Abstract
Generating a defined set of genetic constructs within a large combinatorial space provides a powerful method for engineering novel biological functions. However, the process of assembling more than a few specific DNA sequences can be costly, time consuming and error prone. Even if a correct theoretical construction scheme is developed manually, it is likely to be suboptimal by any number of cost metrics. Modular, robust and formal approaches are needed for exploring these vast design spaces. By automating the design of DNA fabrication schemes using computational algorithms, we can eliminate human error while reducing redundant operations, thus minimizing the time and cost required for conducting biological engineering experiments. Here, we provide algorithms that optimize the simultaneous assembly of a collection of related DNA sequences. We compare our algorithms to an exhaustive search on a small synthetic dataset and our results show that our algorithms can quickly find an optimal solution. Comparison with random search approaches on two real-world datasets show that our algorithms can also quickly find lower-cost solutions for large datasets.
INTRODUCTION
Forward engineering of biological systems has yielded some interesting and useful results (1–3), but a general process for designing DNA sequences that reliably produce specific system-level functionality remains elusive. The key challenge is simply our incomplete knowledge of every relevant biological mechanism and parameter. Several analytical tools help engineers mitigate this biological uncertainty including biophysical modeling to predict unknown quantities (4), network analysis to select designs robust to uncertain parameters (5) and genome-wide topology inference and optimization to bypass poor parameterization (6,7). These analytical approaches are in contrast to experimental assays that attempt to overcome biological uncertainty by evaluating millions or even billions of variants, often short sequences encoding for a single biological function, from a prescribed solution space. For example, panning phage display libraries regularly yields peptides that bind a defined target (8), selecting RNA aptamers using selective immobilization on solid matrices yields sequences with desired binding specificity (9) and statistically guided screening of enzyme variants yields high-performance proteins (10). There has been a recent trend towards combining these analytical and experimental approaches in an effort to build much larger biological systems. Analytical tools can narrow the design space to hundreds or thousands of promising variants, and experimental evaluation can identify the variant that best meets the system-level specifications. This trend has motivated the use of ‘automated DNA assembly’ to build large sets of multi-kilobase DNA variants, each encoding a similar yet complex system-level functionality.
Although many methods have been described for joining existing pieces of DNA (11–19), in this work, we primarily focus on binary DNA assembly methods. In a binary assembly method, two pieces of DNA are combined together in one stage, and multiple stages are used to gradually assemble the final desired DNA sequence. The original set of DNA pieces can be from a previously constructed library or be specially synthesized for use in assembling a specific DNA sequence. Each ‘piece’ of DNA can also be a mixture of DNA sequences for the construction of combinatorial libraries or compounds in diversity-oriented synthesis for medicinal chemistry. Examples of binary assembly include ligation-by-selection (15), BioBrick™ assembly, overlap extension elongation (18) and specific planned ligation (19). There are different approaches to BioBrick™ assembly, including 3A assembly (16) and 2ab assembly (17). Theoretically, these methods could be used to construct many variants of single-gene sequences, multi-gene cassettes or even complete genomes. Unfortunately, traditional DNA assembly protocols are a relatively laborious manual process, and this significantly limited the size and number of variants that could be realistically constructed. However, engineers have recently started to make use of high-throughput robotic platforms that can help automate the process of assembling many multi-kilobase DNA variants.
A robotic platform is just the first step towards fully automated DNA assembly. The automated hardware needs to be complemented with automated software that can quickly determine the most efficient way to assemble the desired final DNA sequences. As the number of final sequences and the size of each sequence grow, so does the number of ways in which we might assemble these variants. Algorithms for determining how to assemble chimaeric genes exist (19), but they focus on the creation of highly similar DNA sequences and assume that all of the sequences are assembled in the exact same way. In this article, we introduce new algorithms that quickly determine a low-cost way to assemble an arbitrary set of final DNA sequences. Our algorithms minimize the assembly time by determining the optimum number of assembly stages and minimize the assembly work by exploiting common sub-sequences across variants. While our algorithms can be applied to small sets of manually assembled short DNA sequences, their true potential is in future automated DNA assembly platforms where hundreds to thousands of very long DNA sequences can be quickly and efficiently assembled.
MATERIALS AND METHODS
This section presents an overview of the proposed algorithms for automated DNA assembly. The Supplementary Data provide additional details for each algorithm including pseudocode and run-time complexity analysis. We begin by briefly reviewing the two binary DNA assembly methods, 3A and 2ab, for which we are currently using our algorithms. We then introduce the basic terminology used in the rest of the article and more formally define the problem we are trying to solve. This section then describes an algorithm for assembling a single long sequence of DNA and explains how this algorithm can be extended for use in assembling hundreds to thousands of DNA variants. Although these algorithms are directly applicable to the 3A assembly method, we also describe how they can be modified to support the more complicated 2ab assembly method.
