reference channel. All measurements were done in a 1 × 1 cm optical quartz cuvette and in the linear range of the instrument. For a better signal-to-noise ratio, the absorbance of the cells at the maximum excitation wavelength of the fluorescent proteins Cerulean, GFPmut3B, or EYFP, respectively, was measured with a Cary 5000 UV-Vis-NIR spectrophotometer (Varian) and set to 0.3 by dilution with BG11 medium. The signal-to-noise ratio was further improved 2-fold by averaging the scans of each sample four times. Fluorescence measurements of Synechocystis cells only containing the empty shuttle vector pPMQAK1 were subtracted as background; the spectra were normalized at the highest peak of the spectrum to 100%.
To determine the optimal excitation and emission wavelengths for each fluorescent protein, three spectra had to be measured in the following order: one spectrum for the determination of a preliminary emission maximum, one to get the optimal excitation wavelength and one to get the optimal emission wavelength.
Promoter and degradation tag characterization
The absorbance and fluorescence measurements for promoter and degradation tag characterization were carried out, if not otherwise stated, using a Plate Chameleon V Microplate Reader (Hidex, referred to as Chameleon). A 595 nm filter (595 ± 10 nm) was used for the absorbance measurements of E. coli cultures, and a 750 nm filter (750 ± 10 nm) for Synechocystis cultures, respectively. For the measurement of the GFPmut3B or EYFP fluorescence a 485 nm excitation filter (485 ± 10 nm) and a 540 nm emission filter (540 ± 20 nm) were chosen. Microtest 96-well Optilux Black Assay Plates with clear flat bottom (Ref 353293, BD Falcon) were used for the measurements in order to minimize optical interference between wells. For data analysis, the background (M9 or BG11 medium) was subtracted from the measured values, and the fluorescence was divided by the absorbance, representing the average fluorescence per cell. Also, the fluorescence per cell values of cells only containing the empty shuttle vector pPMQAK1 were subtracted from the values of the cells carrying the reporter constructs. These values represent either the specific promoter activity, i.e. promoter activity by means of GFPmut3B fluorescence intensity and per cell, or specific EYFP fluorescence intensity, i.e. EYFP fluorescence intensity per cell. All the measurements were in the linear range of the instrument.
For the measurement of the GFPmut3B fluorescence intensities of the Synechocystis cultures in Figure 6, the Horiba Fluorolog-3 fluorometer was used instead of the Chameleon because of better sensitivity. The absorbances of the Synechocystis cultures were measured at 750 nm with the Cary 5000 UV-Vis-NIR spectrophotometer and adjusted to 0.1 with BG11 medium. The excitation wavelength of the fluorometer was set to 500 nm and the emission wavelength to 514 nm, otherwise the same instrument settings were used as for the excitation and emission spectra measurements.
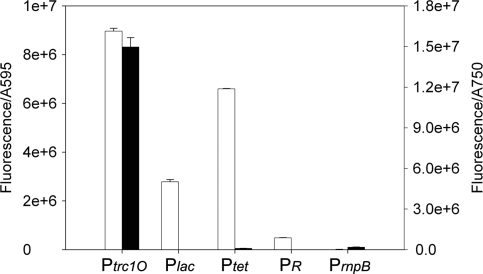
Figure 6.
Specific promoter activities of Ptrc1O, Plac, Ptet, PR and PrnpB in E. coli DH5α (white bar) and Synechocystis sp. strain PCC 6803 cells (black bar). The activities were measured by means of GFPmut3B fluorescence and divided by the absorbance of the cultures at 595 nm (E. coli) or 750 nm (Synechocystis), respectively. The data represent mean ± SD of triple measurements of three independent cultivations.
A seed culture of E. coli DH5α cells containing the GFPmut3B (BBa_E0040) reporter constructs for promoter characterization or an empty shuttle vector pPMQAK1 were grown in M9 medium supplemented with 0.4% (w/v) glucose and 50 µg/ml kanamycin for 16 h, at 37°C, and shaking at 250 rpm. Three tubes containing 5 ml fresh M9 medium supplemented with 0.4% (w/v) glucose and 50 µg/ml kanamycin were inoculated with the seed culture to an initial absorbance Abs595 nm = 0.01, and grown under the same conditions as the seed culture. After 5 h, 100 µl of each culture were collected, diluted with the same volume of M9 medium, and absorbance and fluorescence were measured as triplicates for each sample. For induction experiments, 1 mM IPTG was added to the medium, if not otherwise stated.
A seed culture of Synechocystis cells containing the GFPmut3B (BBa_E0040) reporter constructs for promoter characterization or an empty shuttle vector pPMQAK1 were grown in 20 ml BG11 medium supplemented with 50 µg/ml kanamycin for 4 days, at 30°C, shaking at 120 rpm, and under continuous illumination of approximately 50 µmol photons m–2 s–1. Three E-flasks containing 20 ml fresh BG11 medium with 50 µg/ml kanamycin were inoculated with the seed culture to an initial absorbance Abs750nm = 0.01, and grown under the same conditions as the seed culture. After 48 h, 50 µl of each culture were collected, diluted with 150 µl of BG11 medium, and absorbance and fluorescence were measured as triplicates for each sample. For induction experiments, 2 mM IPTG was added to the medium, if not otherwise stated.
Total protein extraction and western blot analysis
A volume of 25 ml Synechocystis culture containing the GFPmut3B (BBa_E0040) reporter constructs for promoter characterization or an empty shuttle vector pPMQAK1 were collected and re-suspended in extraction buffer containing 50 mM Tris–HCl pH 7.8, 0.1% Triton X-100, 0.02% SDS, 1.42 mM β-mercaptoethanol and one tablet of a protease inhibitor cocktail (Complete Mini, EDTA-free, Roche). The cells were disrupted using 0.6 mm diameter sterile acid washed glass beads in a Precellys 24 homogenizator (Bertin Technologies), and the total protein extract was collected from the supernatant after centrifugation and stored at –20°C until further analysis. Twenty-five micrograms total protein extract of each sample was separated on a SDS (0,1%, w/v) polyacrylamide (12%, w/v) gel by electrophoresis and the resulting gel was stained with Coomassie Blue or transferred to a Hybond ECLTM nitrocellulose membrane (GE Healthcare) with the transfer buffer containing 39 mM glycine, 48 mM Tris, 0.0375% (w/v) SDS and 20% methanol in a TE 22 tank transfer unit (Amersham Bioscience). After the transfer, the membrane was blocked with milk T-TBS [TBS with 0.1% (v/v) Tween 20 and 5% (w/v) Blotting grade non-fat dry milk powder (Bio-Rad, Inc.)] for 1 h. The membrane was incubated with the primary anti-GFP, N-terminal antibody produced in rabbit (Sigma G1544) diluted 1:4000 in T-TBS, for 1 h and washed once with a generous amount of T-TBS then washed three times 10 min with T-TBS. After washing, the membrane was incubated with ECLTM donkey-anti-rabbit IgG linked to horseradish peroxidase as the secondary antibody, diluted 1:5000 in T-TBS, for 1 h. Then, the membrane was washed three times 10 min with T-TBS before detection with the ECLTM western blotting analysis system (GE Healthcare).
