Abstract
The cohesin protein complex holds sister chromatids in dividing cells together and is essential for chromosome segregation. Recently, cohesin has been implicated in mediating transcriptional insulation, via its interactions with CTCF. Here, we show in different cell types that cohesin functionally behaves as a tissue-specific transcriptional regulator, independent of CTCF binding. By performing matched genome-wide binding assays (ChIP-seq) in human breast cancer cells (MCF-7), we discovered thousands of genomic sites that share cohesin and estrogen receptor alpha (ER) yet lack CTCF binding. By use of human hepatocellular carcinoma cells (HepG2), we found that liver-specific transcription factors colocalize with cohesin independently of CTCF at liver-specific targets that are distinct from those found in breast cancer cells. Furthermore, estrogen-regulated genes are preferentially bound by both ER and cohesin, and functionally, the silencing of cohesin caused aberrant re-entry of breast cancer cells into cell cycle after hormone treatment. We combined chromosomal interaction data in MCF-7 cells with our cohesin binding data to show that cohesin is highly enriched at ER-bound regions that capture inter-chromosomal loop anchors. Together, our data show that cohesin cobinds across the genome with transcription factors independently of CTCF, plays a functional role in estrogen-regulated transcription, and may help to mediate tissue-specific transcriptional responses via long-range chromosomal interactions.
First characterized in Saccharomyces cerevisiae and Xenopus laevis, the cohesin protein complex holds sister chromatids in dividing cells together and is essential for chromosome segregation from its establishment in S phase through metaphase (Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998). Cohesin consists of four evolutionarily conserved core subunits: SMC1A, SMC3, RAD21, and STAG1 (or STAG2). Biochemical and electron microscopy studies have indicated that the complex functions by physically linking sister chromatids in a ring structure (Gruber et al. 2003; Haering et al. 2008). A number of ancillary proteins associate with cohesin, including the NIPBL/KIAA0892 (in budding yeast, Scc2/Scc4) adherin complex, which mediates cohesin's loading onto chromatin (Ciosk et al. 2000).
Vertebrate cohesin is continuously associated with chromosomes, except for a short period of time from anaphase to early telophase (Sumara et al. 2000), and is also expressed in post-mitotic cells such as neurons, where chromatid cohesion is not required. This suggests functional roles beyond sister chromatid assembly (Zhang et al. 2007; Wendt et al. 2008). In support of this hypothesis, the complex has been shown to be involved in DNA damage repair (Watrin and Peters 2006) and transcriptional termination (Gullerova and Proudfoot 2008). Results from model organisms and the identification of cohesin mutations associated with human disease suggest cohesin may play a more complex role in regulating gene expression (Krantz et al. 2004; Tonkin et al. 2004; Strachan 2005; Vega et al. 2005; Musio et al. 2006; Horsfield et al. 2007). In Drosophila, the cohesin loading factor Nipped-B partially regulates the expression of the cut gene (Rollins et al. 2004; Dorsett et al. 2005). In humans, mutations in the Nipped-B ortholog NIPBL or the cohesin subunits SMC1A and SMC3 lead to a constellation of severe developmental defects known as Cornelia de Lange syndrome (CdLS, OMIM 122470), which appear to be independent of cohesin's canonical role in chromatid cohesion (Krantz et al. 2004; Tonkin et al. 2004; Strachan 2005; Vega et al. 2005; Musio et al. 2006). Indeed, it has been shown that STAG1 and STAG2 may act as transcriptional coactivators in human cells (Lara-Pezzi et al. 2004). How alterations in cohesin function give rise to pervasive developmental abnormalities and gene expression in the absence of alterations in chromatid cohesion has yet to be elucidated (Kaur et al. 2005; Vrouwe et al. 2007).
Several reports have shown that cohesin localizes to CTCF binding sites in human and mouse cell lines, and that the complex is essential for the insulator function of CTCF (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008). A mechanistic insight into cohesin's role at these sites was given by a recent study proposing that the complex forms the topological basis for cell-type-specific intrachromosomal interactions at the developmentally regulated cytokine IFNG locus (Hadjur et al. 2009). Prior studies have reported the existence of limited cohesin binding that is independent of CTCF, yet these cohesin interactions remain unexplored (Rubio et al. 2008; Wendt et al. 2008).