Background on 3A and 2ab assembly methods
Our algorithms can be extended for use with a variety of binary DNA assembly methods, but in this article we focus on the two assembly methods that we have been actively using in our research: the 3A assembly method (16) and the 2ab assembly method (17). The 3A assembly method relies on restriction digesting the construction vector, the 5’ part, and the 3’ part; gel purifying the parts and then ligating them together. Our algorithms can handle this assembly method without modification. The 2ab strategy is a single-pot method that requires no gel purification steps. DNA sequences can reside in one of six assembly plasmid types; each type contains two of three different antibiotic resistance markers separated by a common restriction site. The most commonly used antibiotics are kanamycin (K), ampicillin (A) and chloramphenicol (C). Two other restriction sites flank the desired DNA sequence and are selectively digested to generate compatible ends for assembly. To join two DNA sequences x and y to yield xy, they must have the correct permutation of antibiotic markers, i.e. if x were in K/A, y would have to be in A/C while the product xy would be in K/C. The additional constraint on plasmids with compatible antibiotic markers will require extensions to our basic algorithms. The primary advantage of the 2ab assembly method is its lack of gel purification steps, which makes it more robust and particularly well suited to automation with a high-throughput robotic platform. Note that in both methods, assembling a longer DNA sequence from two shorter sequences has an associated cost in terms of time and wetlab work. The time cost is related to the delay required to actually complete the assembly protocol, which can range from one to two days for each assembly due to the need for plasmids to propagate, while the wetlab work cost is related to the required amount of reagents or researcher effort.
Terminology and problem statement
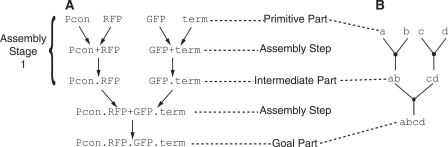
Figure 1A illustrates the assembly of a cassette that expresses both a green and red fluorescent protein. Assembly methods work with ‘parts’, where each part is a sequence of DNA that adheres to the requirements of the assembly method. An engineer begins with a set of ‘primitive parts’ that are gradually assembled to form the final DNA sequence, which is also called the ‘goal part’. The example in Figure 1A uses four primitive parts: a constitutive promoter (Pcon), a red fluorescent protein coding sequence (RFP), a green fluorescent protein coding sequence (GFP) and a transcriptional terminator (term). Primitive parts are assumed to have been created through a separate process such as DNA synthesis. The desired goal part in this example is named Pcon.RFP.GFP.term and includes all four primitive parts. Figure 1A is an example of an ‘assembly graph’, which represents one way of assembling the desired goal part. An assembly graph is made up of ‘assembly steps’ where each step assembles two smaller parts into a longer ‘composite part’. For example, the Pcon and RFP primitive parts are combined using one assembly step to create the new composite part named Pcon.RFP. The 5′ part, Pcon, may be referred to as the left part and the 3′ part, RFP, may be referred to as the right part. The composite part Pcon.RFP is also called an ‘intermediate part’ since it is constructed as an intermediate step in assembling the desired goal part. Multiple independent assembly steps can be performed in parallel in what we call an ‘assembly stage’. For example, the assembly steps that create the Pcon.RFP and GFP.term intermediate parts can be performed in parallel and are part of the first assembly stage. The example assembly graph in Figure 1A requires a total of three assembly steps and two assembly stages. In addition to a selection of primitive parts, an engineer may have access to a ‘part library’ containing previously assembled composite parts. For example, if the composite part Pcon.RFP is already present in a part library, then an engineer can make use of this previously assembled part and eliminate one assembly step from the graph in Figure 1A.
Figure 1.
Example assembly graph. Example of an assembly graph for the goal part Pcon.RFP.GFP.term with real world (A) and abstract (B) representations. The primitive parts include a constitutive promoter (Pcon), red fluorescent protein coding sequence (RFP), green fluorescent protein coding sequence (GFP) and transcriptional terminator (term). These primitive parts are joined in two assembly steps to form composite parts Pcon.RFP and GFP.term during the first assembly stage. A second assembly stage, consisting of a single assembly step, completes construction of the goal part.
In the rest of this article we use a slightly simplified assembly graph notation as illustrated in Figure 1B. Since our algorithms are independent of each part’s precise biological function, we use single letters to represent primitive parts (e.g. a and b) and multiple letters to represent composite parts (e.g. ab). Also notice that arrowheads are omitted but directionality is still assumed.
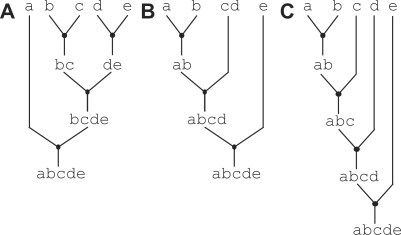
An assembly graph abstractly captures the cost of assembling the desired goal part. The number of assembly stages corresponds to the time cost, while the number of assembly steps corresponds to the wetlab work cost. Any given goal part can be assembled with many different assembly graphs, and each graph can have a very different assembly cost. Figure 2 illustrates three different ways of assembling the goal part abcde. Figure 2A assembles the goal part from five primitive parts and requires three assembly stages and four assembly steps. Figure 2B assembles the goal part from three primitive parts and one composite part (cd) already present in the part library. This alternative approach still requires three stages but is able to save one assembly step. Figure 2C assembles the goal part sequentially from the five primitive parts and requires four stages and four steps. In this example, Figure 2B has the lowest cost since it has the minimum number of stages and/or steps. This assembly graph will result in less time and wetlab work as compared to the other two assembly graphs. In general, some graphs may have more stages but less steps while other graphs will have less stages but more steps, so an algorithm needs to precisely capture how to trade-off time versus wetlab work in a ‘cost function’. Note that, unlike Figure 2B, using a part from the part library can sometimes result in higher cost than an alternative assembly graph that avoids using any parts from the part library. Cost functions can also include other factors such as the failure rate of a certain assembly step, the difficulty in synthesizing a specific primitive part or the likelihood that an intermediate part will be used in a different context.
Figure 2.