RESULTS
Construction of the broad-host-range vector pPMQAK1 and determination of pPMQAK1 replicative ability in cyanobacteria
The DNA fragment corresponding to the replicon of RSF1010 from plasmid pAWG1.1 and the fragment corresponding to the BioBrick cloning site including part BBa_B0015, ampicillin and kanamycin resistance cassettes, from plasmid pSB1AK3 were amplified by PCR, digested and ligated to form plasmid pPMQAK1-BBa_B0015. Subsequently, part BBa_B0015 was excised and part BBa_P1010 was inserted to form plasmid pPMQAK1 (Figure 1). The sequence of the vector pPMQAK1 was determined by sequencing and submitted to GenBank (GU933126). The sequencing revealed an insertion and deletion in the gene coding for replication protein C, repC, which lead to a short frameshift and causing three amino acids to be mutated.
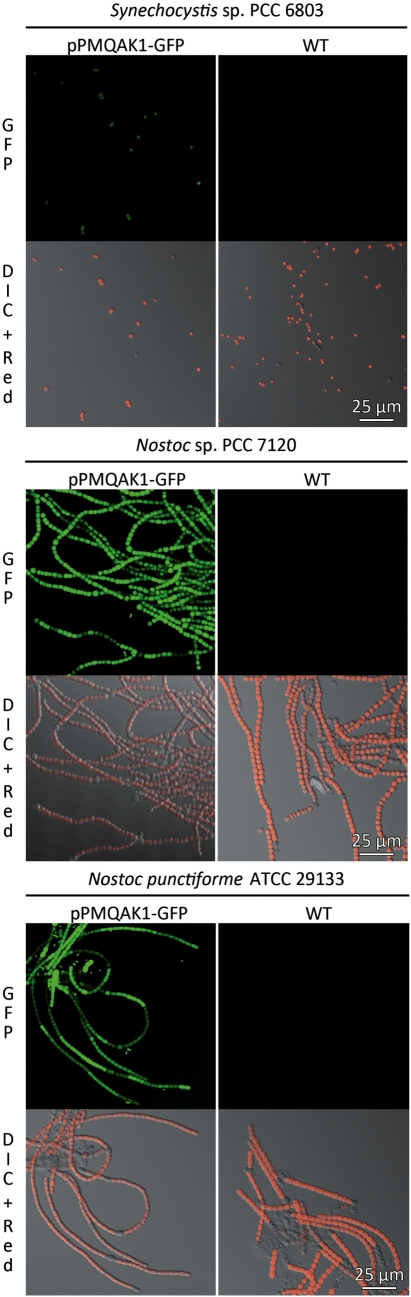
For investigation of the replicative ability of pPMQAK1 in different cyanobacteria, the reporter construct consisting of pPMQAK1-Ptrc1-GFP was transferred into Synechocystis, Nostoc 7120 and N. punctiforme by triparental mating. To determine if the plasmid could successfully replicate inside the cells the transconjugates were imaged by laser scanning confocal microscopy to detect GFPmut3B fluorescence caused by the reporter construct. As a focus and cell integrity control, red auto-fluorescence from the phycobilisomes and photosystem II complexes was imaged in the same focal plane as the GFPmut3B fluorescence. To show all cells independently of fluorescence, a non-confocal DIC image was taken. GFPmut3B fluorescence was clearly detected in all three species, whereas no or very little fluorescence in the GFPmut3B emission wavelength range could be observed in any of the wild-type controls (Figure 3). pPMQAK1-Ptrc1-GFP could be successfully maintained by all three cyanobacterial strains for at least three months (longest time tested) as judged by the continuous presence of GFPmut3B fluorescence, when cultures were grown under antibiotic selection pressure.
Figure 3.
Replicative ability of pPMQAK1 in cyanobacteria. To determine the ability of pPMQAK1 for replication in the cyanobacteria Synechocystis sp. strain PCC 6803, Nostoc sp. strain PCC 7120 and N. punctiforme strain ATCC 29133, a Ptrc1O-GFP (GFP) reporter construct was inserted into the vector which was subsequently transferred to the cyanobacterial cells by the means of triparental mating. GFP: GFPmut3B fluorescence of wild-type cultures or cultures containing the pPMQAK1-GFP reporter constructs. DIC + Red: Control, red autofluorescence stemming from phycobilisomes and photosystem II complexes superimposed with a transmission image in DIC mode. Abbreviations: WT, wild-type; GFP, green fluorescent protein; DIC, differential interference contrast; Red, red auto-fluorescence.
Fluorescent proteins in Synechocystis
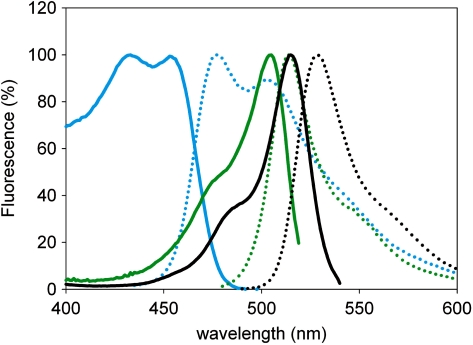
The excitation and emission spectra of three replicates of the three fluorescent proteins Cerulean, GFPmut3B and EYFP expressed by Ptrc1O in Synechocystis were recorded (Figure 4). The maximum excitation wavelengths of Cerulean, GFPmut3B and EYFP were measured as 433, 505 and 515 nm, respectively. The maximum emission wavelengths of Cerulean, GFPmut3B and EYFP were measured as 477, 514 and 529 nm, respectively. At the maximum excitation and emission wavelength, the ratios of the fluorescence intensities per cell of Cerulean, GFPmut3B and EYFP were 1:1.2:3.
Figure 4.
Comparison of excitation and emission spectra of the fluorescent proteins Cerulean, GFPmut3B and EYFP in cyanobacterial background. Background-subtracted excitation (solid line) and emission (dotted line) spectra of the fluorescent proteins Cerulean (blue), GFPmut3B (green) and EYFP (black) expressed in Synechocystis sp. strain PCC 6803.