Here, we report that cohesin regulates global gene expression and modulates cell-cycle re-entry via extensive cobinding of the human genome with tissue-specific transcription factors in multiple cell types, independently of CTCF.
Results
Cohesin occupies thousands of regions in breast cancer cells not bound by CTCF
We first confirmed that cohesin and CTCF cobind across the genome of estrogen-dependent MCF-7 breast cancer cells, thereby acting together to insulate transcriptionally distinct regions, as previous studies have reported in numerous cell lines (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008). We performed chromatin immunoprecipitation experiments followed by high-throughput sequencing (ChIP-seq) against CTCF and two cohesin subunits, STAG1 and RAD21. We identified TF-bound regions using SWEmbl, a dynamic programming algorithm (S Wilder, D Thybert, D Sobral, B Ballester, P Flicek, in prep.), and our results were robust to different peak-calling algorithms (Methods) (Supplemental Fig. S2).
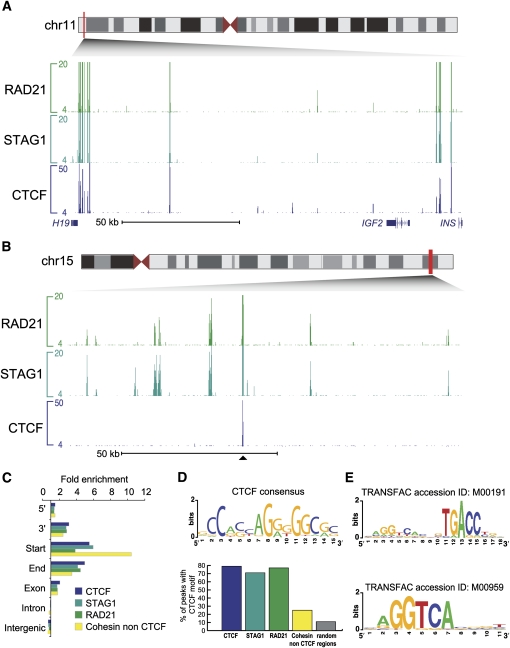
Separate analysis of STAG1 and RAD21 afforded largely indistinguishable results. This observation was expected, as RAD21 binding events were an almost perfect subset of STAG1 binding events (Supplemental Fig. S1A). The apparent presence of RAD21 negative STAG1 binding events appears to be the result of modestly lower enrichment by the RAD21 antibody, and rank order analysis indicates that the two proteins are found at almost identical regions across the genome (Supplemental Fig. S1B). We therefore merged the binding of STAG1 and RAD21 and refer to their collective binding as cohesin-bound. As expected, we found that cohesin cobinds with CTCF at 80% (39,444) of the CTCF binding events (49,243) (Fig. 1A).
Figure 1.
Identification of CTCF-independent cohesin binding events in the human genome. (A) Genomic binding of the cohesin subunits RAD21 and STAG1 as well as CTCF shows colocalization at the H19/IGF2 locus in MCF-7 cells. (B) Illustrative CTCF-independent binding events are shown at a 120-kb region on chromosome 15 in MCF-7 cells; this genomic track is centered around a single shared cohesin-CTCF site. (C) Distribution of cohesin-, CTCF-, and cohesin–non-CTCF-binding (CNC) events in the human genome: 5′, 3′, start, end, exon, intron, and intergenic regions of Ensembl genes are shown. The number of sites was normalized and displayed as fold-enrichment relative to genome background. (D) CTCF consensus DNA motif de novo derived from the MCF-7 CTCF binding events. The occurrence of the CTCF consensus within the CTCF, STAG1, RAD21, CNC events and random genomic regions are presented. (E) Estrogen response elements enriched within the CNC events.
Previous studies have reported that only a small minority of cohesin binding events seem to be independent of CTCF (Parelho et al. 2008; Rubio et al. 2008; Wendt et al. 2008), yet we observed 16,509 cohesin binding events that do not overlap with CTCF (cohesin-non-CTCF events [CNCs]). Some of these CNCs showed notably strong binding to chromatin (Fig. 1B) as well as association with promoter regions (Fig. 1C). This discovery was likely facilitated by the higher sensitivity obtained by using high-throughput sequencing to analyze ChIP enrichment, as opposed to previously employed microarrays (Robertson et al. 2007; Schmidt et al. 2008).