Diversity in assembly graphs. Examples of different assembly graphs. Here, assembly graphs are illustrated for the same part abcde yet with different structures and/or costs in A–C. We assume part cd is already present in the part library.
So far we have examined assembling a single goal part, but in this work we are more interested in assembling a ‘goal-part set’ that can include hundreds to thousands of goal parts. Just as we can represent one way to assemble a single goal part with an assembly graph, we can represent one way to assemble a goal-part set with a larger and more complicated assembly graph. The assembly graph for the goal-part set will include subgraphs for each goal part, and these subgraphs are often interconnected when we can share intermediate parts. The number of goal parts, size of each goal part and size of the part library all combine to create many valid assembly graphs for a specific goal-part set. Searching for a minimum-cost assembly graph is a time-consuming and error-prone problem for an engineer but is well-suited for computer optimization. The algorithms presented in this article are a first step towards solving this problem, which can be more formally specified as follows:
Given a goal-part set and a part library, find the minimum cost assembly graph that builds all parts in the goal-part set.
Single-goal-part assembly algorithm
We begin by describing an efficient algorithm for finding the minimum cost assembly graph given a single goal part. In the next subsection, we will extend this algorithm for use in assembling a complete goal-part set. We define the cost of an assembly graph to be the tuple 〈stages,steps〉, and our initial cost function prioritizes assembly stages unless they are equal, in which case we compare the number of steps. Although this cost function strictly prioritizes time over wetlab work, other cost functions with different trade-offs are possible.
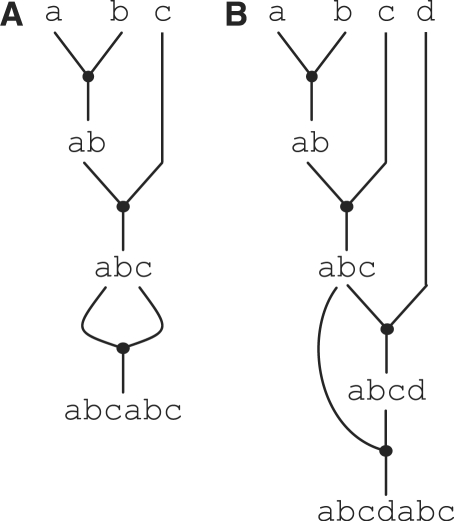
The binary tree structure of assembly graphs for a single goal part enables us to easily leverage several well-known results. The total number of possible assembly graphs for a single goal part is (2n − 1)!/(n − 1)!n! [also known as the Catalan number (20)] where n is the number of primitive parts in the goal part. The minimum number of stages is ⌈log2 n⌉ and the minimum number of steps is n − 1. If n is an even power of two then there is exactly one optimal assembly graph, otherwise there can be many assembly graphs with the minimal number of stages and steps (see Figure 2A for an example with cost 〈3,4〉). Note that these bounds make two important assumptions about the part library and sharing of intermediate parts. First, these bounds assume that the part library is empty. If the library already contains intermediate parts that are in one of the optimal assembly graphs, then these parts can be used to reduce the number of steps and possibly the number of stages (see Figure 2B for an example with cost 〈3,3〉). The second assumption is that the assembly graphs do not exploit ‘intra-goal-part sharing’ (intra-GPS). Intra-GPS occurs when the goal part includes the same sequence of primitive parts multiple times. For example, the goal part abcabc includes the primitive part sequence abc twice. If we do not exploit intra-GPS, then the minimum cost is simply 〈⌈log2 6⌉, 6 − 1〉 = 〈3,5〉. Figure 3A illustrates an alternative assembly graph for this goal part that exploits intra-GPS. The graph assembles the intermediate part abc once and then reuses it in the final step to assemble the goal part abcabc. Exploiting intra-GPS reduces the cost to 〈3,3〉. Unfortunately, exploiting intra-GPS can sometimes lead to assembly graphs with ‘higher’ cost than the straight-forward graph that does not exploit intra-GPS (see Figure 3B for an example). In this work, we assume that intra-GPS will be rare in practice, since it requires long DNA sequences to appear multiple times in a single goal part. We will use real-world datasets in the Results section to help validate this assumption.
Figure 3.
Intra-GPS. Examples of sharing within an assembly graph for a single goal part. Panels A and B illustrate how intra-GPS can either decrease (A) or increase (B) the cost of an assembly graph.
A brute-force algorithm for finding the minimal cost assembly graph would enumerate all (2n−1)!/(n−1)!n! assembly graphs and then exhaustively search for the minimal cost graph. A slightly less naive algorithm can use a simple divide-and-conquer approach. The algorithm begins by examining all possible ways in which we might perform the final assembly step. For example, with the goal part abcde the naive algorithm examines a + bcde, ab + cde, abc + de and abcd + e. For each of these potential assembly steps, the naive algorithm recursively examines all possible assembly steps for forming the left part and the right part. We can view the naive algorithm as searching a tree where each node in the tree is an intermediate part, and the edges represent all possible ways of assembling that intermediate part. Unfortunately, the naive algorithm has an exponential running time as a function of the number of primitive parts in the goal part, and this can make it slow for goal parts containing tens of primitive parts.