Promoter characterization in E. coli and Synechocystis
For the characterization of different promoters in E. coli and the cyanobacterium Synechocystis the promoters were cloned into a reporter construct with GFPmut3B as a reporter protein, and the promoter strength was measured as fluorescence intensity. A western blot analysis of extracted and by SDS–PAGE separated proteins confirmed that the measured fluorescence intensities reflect the actual protein concentration of the reporter protein in the cells (Figure 5).
Figure 5.
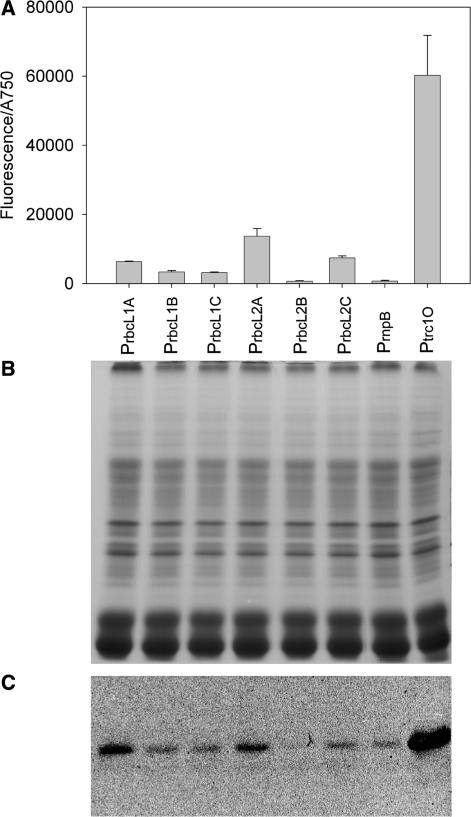
Specific activities of the six rbcL promoter variants and confirmation of fluorescence measurements by SDS–PAGE/western blot analysis (A) Specific activities of the six rbcL promoter variants in comparison to PrnpB and Ptrc1O. The activities were measured by means of GFPmut3B fluorescence and divided by the absorbance of the cultures at 750 nm. The data represent mean ± SD of triple measurements of three independent cultivations. (B) Coomassie-stained SDS–PAGE of total proteins extracted from Synechocystis sp. strain PCC 6803 cultures that expressed the GFPmut3B reporter constructs of the six rbcL promoter variants, PrnpB and Ptrc1O, respectively. (C) Western blot analysis of the same samples as in (B) using anti-GFP, N-terminal, antibodies.
In E. coli DH5α, the specific promoter activities of the promoters Ptrc1O, Ptet, Plac and PR in relation to PrnpB were 881, 649, 274 and 48, respectively (Figure 6 and Table 4). The PrnpB construct in E. coli DH5α showed very low but detectable fluorescence measured by the Chameleon multi plate reader.
Table 4.
Specific promoter activities from Figure 6 in units relative to PrnpB
| Host strain | Promoter activity relative to PrnpB |
||||
|---|---|---|---|---|---|
| Ptrc1O | Plac | Ptet | PR | PrnpB | |
| E. coli | 881 | 649 | 274 | 48 | 1 |
| Synechocystis | 78 | ND | 0.5 | ND | 1 |
ND, not detected.
However, in Synechocystis the Chameleon multi plate reader could not detect the fluorescence of the constructs with the promoters Plac, PR and Ptet. By using the Fluorolog-3 fluorometer, which can excite and detect emission at optimal wavelengths, the fluorescence of the Ptet construct was detectable, but Plac and PR still showed no fluorescence. In Synechocystis, the specific promoter activities of the promoters Ptrc1O and Ptet in relation to PrnpB were 78 and 0.5, respectively (Figure 6 and Table 4).
For the design of genetic circuits the control of protein expression is of great importance. The promoter Ptrc1O shows strong expression in Synechocystis, and can be repressed by the LacI protein and induced by the lactose analog IPTG. Due to the lack of the lacI gene in the Synechocystis genome, lacI was expressed on a plasmid by a constitutive and medium strength promoter. To find a suitable promoter, the promoter PrnpB and six variants of PrbcL, all derived from native Synechocystis genes, were characterized by measuring the fluorescence of the reporter protein GFPmut3B (Figure 5A and Table 5).
Table 5.
Specific promoter activities in Synechocystis from Figure 5A in units relative to PrnpB
| Ptrc1O | PrbcL1A | PrbcL1B | PrbcL1C | PrbcL2A | PrbcL2B | PrbcL2C | PrnpB |
|---|---|---|---|---|---|---|---|
| 83 | 9 | 5 | 4 | 19 | 1 | 10 | 1 |
The specific promoter activities of the promoters PrbcL2A, PrbcL2C, PrbcL1A, PrbcL1B, PrbcL1C and PrbcL2B in relation to PrnpB were 19, 10, 9, 5, 4 and 1, respectively (Figure 5A and Table 5). Based on these results, the concentration of LacI in the cells should be around 20 times higher when using PrbcL2A for lacI expression compared to expression by PrnpB.
In order to obtain a strong, inducible expression system for Synechocystis, the lac repressor, expressed by the promoters PrnpB or PrbcL2A, was cloned into the same plasmid, but in opposite direction, as the GFPmut3B reporter constructs with the promoters Ptrc1O or Ptrc2O (Figure 2C). In addition to the operator O1 in Ptrc1O, Ptrc2O contains a second operator (Oid) that has a higher affinity to the LacI repressor than operator O1. These constructs were characterized in E. coli DH5α (this strain is normally not expressing LacI) and Synechocystis without and with induction by IPTG, and compared to the constructs without repressor expression.
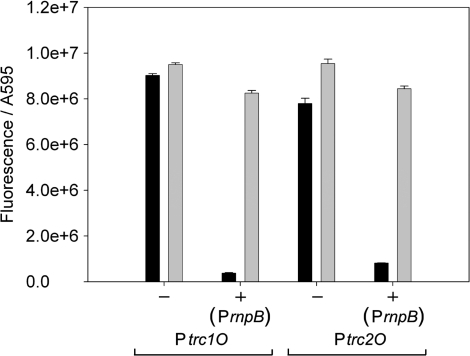
In E. coli, the two promoters show similar expression in constructs without the repressor and under induced conditions when the repressor is expressed. With expressed LacI and without IPTG, Ptrc1O could be repressed 22-fold, whereas Ptrc2O was repressed 10-fold (Figure 7).
Figure 7.