Regions bound by cohesin but not CTCF are enriched for motifs of tissue-specific transcription factors
We asked whether any underlying sequence features corresponded with cohesin binding events, based on the presence or absence of CTCF. We easily identified the known CTCF consensus sequence when all CTCF bound regions were subject to de novo motif discovery, and this motif was present in the vast majority of CTCF bound regions (Fig. 1D; Kim et al. 2007; Chen et al. 2008; Cuddapah et al. 2008). Most cohesin-bound regions showed the presence of the CTCF motif as well, as expected based on the substantial cobinding we and others have observed between CTCF and cohesin. We found that 79% of the CTCF binding events harbored the CTCF motif compared with 71% (STAG1) and 77% (RAD21) of all cohesin binding events.
In contrast, when analyzed as an independent set of bound regions, the sites containing cohesin yet lacking CTCF showed low enrichment for the CTCF motif, similar to the background genome (Fig. 1D). This finding suggests these regions have little if any low-level CTCF binding, since all prior studies indicate that genomic occupancy of CTCF mainly occurs at its consensus motif.
Because cohesin lacks a known DNA binding domain yet binds thousands of CTCF independent regions, we sought to identify sequence motifs enriched in CNCs that could indicate other possible interacting TFs. We found that the estrogen response element (ERE) was present in CNC regions (P < 0.0001) (Fig. 1E). The ERE is the consensus binding motif for estrogen receptor alpha (ER), a master regulator of transcription in breast cancer cells with numerous known target genes including trefoil factor 1 (TFF1) (Brown et al. 1984), the estrogen receptor alpha gene itself (ESR1) (Carroll et al. 2006), and NRIP1 (Carroll et al. 2005). The enrichment of this sequence motif in CNCs suggested that cohesin might co-occupy sites in the human genome bound by ER.
Cohesin–TF cobinding events lacking CTCF are specific to each cell type
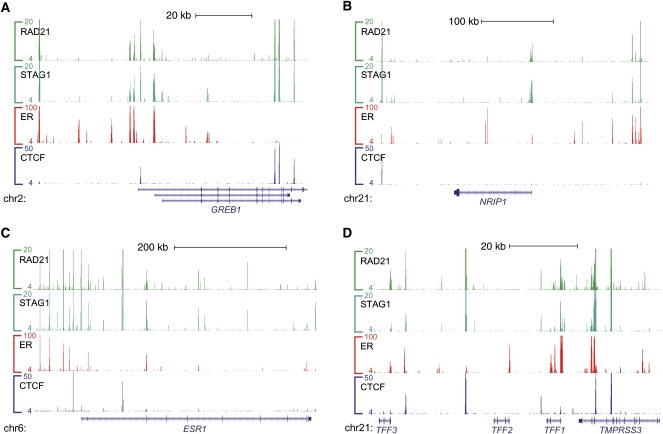
We tested the hypothesis that cohesin and ER colocalize by experimentally identifying the regions bound by ER using ChIP-seq. We found the expected binding of cohesin at sites of CTCF binding, yet cohesin binding also frequently occurred at many of the same locations as ER in MCF-7 cells (Figs. 2A–D, 3). At 70 kb around the GREB1 locus, for example, cohesin was clearly present in all CTCF bound regions and approximately half of those bound by ER (Fig. 2A).
Figure 2.
Estrogen receptor, cohesin, and CTCF binding at known estrogen receptor target genes. The genomic binding profiles for cohesin (RAD21 and STAG1), estrogen receptor alpha (ER), and CTCF at four known ER target genes demonstrate extensive co-occupancy of ER and cohesin in the absence of CTCF.
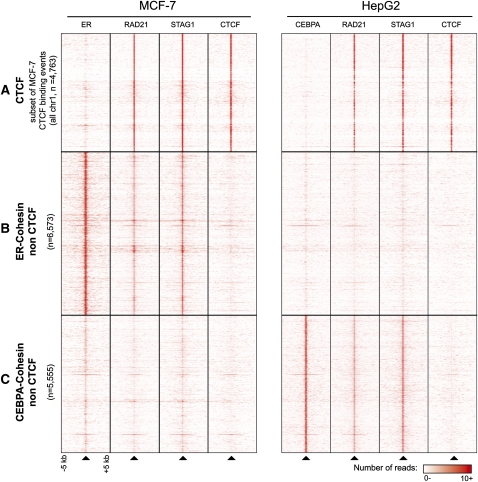
Figure 3.