Dynamic programming is a generic algorithmic technique specifically for these types of problems (20). Dynamic programming is only applicable because our problem has two specific properties: ‘optimal substructure’ and ‘sub-problem reuse’. Our problem has optimal substructure because the minimal-cost assembly graph for a specific intermediate part is optimal regardless of how it is assembled with a second part. For example, assume we are trying to find the minimal-cost assembly graph for the goal part abcdefgh. The optimal assembly graph for the intermediate part cdef is still optimal regardless if it is used in the assembly step ab + cdef, b + cdef, cdef + g or cdef + gh. Note that while this is true for the stage- and step-based cost function described earlier in this subsection, it may not be true for all possible cost functions. The optimal substructure property is, however, still true when using a part library. A part from the library has zero cost regardless of context. The optimal substructure property allows us to calculate the cost for an intermediate part once and reuse this calculation in many different contexts. This leads us to sub-problem reuse. Dynamic programming is only helpful if the same calculation is used many times, which is indeed the case in our assembly problem. We can view dynamic programming as essentially merging some of the nodes in the search tree used by the naive algorithm. This allows us to search a part of the tree once and then reuse the result multiple times. Our dynamic programming algorithm has a polynomial running time and is guaranteed to find an optimal assembly graph ignoring intra-GPS. Exploiting intra-GPS would significantly complicate the algorithm, since intra-GPS essentially violates the optimal substructure property. This is because the cost of assembling a specific intermediate part depends on its context; it depends on whether or not that intermediate part is reused elsewhere. The Supplementary Data provide more detailed pseudocode and run-time analysis for our single-goal-part algorithm based on dynamic programming.
Multiple-goal-part assembly algorithm
In this section, we describe how the single-goal-part algorithm can be used as a basis for determining a near-optimal way to assemble a full goal-part set. Initially, we will consider the cost of an assembly graph for a goal-part set to still be the tuple 〈stages,steps〉 with the same cost function as in the single-goal-part algorithm. Since we can assemble goal parts in parallel, the number of stages for a full goal-part set is equal to the maximum number of stages over all disjoint assembly subgraphs. This implies that a goal-part set with a few longer goal parts will be limited, in terms of the time cost, by the many stages required to assemble these longer goal parts.
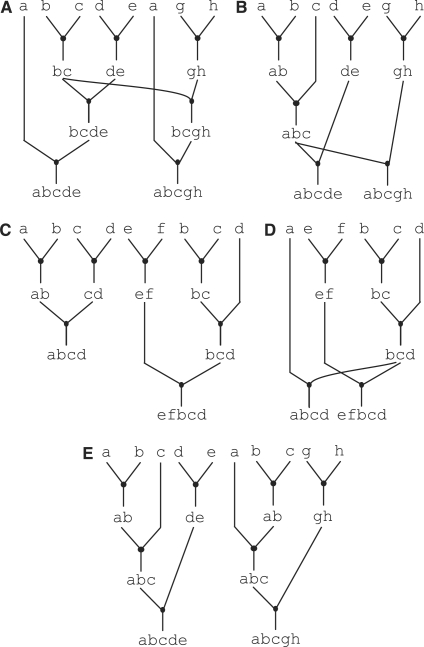
The most straight-forward approach is to use the single-goal-part algorithm as currently described for each goal part in isolation. This approach is guaranteed to find a full assembly graph with the minimum number of assembly stages, but it misses opportunities for reducing the number of assembly steps through ‘inter-goal-part sharing’ (inter-GPS). Inter-GPS occurs when two goal parts share a common sequence of primitive parts. For example, assume the goal-part set includes the goal parts abcde and abcgh. Applying the single-goal-part algorithm on each part in isolation results in two disjoint assembly subgraphs each with a cost of 〈3,4〉 and thus a total cost for the goal-part set of 〈3,8〉. Figure 4A illustrates how we might exploit inter-GPS in this example, by building the intermediate part bc once, and reusing it to build both goal parts. This reduces the total cost to 〈3,7〉. It is actually possible to achieve even greater savings by sharing the longer intermediate part abc as shown Figure 4B. Unlike intra-GPS, exploiting inter-GPS is guaranteed to reduce the number of assembly steps without increasing the number of assembly stages. Intuitively this can be seen by noticing that intra-GPS involves sharing an intermediate part in two contexts that might have been previously connected, while inter-GPS involves sharing an intermediate in two completely disjoint contexts. Also unlike intra-GPS, inter-GPS is quite common in realistic goal-part sets. Engineers often want to experiment with many variants where only one or two parts are different across goal parts. For example, an experiment to tune gene expression levels might use a goal-part set where each goal part has the same promoter, protein coding sequence and terminator, but a different ribosomal binding site. We will use real-world datasets in the Results section to help validate the assumption that inter-GPS is common. The rest of this section illustrates techniques for exploiting inter-GPS to reduce the total number of required assembly steps for a specific goal-part set.
Figure 4.
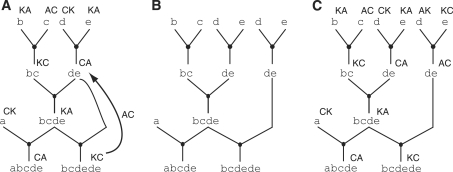
Inter-GPS, slack and iterative refinement. Inter-GPS is shown in (A–B) and allows assembly graphs for different goal parts to share common intermediate parts reducing the assembly cost from 〈3, 8〉 to 〈3, 7〉 in A and 〈3, 6〉 in B. Slack is shown in (C–D) and allows suboptimal solutions to increase the potential for inter-GPS reducing the assembly cost from cost from 〈3, 7〉 in C to 〈3, 5〉 in D. Iterative refinement helps ensure common intermediate parts are constructed in the same way. For example, iterative refinement can turn the assembly graph in E into the assembly graph in B.