Specific promoter activities of Ptrc1O and Ptrc2O in E. coli DH5α without (–) and with ( + ) the expressed lac repressor under non-induced (black bars) and induced (gray bars) conditions. The activities were measured by means of GFPmut3B fluorescence and divided by the absorbance of the cultures at 595 nm. The data represent mean ± SD of triple measurements of three independent cultivations. The lac repressor was expressed on the same plasmid as the GFPmut3B reporter constructs under the control of PrnpB. Induction was done by 1 mM IPTG.
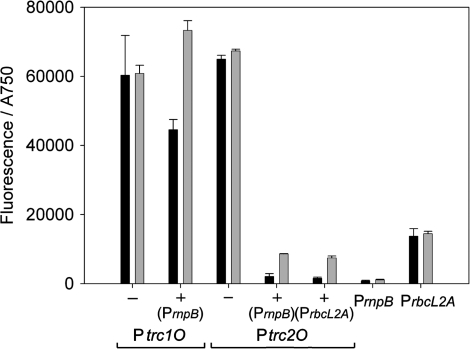
In Synechocystis, the Ptrc2O showed slightly higher expression than Ptrc1O in constructs without the repressor. With the repressor expressed by PrnpB, Ptrc1O was repressed to 75% of the activity without repressor and then induced 1.6-fold. Ptrc2O was repressed to 3% of its activity without repressor and then induced 4-fold. However, this activity under induced conditions corresponds to only 7.5% of its activity without repressor (Figure 8 and Table 6).
Figure 8.
Specific promoter activities of Ptrc1O and Ptrc2O in Synechocystis sp. strain PCC 6803 without (–) and with ( + ) the expressed lac repressor under non-induced (black bars) and induced (gray bars) conditions. The activities were measured by means of GFPmut3B fluorescence and divided by the absorbance of the cultures at 750 nm. The data represent mean ± SD of triple measurements of three independent cultivations. The lac repressor was expressed on the same plasmid as the GFPmut3B reporter constructs under the control of PrnpB or PrbcL2A (as indicated). The specific promoter activities of PrnpB and PrbcL2A GFPmut3B reporter constructs are shown as indication for the lac repressor levels in the cells. Induction was done by 2 mM IPTG.
Table 6.
Specific promoter activities in Synechocystis from Figure 8 in units relative to PrnpB
| IPTG (mM) |
Ptrc1O |
Ptrc2O |
PrnpB | PrbcL2A | |||
|---|---|---|---|---|---|---|---|
| LacI not expressed | LacI expressed by PrnpB | LacI not expressed | LacI expressed by PrnpB | LacI expressed by PrbcL2A | |||
| 0 | 83 | 62 | 90 | 3 | 2 | 1 | 19 |
| 2 | 84 | 101 | 93 | 12 | 10 | 1 | 20 |
These results illustrate two major problems: Even though Ptrc1O showed high expression under induced conditions, the rate of repression is not satisfying. Ptrc2O on the other hand, can be strongly repressed, but could not be induced back to the level without repressor.
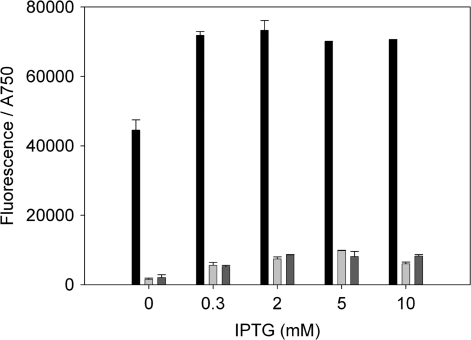
For further investigation, both the repressor and the inducer concentration were varied. In order to get higher repression, the stronger promoter PrbcL2A was used instead of PrnpB for the expression of LacI. However, only a small effect of higher repressor concentrations in the cells was visible (Figures 8, 9 and Table 6). For higher induction, the IPTG concentration in the medium was increased up to 10 mM. Ptrc1O showed maximal expression already at 0.3 mM IPTG, and also the expression of Ptrc2O could not be increased significantly even with high IPTG concentrations (Figure 9).
Figure 9.
Specific promoter activities of Ptrc1O (black bars) and Ptrc2O (light and dark grey bars) in Synechocystis sp. strain PCC 6803 against the IPTG concentration. The activities were measured by means of GFPmut3B fluorescence and divided by the absorbance of the cultures at 750 nm. The data represent mean ± SD of triple measurements of three independent cultivations. The lac repressor was expressed on the same plasmid as the GFPmut3B reporter constructs under the control of PrnpB (black and light grey bars) or PrbcL2A (dark grey bars).
Degradation tags in Synechocystis
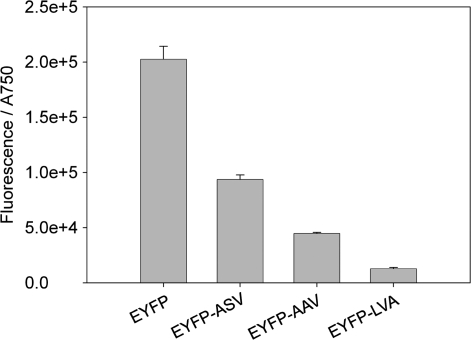
For the characterization of degradation tags, variants of EYFP, tagged with the degradation tags ASV, AAV or LVA, respectively, were expressed in Synechocystis, and the fluorescence of accumulated EYFP was measured. The cultures which expressed EYFP without tag showed highest fluorescence, and the signal of the cultures with tagged EYFP decreased in the following order (Figure 10): EYFP-ASV, EYFP-AAV, EYFP-LVA.
Figure 10.
Specific fluorescence intensities of EYFP and its three different degradation tagged variants in Synechocystis sp. strain PCC 6803. The EYFP fluorescence was measured after 48-h cultivation time and divided by the absorbance of the cultures at 750 nm. The data represent mean ± SD of triple measurements of three independent cultivations. EYFP (BBa_E0030) or the degradation tagged variants EYFP-ASV (BBa_E0036), EYFP-AAV (BBa_E0034) and EYFP-LVA (BBa_E0032) were expressed by the promoter Ptrc1O.