Cohesin binding with CTCF is cell-type invariant, whereas cohesin binding with tissue-specific TFs is cell-type specific. (A) The CTCF binding events on chromosome 1 in MCF-7 cells strongly correspond with both CTCF binding in HepG2 cells and cohesin in both tissues but are not generally shared with tissue-specific master regulators. (B) In contrast, a subset of the ER binding events bound by cohesin in MCF-7 cells independently of CTCF are shown. Showing the tissue specificity of these binding events, in HepG2 cells, these regions are rarely found to be bound by cohesin. (C) Similarly, a subset of the CEBPA binding events that are bound by cohesin in HepG2 cells independently of CTCF do not show cohesin binding in MCF-7 cells.
As expected, CTCF bound regions show consistently strong enrichment of cohesin (Fig. 3A); remarkably, some regions bound in MCF-7 cells by cohesin and ER in the absence of CTCF (ER-CNCs) showed high cohesin enrichment (Figs. 2, 3B). In total, we found 6573 regions that were bound by both ER and cohesin without enrichment of CTCF (P-value = 0.0003, Mann-Whitney U-test) (Fig. 3B). Approximately half that many (3367) ER binding events colocalized with CTCF.
The high overlap of cohesin and ER in breast cancer cells suggested that cohesin could be an integral component of transcriptional regulatory networks in a tissue-specific manner. To explore this cell-type specificity, we identified the genome-wide binding of cohesin (RAD21 and STAG1) and CTCF in HepG2 hepatocellular carcinoma cells. As expected, we found that most (76%) CTCF binding events in HepG2 cells were also enriched for cohesin. Because CTCF binding has been shown to be relatively independent of cell type (Cuddapah et al. 2008; Heintzman et al. 2009), we were not surprised to find that the majority of CTCF binding events were common to both breast cancer and liver cell types (Fig. 3A). Thus, cohesin–CTCF bound regions appear to be largely independent of the cell type.
Next, we confirmed that the CTCF-independent sites in MCF-7 cells are largely absent from HepG2 liver cancer cells. We found that only about 400 of the 6573 ER binding events enriched by cohesin but not CTCF in breast cancer cells showed cohesin binding in HepG2 cells (Fig. 3B).
We then tested whether two well-characterized liver-specific TFs, HNF4A and CEBPA, cobound with cohesin in HepG2 cells. Similar to MCF-7 cells, we identified 4382 and 5555 regions sharing cohesin and either HNFA4 or CEBPA (respectively) but no CTCF. These regions rarely showed cohesin or ER enrichment in breast cancer cells (Fig. 3C). Thus, cohesin associates with cell-type-specific master regulators independently of CTCF in both MCF-7 and HepG2 cells, in regions that generally do not show cohesin binding in both cell types.
MCF-7-specific CNCs increase upon estrogen induction of ER binding
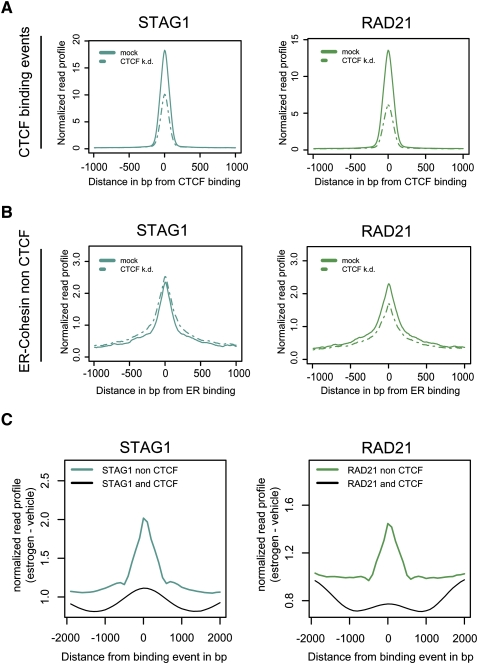
To confirm that cohesin colocalization with transcription factors is entirely independent of CTCF, we reduced the concentration of CTCF using CTCF RNAi in breast cancer cells and performed ChIP-seq to confirm that cohesin remained associated with tissue-specific transcriptional regulators. Western blots confirmed efficient RNAi knockdown (Supplemental Fig. S3). As reported previously (Parelho et al. 2008), cohesin is recruited to CTCF binding events in a CTCF-dependent manner (Fig. 4A). Conversely, the recruitment of cohesin to ER-CNC binding events was largely unaffected by the CTCF RNAi. These results show that cohesin recruitment to CNCs is CTCF-independent (Fig. 4B; Supplemental Fig. S4).