Note that applying the basic single-goal-part algorithm to each goal part may serendipitously result in shared intermediate parts between some of the assembly subgraphs. Unfortunately, since there can be many equally optimal assembly subgraphs for each goal part, there is no guarantee that the single-goal-part algorithm will choose the subgraph that has the most shared intermediate parts. In order to bias the single-goal-part algorithm towards subgraphs with many shared intermediate parts, we need to somehow leverage global information about all goal parts in the single-goal-part algorithm. To this end, our multiple-goal-part algorithm first calculates the ‘sharing factor’ for every possible intermediate part. The sharing factor is the number of redundant times each sequence of primitive parts appears across all goal parts. For the goal parts abcde and abcgh, the sharing factor for all intermediate parts is zero except for the parts ab, bc and abc. These intermediate parts have a sharing factor of one, since they appear in both goal parts. We then pass the sharing factors for all possible intermediate parts into a modified version of the single-goal-part algorithm, and we extend the cost tuple to be 〈stages, steps, sharing〉. We update our cost function so that if the number of stages are equal, we consider the number of steps minus the sharing factor. Essentially, this new cost function optimistically assumes that any intermediate part that can be shared is indeed maximally shared, even though there is no guarantee that this sharing will actually take place. There may be cyclic sharing dependencies between goal parts which prevent maximal sharing, or two goal parts may assemble a shared intermediate in two different ways. Regardless, this approach biases the single-goal-part algorithm towards locally optimal assembly subgraphs that are also likely to result in shared intermediates with the assembly subgraphs for other goal parts. The recursive nature of our dynamic programming algorithm explicitly favors long shared intermediates that can amortize many assembly steps. The Supplementary Data include detailed pseudocode illustrating how we can modify the basic single-goal-part algorithm to include sharing factors.
The modified single-goal-part algorithm described so far will always find a locally optimal assembly subgraph, but we may be able to increase opportunities for shared intermediates by considering locally suboptimal assembly subgraphs. For example, assume our goal-part set includes the parts abcd and efbcd. Figure 4C illustrates one assembly graph where both goal parts use locally optimal assembly subgraphs. Notice, however, that efbcd requires three stages, while abcd only requires two stages. We call the difference between the locally optimal number of assembly stages for a specific goal part and the maximum number of assembly stages across all goal parts the ‘slack factor’. Goal parts with a slack factor greater than zero can potentially use locally suboptimal assembly subgraphs to increase opportunities for shared intermediate parts without impacting the cost of the overall assembly graph. Figure 4D illustrates a different assembly graph where abcd now requires three assembly stages, which is greater than the locally optimal number of two stages, but is now able to reuse the intermediate part bcd. This reduces the overall cost to three stages and five steps. To include the slack factor in our single-goal-part algorithm, we simply add the slack factor to our cost function so that subgraphs with more stages but available slack are not automatically eliminated. The Supplementary Data include detailed pseudocode illustrating how we can modify the basic single-goal-part algorithm to include the slack factor.
Even after modifying the single-goal-part algorithm to include the sharing and slack factors, there is still no guarantee that the single-goal-part algorithm will choose graphs with the maximum number of shared intermediate parts. This is because, even with these additional factors, there will still be many minimal-cost assembly subgraphs for each goal part. For example, assume our goal part set includes the parts abcde and abcgh. We would like to ultimately find the assembly graph shown in Figure 4B, but it is equally likely that we will find the assembly graph shown in Figure 4E. This is because when we are determining the optimal assembly subgraph for the intermediate part abc both subgraphs have equal cost. So the issue is that although the single-goal-part algorithm correctly biased its search based on the sharing factors, it ended up being biased in two equal yet different directions for the intermediate part abc. To address this issue we add ‘iterative refinement’ to our multiple-goal-part algorithm. Iterative refinement first runs the single-goal-part algorithm on each goal part. Then we ‘pin’ the assembly subgraph for one of the goal parts by effectively making all intermediate parts in that subgraph zero cost. We then rerun the single-goal-part algorithm on all non-pinned goal parts. We continue to pin one goal part after each iteration until all goal parts are pinned. Pinning a goal part effectively biases later iterations towards the pinned intermediate parts. We have experimented with various heuristics for determining which goal part to pin including the longest goal part and the goal part with the least degrees-of-freedom (DOF). DOF is the number of minimal-cost assembly subgraphs for a given goal part, so pinning the goal part with the least DOF enables goal parts with more DOF to reconfigure so as to better match the pinned graph. We have found choosing the longest goal part to be simple to implement and to perform well across a range of goal-part sets, and we use this approach throughout the rest of the paper. The Supplementary Data include detailed pseudocode for our final multiple-goal-part algorithm which combines iterative refinement with the sharing and slack factors.
Extensions to support 2ab assembly method
The single-goal-part and multiple-goal-part algorithms described so far are directly applicable for use in the 3A assembly method. The 2ab assembly method requires an additional post-processing step to handle the antibiotic markers. The post-processing consists of assigning antibiotic markers to parts in the graph, identifying any conflicts in this assignment, determining the lowest cost common node of the two conflicting subgraphs, breaking the sharing of the corresponding intermediate part and merging the resulting graphs.