DISCUSSION
The BioBrick compatible broad-host-range vector pPMQAK1 was constructed and shown, by the means of plasmid-bound GFPmut3B expression, to be able to replicate and express GFPmut3B in four different species: E. coli and the cyanobacterial strains Synechocystis, Nostoc 7120 and N. punctiforme. pPMQAK1-Ptrc1-GFP was stably maintained in all three cyanobacterial strains for at least three months (the longest time tested) when cultures were grown under antibiotic selection pressure. The replicative stability of the RSF1010 replicon depends upon its copy number, and it has been shown to become unstable when the copy number is lowered (12,34). Therefore, when cultures harboring pPMQAK1 are cultivated without antibiotic selection pressure, it would be advisable to minimize the metabolic burden introduced by plasmid carried constructs in order to avoid the selection of lower copy number variants and subsequent plasmid instability. Other plasmids derived from the RSF1010 replicon have been shown to possess copy numbers in Synechocystis ranging from 10 (35) to 30 (36) per cell. The copy numbers of these plasmids were obtained using different methods, which may, when differences in cultivation and plasmid sequences are also considered, explain the difference. These average copy number values should be compared with the average value of the Synechocystis chromosome number that was found to be 12 (37). Since both plasmid and chromosome copy numbers are expected to vary with growth conditions and different selection pressures, information about the copy number variance would be useful. However, based on these reported values of RSF1010 derived plasmid copy numbers, the average copy number of pPMQAK1 would be expected to be similar or slightly higher than the average chromosome copy number of Synechocystis. In E. coli the average copy number was found to be 10 (38). Finally, as a RSF1010 derivative, pPMQAK1 is likely to successfully replicate in a wide range of bacteria (11,12), opening up the possibility to utilize the large and growing collection of BioBrick parts found at the Registry of standard biological parts (http://partsregistry.org) in many different species of cyanobacteria (14–16), or other model organisms (13,17), for use in biotechnological applications or genetic studies.
Fluorescent proteins are widely used as reporter proteins for either expression or localization studies. As cyanobacterial cells are filled with a huge amount of pigments for utilizing light as energy source these molecules could influence the measurement of the emitted light of the fluorescent proteins. In this study, the spectra of the three fluorescent proteins Cerulean, GFPmut3B and EYFP, expressed in cells of the cyanobacterium Synechocystis, have been measured and compared to the spectra of wild-type cells without expressing the fluorescent proteins. Cerulean, which was originally developed as an improved donor fluorophore for FRET measurements replacing ECFP, has similar spectral properties as ECFP but is 2.5-fold brighter (39). The maximum excitation and emission wavelengths of the Cerulean expressed in Synechocystis are only slightly different from the wavelengths reported for the pure protein. GFPmut3B folds more efficiently and is better soluble in the cell compared to the wild-type GFP (40). The maximum excitation and emission wavelengths of GFPmut3B expressed in Synechocystis fit well with the reported maxima for the pure protein. Also the excitation and emission peaks of EYFP expressed in Synechocystis are close to the reported maxima for the pure protein (41). These data show that the fluorescent proteins can be used as reporter proteins in cyanobacteria, despite the background of photosynthetic active proteins. However, it has still to be investigated to which extent the photosynthetic pigments can influence the fluorescence intensities by e.g. quenching effects. When two fluorescent proteins should be used simultaneously, e.g. for promoter characterization with internal standard, the measured spectra show that Cerulean should preferably be used with EYFP as the emission spectrum of Cerulean overlaps less with the excitation spectrum of EYFP than that of GFPmut3B. Förster resonance energy transfer (FRET) between the fluorescent proteins is not likely because for FRET the distance between the donor and the acceptor molecules should not exceed 20–60 Å (42).
For the design of regulated genetic circuits well-characterized promoters are crucial biological parts. Standardized promoter characterization has been done in E. coli (18), but so far not in cyanobacteria. When measuring the activity of the promoters Plac, Ptet, PR and Ptrc1O in E. coli and Synechocystis, only Ptrc1O showed similar results in both strains. The hybrid promoter Ptrc1O contains the consensus –35 sequence from Ptrp and the consensus –10 sequence from PlacUV5 (both from E. coli), which are separated by a 17-bp spacer, and the LacI-binding operator O1 (28). The ideal consensus sequences for type 1 promoters (43) of this promoter could explain why it functions well in both E. coli and Synechocystis. However, in Synechocystis the activity of Ptet was very low and activities of the promoters Plac and PR were not detectable. These very different results for the activities of the latter promoters in E. coli and Synechocystis may have different reasons.
The promoter Plac belongs to the class I cAMP receptor protein (Crp) dependent promoters in E. coli, which require Crp for transcription activation and have a single Crp-binding site centered at position –61.5 relative to the transcription start point (44). For initiation of transcription the Crp has to be on the same side of the DNA molecule as the RNAP, which limits the spacing from the middle of the –10 sigma factor-binding site to the middle of the Crp site in intervals of 10.5-bp α-helical turns (45). By definition class I Crp dependent promoters have five turns or more. In Synechocystis a cAMP receptor protein, SYCRP1, was identified (46) and a putative SYCRP1-binding site was proposed (47). The promoter regions of further target genes of SYCRP1 were identified in vitro and characterized in vivo (45). The analysis of the murF P3 promoter region revealed spacing from the middle of the –10 sigma factor binding site to the middle of the Crp site of 5.7 α-helical turns, which differs from the conserved spacing of 5.0 turns in E. coli. This could be one explanation why Plac, which shows the typical spacing of 5.0 turns, did not show detectable activity in Synechocystis. In in vitro studies of Calothrix sp. strain PCC 7601 the cyanobacterial RNAP could initiate transcription at the Crp-independent PlacUV5, but not at the wild-type Crp-dependent Plac (19). Additionally, enteric and cyanobacterial RNAP have different architectures. Instead of one β′ subunit (RpoC) in the enteric RNAP, the cyanobacterial RNAP possesses two subunits γ (RpoC1) and β′ (RpoC2), and also a large insertion between the domains G and H. This may influence the protein/DNA interaction ability (19,43).
In the Ptet promoter sequence two DNA-binding sites for the TetR repressor overlap the –10 and –35 elements. In Synechocystis three potential members of the TetR family were predicted from high structural similarities to the helix-turn-helix motif of the DNA-binding domain of TetR (48). Additionally, the transcriptional regulator PrqR of the TetR family is known as a repressor of the pqrRA operon (49). Repression of Ptet by PrqR or one of the other two predicted TetR family members could explain the very low activity of this promoter in Synechocystis.
In the bacteriophage λ PR promoter sequence two DNA-binding sites for the λ-encoded cI repressor overlap the –10 and –35 elements, which show high similarity to the consensus sequence for the E. coli σ70 and the Synechocystis group 1σ factors. As no λ-encoded cI repressor is expressed in Synechocystis the missing activity of this promoter in Synechocystis cannot be explained by repression through a homologue of this protein. However, in a recent study of E. coli the binding of the protein DksA directly to the secondary channel of RNAP and consecutively stimulating PR-initiated transcription was proposed (50,51). This investigation broadens the general picture of promoter control by transcription factors binding to DNA sequences near the RNAP-binding site by transcription factors binding directly to the RNAP without binding to DNA (50). As the protein DksA is not distributed widely among bacteria and not yet found in cyanobacteria (52,53), the lack of DksA might explain the missing activity of PR in Synechocystis.