The assignment of antibiotic markers is termed ‘coloring’ for short. The coloring algorithm considers the assembly graph for each goal part and assigns the markers to parts in a deterministic fashion (K/A and A/C combine to form K/C, A/K and K/C combine to form A/C and so on). The algorithm is analogous to a breadth-first search in that it colors both the parents and the children of a part; if two goal parts share subparts, their assembly subgraphs are connected and colored at the same time. Sometimes, the algorithm attempts to recolor a previously-colored part and a coloring conflict arises if the two colors disagree (see Figure 5A). If we consider the assembly graph as an undirected graph, a conflict can only arise from a cycle in the graph (a cycle being a closed path with no repeated nodes or edges other than the starting and ending nodes). An example of a cycle, abcde − bcde − de − bcdede, is shown in Figure 5A.
Figure 5.
Assembly graph coloring. Breaking assembly graph ‘cycles’ allows for 2ab coloring extensions. In A, an assembly graph is presented with a coloring conflict from bcdede to de. In B, a cycle is broken at part de. Now the graph can be colored as shown in C. However, upon inspection, there may be a number of duplicate parts once coloring is taken into account. These are then merged in the final step of post-processing.
For the sake of brevity, the example is of an intra-GPS cycle, but the concept remains the same for inter-GPS cycles. All cycles must have a lowest-cost part because edges only connect two parts of unequal cost. In order to resolve the coloring conflict in the most economical manner, we find the lowest-cost part in the cycle and duplicate its graph and subgraphs as shown in Figure 5B. The coloring algorithm is then called again to color the duplicated subgraphs and to discover if coloring conflicts still exist as shown in Figure 5C. Once all coloring conflicts are resolved, we make the final assembly graph by traversing the graph, making a record of each unique graph and attempting to recapture any potential subpart sharing with coloring taken into account by merging the duplicate nodes (same part and same color).
RESULTS
In this section, we demonstrate the effectiveness of our algorithms by running them on two real-world goal-part sets. The first goal-part set is for a phagemid project from the Anderson lab and contains 131 goal parts. In order to screen for a correctly functioning device, this ‘phagemid’ dataset encoded three specific biochemical functions into a device and varied the transcriptional regulation. These three biochemical functions were distributed into one or more operons and each operon was under control of one of four promoters. The order of these operons was also varied. The second goal-part set was designed by a group of students at the University of California at Berkeley as part of the 2008 International Genetically Engineering Machines competition (17). This ‘iGEM-2008’ dataset contains 397 goal parts that were used as devices for simplifying in vivo cloning reactions. Part lengths in these datasets range from 2 to 14 primitive parts. Preliminary analysis of both datasets found that there were no instances of intra-GPS and many instances of inter-GPS, confirming our earlier assumptions. For example, in the ‘iGEM-2008’ dataset the average sharing factor for a subpart was four with a maximum of 238. For the ‘phagemid’ data set the average sharing factor was 6 with a maximum of 130. Both datasets are available in the Supplementary Data.
We have implemented our algorithms using Java as part of the Clotho framework (21). Clotho is a ‘platform-based design’ environment for the engineering of biological systems. It enables the development of independent tools that communicate in well-structured ways and provides a rich data-interface for the retrieval of biological information. Clotho allows our assembly algorithms to cleanly communicate with high-throughput robotic platforms and easily access standard part libraries. The source code for our assembly algorithms and the rest of the Clotho framework and documentation are available at http://www.clothocad.org.
Results for the single-goal-part algorithm
The polynomial run-time of the single-goal-part algorithm allows it to efficiently find the optimal assembly graph for a single goal part. For example, finding a solution for a goal part with 50 primitive parts requires milliseconds with our dynamic programming approach, but requires 5–10 min for the naive exponential-time method on a standard general-purpose workstation. Since the single-goal-part algorithm is used many times by the multiple-goal-part algorithm, this difference in run-time can result in significant speed-up when working with large goal-part sets.
Results for the multiple-goal-part algorithm
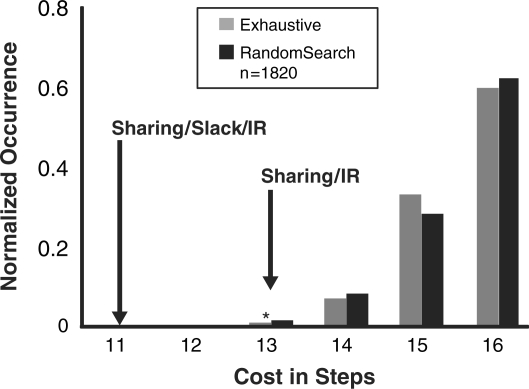
To evaluate our multiple-goal-part algorithm, we measure not only the approximate run-time but also the quality of the final assembly graph. We begin by examining the performance of our algorithm on a small synthetic goal-part set. Figure 6 shows the results for three different algorithmic approaches: exhaustive search, random search and our multiple-goal-part algorithm. Note that all three algorithmic approaches will always produce assembly graphs with the optimal number of stages. The primary difference is in how well the algorithmic approaches minimize the total number of steps. In the ‘exhaustive search approach’, we enumerate all possible assembly subgraphs that are locally optimal for each goal part. We then examine all combinations of these assembly subgraphs to exhaustively search all possible graphs for the entire goal-part set. Figure 6 shows the distribution of the number of assembly steps for the final combined assembly graphs. Notice that most of the graphs have 15–16 steps, but graphs that better exploit inter-GPS are able to reduce the cost to a minimum of 13 steps. In the ‘random search approach’, we also enumerate all possible assembly subgraphs that are locally optimal for each goal part. We then randomly pick a subgraph for each goal part and combine them to take advantage of shared intermediate parts. Since the random search approach does not examine all possible combinations, it is much faster than the exhaustive search. Figure 6 shows that the random search approach still produces a similar distribution of assembly graphs as the exhaustive search approach. Notice that very few assembly graphs actually contain the minimum number of assembly steps. This implies that while serendipitously shared intermediate parts are possible, a more targeted algorithm is needed to quickly find the optimum solution. Figure 6 also shows the solutions found with our multiple-goal-part algorithm. With the sharing factors and iterative refinement, our algorithm is able to quickly find the optimal solution with 13 assembly steps. With the addition of the slack factor, our algorithm is actually able to find a solution that requires only 11 assembly steps. This solution is not found by the other approaches, since they rely on locally optimal subgraphs, but the slack factor enables locally suboptimal subgraphs to be used when they produce a more optimal global result.