In order to find a suitable, constitutive but not too strong promoter for the expression of the lac repressor, PrnpB and six different variants of PrbcL have been characterized. All promoter constructs showed detectable but compared to Ptrc1O not very strong expression. The rnpB gene is widely used as a housekeeping gene probe. The rnpB mRNA transcript level was not affected upon transfer of dark-adapted cells to light and did not decrease upon addition of the electron transport inhibitors DCMU or DBMIB (54,55). Therefore, the rnpB promoter could be used as a stable constitutive promoter in Synechocystis.
The six different variants of PrbcL showed different expression depending on their architecture. A possible ribosome-binding site (GGAGGA) upstream the start codon of the rbcL gene and a 25-bp spacer between the putative –35 element (TAATAA) and the –10 element (TATATT), which is located 60-bp upstream of the start codon, were proposed in the intergenic region between ctpA and rbcL (56). To enable the comparison of promoter activities, the putative rbcL ribosome-binding site (RBS) was replaced by a BioBrick RBS (BBa_B0034) in all versions of PrbcL. When comparing the activities of PrbcL2A with PrbcL1A, and PrbcL2C with PrbcL1C, the presence of the AT-rich element enhances the activity by about 2-fold. Previously, a putative-binding site for the transcriptional regulator NtcA was discovered within this AT-rich element, and a –10 like element was found just downstream (57). This predicted result fits with the structure of an NtcA-activated promoter similar to the class II Crp-dependent promoters (58). NtcA, which responds to 2-oxoglutarate, a signaling molecule connected to the carbon/nitrogen balance of the cells, belongs to the Crp-FNR family of transcriptional regulators (59,60). The deletion of this AT-rich element would remove the putative NtcA-binding site, and hence explain the reduced promoter activity of PrbcL1A and PrbcL1C compared to PrbcL2A and PrbcL2C, respectively. However, comparing the results of PrbcL2B with PrbcL1B the AT-rich element is not enhancing promoter activity. PrbcL2B showed the lowest activity of all the rbcL promoter variants. Unexpectedly, a presumptive LysR-type T(N11)A motif was found in the PrbcL1B and PrbcL2B sequences from –22 to –10-bp upstream of the rbcL start codon. This motif was maintained in the PrbcL1A and PrbcL2A sequences, but in the PrbcL1B and PrbcL2B sequences this motif was mutated due to the insertion of the scar sequence during assembly. Two DNA-binding protein candidates belonging to the LysR-type transcriptional regulators were proposed: Sll1594 (NdhR) (61,62), whose consensus-binding sequence is TCAATG(N10)ATCAAT (the underlined T and A are the conserved nucleotides of the LysR-type motif), and Sll0998 (RbcR) (63) for which there is no specific consensus-binding site motif information other than the general LysR T(N11)A motif. The observed motif TCAATG(N10)AgaAAT of the PrbcL1A and PrbcL2A is highly conserved to the consensus-binding sequence of Sll1594 and there was a dramatic reduction in promoter activity in the PrbcL2B due to the motif mutation. However, it has been proposed that Sll1594 is not involved in the regulation of the rbcLS operon in Synechocystis (61). Further investigations into the putative LysR motif and its transcriptional factor are needed. Importantly, when the putative NtcA motif is present simultaneously as the presumptive LysR motif, such as in PrbcL2A, the highest activity is observed, whereas when the NtcA motif is present but the LysR motif is absent, such as in PrbcL2B, a dramatic decrease in promoter activity is observed. When NtcA is missing and the LysR site is present, as in PrbcL1A, a small decrease is observed. Interestingly, the PrbcL1B and PrbcL1C constructs, without both the putative NtcA-binding site and the presumptive LysR motif, had similar activities. For a typical LysR-type transcriptional regulated promoter, coinducer-dependent cooperative protein-protein interactions seem to be required for occupancy of the activation site (64). There could be a binding site for a LysR interaction partner required for efficient activation by LysR in the promoter sequence, e.g. in the sequence missing from the promoters Prbcl1C and Prbcl2C, i.e. from –48 to –18 bp from the rbcL start codon. This agrees with the observation that the strongest activation is observed for PrbcL2A, which contains the binding sites for NtcA and LysR plus its possible interaction partner. Finally, an interaction between the presumptively bound transcription factors NtcA and LysR is also possible.
The promoters Ptrc1O and Ptrc2O were both successfully repressed in E. coli when LacI was expressed using the promoter PrnpB. Unexpectedly, Ptrc1O was about two times more effectively repressed than Ptrc2O in E. coli. The anticipated higher repressibility of Ptrc2O compared to Ptrc1O depends on DNA looping (65), which is mediated by the binding of the LacI tetramer to two lac operators (66). Perhaps the low amounts of LacI expressed from the weak promoter PrnpB leads to an insufficient amount of the LacI tetramers required for efficient repression of the Ptrc2O by DNA looping, causing Ptrc1O and Ptrc2O to seem similar in repressibility. However, in Synechocystis, Ptrc1O is barely repressed whereas Ptrc2O is strongly repressed compared to the promoter activity without the lac repressor. Evidently, in Synechocystis there is enough LacI present to provide efficient repression of Ptrc2O through DNA looping, whereas Ptrc1O cannot not be efficiently repressed, which seems paradoxical, also considering its much more efficient repression in E. coli. This leakiness of Ptrc1O has already been reported in Synechocystis (36) and, in a version with a twin LacI operator downstream, also in Synechococcus (67). Possibly, explanations could be found in the differences of E. coli and Synechocystis RNAPs. Both the Ptrc1O and the Ptrc2O promoter could be fully induced with 1 mM IPTG in E. coli. In Synechocystis, the weakly repressed Ptrc1O promoter was successfully induced with 2 mM IPTG to levels comparable to its transcriptional activity without LacI repression. However, the more powerfully repressed Ptrc2O could not be induced to levels corresponding to its activity without repression. Increasing the IPTG concentration from 2 to 10 mM did not result in a significant increase of expression from the Ptrc2O construct. Possible limiting factors such as an exhausted metabolic state of the cell or a saturated translational system cannot be the reason for this limited induction since the twin construct of Ptrc2O, Ptrc1O, with a much higher rate of expression, could be fully induced, and had a very similar growth rate. A reason for the limited induction of Ptrc2O could be a very limited diffusion or import rate of IPTG into the cells, resulting in a significantly lower intracellular than extracellular IPTG concentration. As the two lac operators of Ptrc2O lead to a much stronger repression than the single operator of Ptrc1O, Ptrc2O would not be fully induced at these lower intracellular IPTG concentrations, whereas Ptrc1O could be induced. It is not, to our knowledge, established whether Synechocystis can actively transport lactose, and hence IPTG, into the cells, but there are candidate genes such as slr1723 and slr1202 (permeases) (68) and sll1374 (putative sugar transporter) (69). The low induction problem caused by limited diffusion of IPTG has been previously recognized (70) and solved in Pseudomonas fluorescens by the introduction of the E. coli lactose permease gene lacY. Another approach to enhance IPTG induction of a LacI repressed promoter was to develop mutants of the lac repressor with a higher sensitivity to IPTG. By using a directed evolution method based on error-prone PCR, a mutant LacI was acquired that allowed a LacI repressed promoter to be induced with as little as 1 µM of IPTG (71). Using a combination of improved IPTG import through the introduction of a lactose permease and a LacI mutant with higher IPTG sensitivity, a system for very efficient induction of LacI repressed promoters could be acquired. For producing lac repressor controlled promoters with maximized repressibility, the lac inter-operator helical phase and inter-operator distance could be optimized (72). Also, all lac operators could be exchanged for the ideal lac operator that binds the lac repressor tighter than all natural operators (73), and finally, a third or more lac operators could be introduced.
For preventing proteins from accumulating within the cell, degradation tags, recognized by the ClpXP and ClpAP cytoplasmic proteases (20,21), can be utilized. When measuring the fluorescence of Synechocystis cultures expressing tagged EYFP, a tag depending decrease of accumulated protein in the order EYFP, EYFP-ASV, EYFP-AAV and EYFP-LVA was observed, which implicates a reduced stability of the tagged proteins. The LVA tag decreases the protein stability most, followed by AAV and ASV, which resembles the results found in E. coli (20). A high fluorescence intensity of EYFP together with the efficient degradation tag LVA makes EYFP-LVA a sensitive probe giving a good signal-to-noise ratio for the gene expression in genetic circuits of dynamic processes.
Our results show that the biological parts of the Registry of Standard Biological Parts must be characterized in the host organism of choice since their function outside the organism of original use may differ significantly. For the use of Synthetic Biology approaches in cyanobacteria we developed and demonstrated the usefulness of various genetic tools, i.e. a broad-host-range BioBrick shuttle vector, fluorescent proteins, constitutive and repressible/inducible promoters and degradation tags, in the cyanobacterium Synechocystis sp. strain PCC 6803. These tools facilitate the implementation of dynamic regulatory systems such as toggle-switches (7) and oscillators (6,74) that could be used for enhanced control of cyanobacterial production processes. Cyanobacteria have evolved circadian rhythms to synchronize some of their activities to the cycles of light and darkness (75), which may be important to consider for the implementation of any photobiological production system. Artificial genetic oscillators inspired by circadian rhythms may therefore be regulative circuits of special importance for the design of cyanobacterial production strains. Such dynamic systems require inducible promoters with a low level of leakiness and degradation tags for transient protein expression (6). The Ptrc2O promoter herein presented may partly fulfill the inducible promoter demand, especially if the inducibility is improved, and the degradation tags, for use in transient expression, were demonstrated to be functional. In conclusion, we aim for implementing these molecular tools to accelerate the development of cyanobacterial biotechnology, helping to open up important routes towards sustainable production and biofuels.
FUNDING
Swedish Energy Agency; Knut and Alice Wallenberg Foundation; EU/NEST FP6 project BioModularH2 (043340); EU/Energy FP7 project SOLAR-H2 (212508). Funding for open access charge: EU/NEST FP6 project BioModularH2 (043340).
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank M. Rögner (Ruhr-Universität Bochum, Germany) for the plasmid pAWG1.1 and E. Flores (CSIC-Universidad de Sevilla, Spain) for the plasmid pTrc99A. BioBrick parts and plasmids were kindly distributed by the Registry of Standard Biological Parts (MIT, USA).
REFERENCES
- 1.Chisti Y. Microalgae as sustainable cell factories. Environ. Engineer. Management J. 2006;5:261–274. [Google Scholar]
- 2.Tamagnini P, Leitao E, Oliveira P, Ferreira D, Pinto F, Harris DJ, Heidorn T, Lindblad P. Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- 3.Koksharova O, Wolk C. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 2002;58:123–137. doi: 10.1007/s00253-001-0864-9. [DOI] [PubMed] [Google Scholar]
- 4.Ow SY, Wright PC. Current trends in high throughput proteomics in cyanobacteria. FEBS Lett. 2009;583:1744–1752. doi: 10.1016/j.febslet.2009.03.062. [DOI] [PubMed] [Google Scholar]
- 5.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 6.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 7.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 8.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat. Biotech. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight T. MIT Synthetic Biology Working Group Technical Reports. Cambridge: MIT; 2003. Idempotent vector design for standard assembly of biobricks. [Google Scholar]
- 10.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 11.Davison J. Genetic tools for pseudomonads, rhizobia, and other gram-negative bacteria. Biotechniques. 2002;32:386–401. doi: 10.2144/02322rv02. [DOI] [PubMed] [Google Scholar]
- 12.Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 14.Marraccini P, Bulteau S, Cassierchauvat C, Mermetbouvier P, Chauvat F. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 1993;23:905–909. doi: 10.1007/BF00021546. [DOI] [PubMed] [Google Scholar]
- 15.Muhlenhoff U, Chauvat F. Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 1996;252:93–100. doi: 10.1007/BF02173209. [DOI] [PubMed] [Google Scholar]
- 16.Tolonen AC, Liszt GB, Hess WR. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 2006;72:7607–7613. doi: 10.1128/AEM.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Chen Y, Wood DW, Nester EW. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 2002;184:4838–4845. doi: 10.1128/JB.184.17.4838-4845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schyns G, Jia L, Coursin T, Tandeau de Marsac N, Houmard J. Promoter recognition by a cyanobacterial RNA polymerase: in vitro studies with the Calothrix sp. PCC 7601 transcriptional factors RcaA and RcaD. Plant Mol. Biol. 1998;36:649–659. doi: 10.1023/a:1005983320006. [DOI] [PubMed] [Google Scholar]
- 20.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 2001 [Google Scholar]
- 24.Stanier RY, Kunisawa R, Mandel M, Cohenbaz G. Purification and Properties of Unicellular Blue-Green Algae (Order Chroococcales) Bacteriol. Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rippka R, Neilson A, Kunisawa R, Cohen-Bazire G. Nitrogen fixation by unicellular blue-green algae. Arch Mikrobiol. 1971;76:341–348. doi: 10.1007/BF00408530. [DOI] [PubMed] [Google Scholar]
- 26.Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M. Interactions of CcdB with DNA gyrase – inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J. Biol. Chem. 1999;274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 27.Yanischperron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains – nucleotide-sequences of the M13mp18 and Puc19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 28.Brosius J, Erfle M, Storella J. Spacing of the -10 and -35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- 29.Kato K, Marui T, Kasai S, Shininyo A. Artificial control of transgene expression in Chlamydomonas reinhardtii chloroplast using the lac regulation system from Escherichia coli. J. Biosci. Bioeng. 2007;104:207–213. doi: 10.1263/jbb.104.207. [DOI] [PubMed] [Google Scholar]