Figure 6.
Comparison of solutions found for the small synthetic goal-part set. A comparison of exhaustive search, random search, and our algorithm’s output for a small set of goal parts. The x-axis provides the number of steps required and the y-axis shows the normalized occurrence of assembly graphs of that size. As shown, a random search with 1820 graphs compares favorably with locally optimal exhaustive search, with a best solution of cost 13 (denoted by an asterisk), while our algorithm is able to provide an assembly graph with lower cost due to the slack factor. This result also demonstrates we can use random search as a point of comparison with our algorithm.
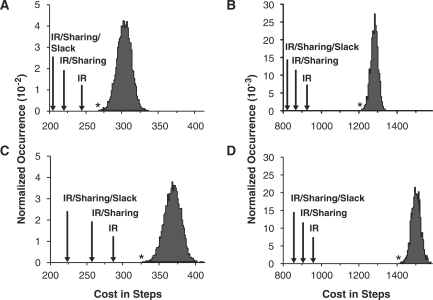
For real-world datasets an exhaustive search of all possible solutions is not feasible, so we only compare our multiple-goal-part algorithm to the random search approach. Figure 7A–B shows the results for the ‘phagemid’ and ‘iGEM-2008’ datasets. We show the solution chosen by three variants of our multiple-goal-part algorithm. The ‘IR’ solution corresponds to our algorithm with iterative refinement but without using sharing or slack factors. The ‘IR/sharing’ solution correspond to our algorithm with iterative refinement augmented with sharing factors, and the ‘IR/sharing/slack’ solution corresponds to our complete multiple-goal-part algorithm. Notice that the random search approach produces roughly Gaussian distributions, meaning that very few solutions are near the lower-cost tail. Even with a large random search, the best solution is still significantly worse than our ‘IR’ solution. Adding the sharing and slack factors further reduces the required number of steps. Ultimately, our algorithm is able to reduce the number of assembly steps by 23% for the ‘phagemid’ dataset and by 37% for the ‘iGEM-2008’ dataset. This reduction in assembly steps directly corresponds to reduced assembly work in terms of engineer effort and reagents. The running time for our algorithm on the ‘phagemid’ and ‘iGEM-2008’ datasets was on the order of minutes while the random search approach took many hours on standard general-purpose workstation. Replacing our dynamic programming single-goal-part algorithm with one based on the naive exponential approach increased the running time of our multiple-goal-part algorithm by several orders-of-magnitude.
Figure 7.
Comparison of solutions found for the phagemid and iGEM datasets. These figures show the assembly cost (in terms of assembly steps) for the ‘phagemid’ (A, C) and ‘iGEM-2008’ (B, D) datasets for 3A assembly (A–B) and 2ab assembly (C–D). The arrows indicate the number of steps for the solution found with just iterative refinement (‘IR’), for iterative refinement with sharing factors (‘IR/sharing’), and our complete multi-goal-part algorithm (‘IR/sharing/slack’). The asterisk indicates the lowest cost solution found by the random search.
Results for extensions to support 2ab assembly method
The Materials and methods section introduced a coloring post-processing step for determining the correct antibiotic markers in the 2ab assembly method. This post-processing step can be applied to solutions generated by both the random search approach and our multiple-goal-part algorithm. Unfortunately, coloring conflicts can cause an increase in the number of steps. Figure 7C and D illustrates the impact of these conflicts for the two real-world datasets. As expected, the total number of steps is increased due to coloring conflicts, but our algorithms still find lower cost solutions than the random search approach. For the 2ab assembly method, our algorithm is able to reduce the number of assembly steps by 31% for the ‘phagemid’ dataset and by 39% for the ‘iGEM-2008’ dataset.
DISCUSSION
This article has presented algorithms for automated binary DNA assembly that are particularly relevant when constructing large goal-part sets with a high degree of inter-GPS. Our basic algorithm uses dynamic programming to efficiently find optimal assembly graphs for a single goal part. We augment this basic algorithm for use in our multiple-goal-part algorithm with three techniques to bias the search towards graphs with many shared intermediate parts: ‘sharing factors’ capture global sharing information when performing local optimization, ‘slack factors’ enable locally suboptimal solutions that can benefit the global result and ‘iterative refinement’ ensures that common intermediate parts are assembled in the same way. We have used two real-world datasets to demonstrate that our algorithms efficiently generate cost-saving assembly graphs.
Analogous problems have been explored for assembly plan optimization when manufacturing goods (21). However, such approaches have to deal with scheduling multiple different operations in order to build one product. Manufacturing operations also have ordering constraints, e.g. for the construction of a pen ink, cartridges must be placed inside the bodies before end caps are attached. In contrast, for binary assembly of DNA sequences, a single ligation operation is repeated in order to build multiple goal parts. In general binary assembly there are no ordering constraints. It does not matter which junctions are made first, but planning becomes non-trivial when an optimal assembly graph for multiple goal parts is desired.