- 30.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elhai J, Wolk CP. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 32.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardona T, Battchikova N, Zhang P, Stensjo K, Aro EM, Lindblad P, Magnuson A. Electron transfer protein complexes in the thylakoid membranes of heterocysts from the cyanobacterium Nostoc punctiforme. Biochim. Biophys. Acta. 2009;1787:252–263. doi: 10.1016/j.bbabio.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Becker EC, Meyer RJ. Acquisition of resistance genes by the IncQ plasmid R1162 is limited by its high copy number and lack of a partitioning mechanism. J. Bacteriol. 1997;179:5947–5950. doi: 10.1128/jb.179.18.5947-5950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marraccini P, Bulteau S, Cassier-Chauvat C, Mermet-Bouvier P, Chauvat F. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 1993;23:905–909. doi: 10.1007/BF00021546. [DOI] [PubMed] [Google Scholar]
- 36.Ng WO, Zentella R, Wang Y, Taylor JS, Pakrasi HB. PhrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch Microbiol. 2000;173:412–417. doi: 10.1007/s002030000164. [DOI] [PubMed] [Google Scholar]
- 37.Labarre J, Chauvat F, Thuriaux P. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 1989;171:3449–3457. doi: 10.1128/jb.171.6.3449-3457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frey J, Bagdasarian M. The molecular biology of IncQ plasmids. In: Thomas CM, editor. Promiscuous Plasmids of Gram-negative Bacteria. London: Academic Press; 1989. pp. 79–94. [Google Scholar]
- 39.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 40.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 41.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 42.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2006. Springer Science + Business Media, LLC: New York. [Google Scholar]
- 43.Imamura S, Asayama M. Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Bio. 2009;3:65–87. doi: 10.4137/grsb.s2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 45.Hedger J, Holmquist PC, Leigh KA, Saraff K, Pomykal C, Summers ML. Illumination stimulates cAMP receptor protein-dependent transcriptional activation from regulatory regions containing class I and class II promoter elements in Synechocystis sp. PCC 6803. Microbiology. 2009;155:2994–3004. doi: 10.1099/mic.0.028035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimura H, Hisabori T, Yanagisawa S, Ohmori M. Identification and characterization of a novel cAMP receptor protein in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2000;275:6241–6245. doi: 10.1074/jbc.275.9.6241. [DOI] [PubMed] [Google Scholar]
- 47.Yoshimura H, Yanagisawa S, Kanehisa M, Ohmori M. Screening for the target gene of cyanobacterial cAMP receptor protein SYCRP1. Mol. Microbiol. 2002;43:843–853. doi: 10.1046/j.1365-2958.2002.02790.x. [DOI] [PubMed] [Google Scholar]
- 48.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirik IA, Nefedova LN, Fantin Iu S, Babykin MM. Inversion of phototaxis in cells of Synechocystis sp. PCC 6803 determined by a mutation in the regulatory gene prqR. Genetika. 2008;44:474–482. [PubMed] [Google Scholar]
- 50.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyzen R, Kochanowska M, Wegrzyn G, Szalewska-Palasz A. Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 2009;37:6655–6664. doi: 10.1093/nar/gkp676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Alfonso M, Perewoska I, Kirilovsky D. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b(6)/f complex. Plant Physiol. 2000;122:505–516. doi: 10.1104/pp.122.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alfonso M, Perewoska I, Kirilovsky D. Redox control of ntcA gene expression in Synechocystis sp. PCC 6803. Nitrogen availability and electron transport regulate the levels of the NtcA protein. Plant Physiol. 2001;125:969–981. doi: 10.1104/pp.125.2.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amichay D, Levitz R, Gurevitz M. Construction of a Synechocystis PCC6803 mutant suitable for the study of variant hexadecameric ribulose bisphosphate carboxylase/oxygenase enzymes. Plant Mol. Biol. 1993;23:465–476. doi: 10.1007/BF00019295. [DOI] [PubMed] [Google Scholar]
- 57.Su Z, Olman V, Mao F, Xu Y. Comparative genomics analysis of NtcA regulons in cyanobacteria: regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res. 2005;33:5156–5171. doi: 10.1093/nar/gki817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 60.Vega-Palas MA, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 61.Figge RM, Cassier-Chauvat C, Chauvat F, Cerff R. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol. Microbiol. 2001;39:455–468. doi: 10.1046/j.1365-2958.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 62.Wang HL, Postier BL, Burnap RL. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004;279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 63.Gibson JL, Tabita FR. The molecular regulation of the reductive pentose phosphate pathway in Proteobacteria and Cyanobacteria. Arch. Microbiol. 1996;166:141–150. doi: 10.1007/s002030050369. [DOI] [PubMed] [Google Scholar]
- 64.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 65.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 66.Lewis M. The lac repressor. C. R. Biol. 2005;328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 2003;278:19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 68.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 69.Vandenbroucke K, Robbens S, Vandepoele K, Inze D, de Peer YV, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol. Biol. Evol. 2008;25:507–516. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 70.Hansen LH, Knudsen S, Sorensen SJ. The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Curr. Microbiol. 1998;36:341–347. doi: 10.1007/s002849900320. [DOI] [PubMed] [Google Scholar]
- 71.Satya Lakshmi O, Rao NM. Evolving Lac repressor for enhanced inducibility. Protein Eng. Des. Sel. 2009;22:53–58. doi: 10.1093/protein/gzn069. [DOI] [PubMed] [Google Scholar]
- 72.Goyal S, Lillian T, Blumberg S, Meiners JC, Meyhofer E, Perkins NC. Intrinsic curvature of DNA influences LacR-mediated looping. Biophys. J. 2007;93:4342–4359. doi: 10.1529/biophysj.107.112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muller J, Oehler S, MullerHill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 74.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong G, Golden SS. How a cyanobacterium tells time. Curr. Opin. Microbiol. 2008;11:541–546. doi: 10.1016/j.mib.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]