There are many directions for future work. Other heuristic algorithms are possible for searching how to combine the assembly subgraphs for each goal part. For example, one might employ a genetic algorithm that keeps highly shared subgraphs while exploring other assembly subgraphs for remaining goal parts. Another interesting direction for future work is to factor assembly risk into our assembly algorithm. In our experience, assembly failure arises from two sources: randomly distributed failure and part-specific failure. Randomly distributed failure affects all parts equally; to combat this type of error, the assembly would be duplicated in a post-processing step to increase the chances of having at least one correct assembly for each part. Duplicating the assembly graph is a necessary consequence of the error rates that still pervade sequence construction and would be obviated only if the efficiency of the assembly step is improved drastically. Part-specific failure is due to cloning difficulties, known or unknown, associated with a certain sequence. For known problematic sequences, the cost function can be used to make the assembly algorithms avoid combinations of parts, but this may impact the optimal substructure property. For example, enzymes that synthesize toxic intermediates should not be completed before downstream, toxicity-relieving components. Errors due to unknown problematic sequences can only be avoided by speculative pursuit of different assembly trajectories and a subsequent loss of savings.
Design automation in synthetic biology is not only going to depend on the development of algorithms such as those outlined here, but also on the development of efficient design flows that make use of these algorithms. If these design flows are not robust, then any performance advantages gained by the algorithmic work are lost. It can be argued that the design flows are as critical to actual users as the algorithms themselves. Key to making this process robust is that design flows be modularized around specific design activities. Future work should focus on rules that generate appropriate goal parts to test for a biological function and recapturing assembly failure information to identify cloning issues with specific DNA sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation Graduate Research Fellowships (to T.H.H. and J.T.K.); Department of Defense National Defense Science and Engineering Graduate Fellowship (to T.H.H.); Amgen Scholars Program (to W.D.) and the Center for Hybrid and Embedded Software Systems (CHESS) at University of California, Berkeley. CHESS receives support from the National Science Foundation [NSF awards #CCR-0225610 (ITR), #0720882 (CSR-EHS: PRET), #0647591 (CSR-SGER) and #0720841 (CSR-CPS)]; US Army Research Office (ARO #W911NF-07-2-0019); US Air Force Office of Scientific Research (MURI #FA9550-06-0312 and AF-TRUST #FA9550-06-1-0244); Air Force Research Lab (AFRL), the State of California Micro Program and the following companies: Agilent, Bosch, Lockheed Martin, National Instruments, Thales and Toyota. Funding for open access charge: The Synthetic Biology Engineering Research Center (SynBERC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Ann Van Devender for her early contributions to the development of the Clotho algorithm manager and associated support for importing algorithms into Clotho. Nathan Hillson provided feedback on the paper. Nina Revko, Thien Nguyen and Bing Xia provided software tools which made the testing of the algorithms presented here possible. Finally, the Synthetic Biology Engineering Research Center (SynBERC) was vital in making the interdisciplinary nature of this work possible.
REFERENCES
- 1.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 2.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 3.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batt G, Yordanov B, Weiss R, Belta C. Robustness analysis and tuning of synthetic gene networks. Bioinformatics. 2007;23:2415–2422. doi: 10.1093/bioinformatics/btm362. [DOI] [PubMed] [Google Scholar]
- 6.Carrera J, Rodrigo G, Jaramillo A. Towards the automated engineering of a synthetic genome. Mol. Biosyst. 2009;5:733–743. doi: 10.1039/b904400k. [DOI] [PubMed] [Google Scholar]
- 7.Lun DS, Rockwell G, Guido NJ, Baym M, Kelner JA, Berger B, Galagan JE, Church GM. Large-scale identification of genetic design strategies using local search. Mol. Syst. Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brissette R, Goldstein NI. The use of phage display peptide libraries for basic and translational research. Methods Mol. Biol. 2007;383:203–213. doi: 10.1007/978-1-59745-335-6_13. [DOI] [PubMed] [Google Scholar]
- 9.Collett JR, Cho EJ, Ellington AD. Production and processing of aptamer microarrays. Methods. 2005;37:4–15. doi: 10.1016/j.ymeth.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S, et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat. Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 11.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 12.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 13.Shao Z, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc. Natl Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Layered Assembly. 2008. [(13 March 2010, date last accessed)]. http://2008.igem.org/Team:UC_Berkeley/LayeredAssembly. [Google Scholar]
- 18.Linshiz G, Yehezkel TB, Kaplan S, Gronau I, Ravid S, Adar R, Shapiro E. Recursive construction of perfect DNA molecules from imperfect oligonucleotides. Mol. Syst. Biol. 2008;4:191. doi: 10.1038/msb.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saftalov L, Smith PA, Friedman AM, Bailey-Kellogg C. Site-directed combinatorial construction of chimaeric genes: general method for optimizing assembly of gene fragments. Proteins. 2006;64:629–642. doi: 10.1002/prot.20984. [DOI] [PubMed] [Google Scholar]
- 20.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. 3rd edn. Cambridge, MA: The MIT Press; 2009. [Google Scholar]
- 21.Densmore D, Devender AV, Johnson M, Sritanyaratana N. Proceedings of The Fifth Richard Tapia Celebration of Diversity in Computing Conference: Intellect, Initiatives, Insight, and Innovations. Portland, Oregon, New York, NY: ACM; 2009. A platform-based design environment for synthetic biological systems; pp. 24–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.