Figure 4.
Cohesin binding can be independent of CTCF. (A) Cohesin (STAG1 and RAD21) enrichment at cohesin-CTCF sites is reduced upon CTCF removal by RNAi knockdown (CTCF k.d.). (B) Cohesin enrichment at ER-CNCs is largely unaffected by CTCF knockdown. (C) Cohesin binding increases upon estrogen treatment at sites not shared with CTCF (Kolmogorov-Smirnov, P-value < 10−9 [STAG1]; P-value < 10−15 [RAD21]).
To test whether ER was required for cohesin recruitment to CNCs in MCF-7 cells, we carried out ChIP-seq experiments of cohesin in hormone-deprived MCF-7 cells after 45 min of vehicle (ethanol) treatment and compared the cohesin binding profile to our data after 45 min of 17β-estradiol (estrogen, E2) treatment. It has previously been shown that ER binding is significantly reduced in vehicle compared with estrogen treatment (Carroll et al. 2005). Indeed, under vehicle conditions we find ER binding heavily reduced compared with E2 treatment. We observed that cohesin binding significantly increases at CNCs after estrogen treatment compared with the binding events shared between CTCF and cohesin, thus demonstrating that cohesin binding at CNCs must be in part dependent on ER binding in MCF-7 cells (Fig. 4C; Supplemental Fig. S5).
Taken together, our data indicate that cohesin binding events lacking CTCF appear to be highly specific for each cell type, are independent of CTCF presence, and associate with a substantial subset of binding events for tissue-specific TFs. Given that cohesin is known to be recruited to actively transcripted coding regions in Drosophila (Misulovin et al. 2008), we then asked whether cohesin preferentially binds functional transcriptional targets of tissue-specific master regulators.
Genes cobound by cohesin and ER are preferentially regulated by estrogen
The ER-mediated transcriptional response of MCF-7 cells to estrogen treatment is a well-studied system for dissecting the functional roles of transcriptional complexes (Prall et al. 1997). If cohesin were required for ER-dependent transcription, then estrogen transcriptional targets would be expected to show preferential binding by cohesin.
To determine the functional significance of the colocalization of ER and cohesin, we established whether the distribution of cohesin and ER binding sites varied around estrogen-regulated and nonregulated genes (Fig. 5). We compared the fraction of E2-regulated versus nonregulated genes in breast cancer cells with at least one binding site specific to ER or shared by cohesin and ER within 20 kb of their transcriptional start sites. This distance was chosen because a disproportionate fraction of estrogen-regulated genes show ER binding within these regions (Lupien et al. 2008), despite the fact that many ER functional enhancers are located at great distances from regulated genes (Carroll et al. 2005).
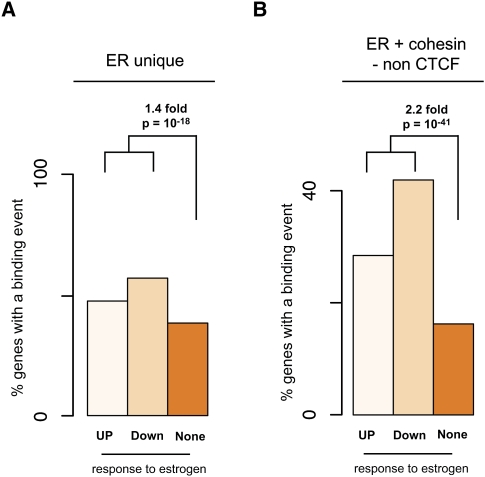
Figure 5.
Correlation of estrogen receptor and cohesin binding events with estrogen gene regulation. (A) Estrogen binding alone enriches for functional targets. The genes that have an ER binding event are 1.4 times more likely to have changed their gene expression than expected by random chance (Fisher's exact test, P-value = 10−18). (B) Genes bound by both ER and cohesin (but not CTCF) are 2.2 times more likely to have altered gene expression upon estrogen treatment than by random chance, further enriching functional targets (Fisher's exact test, P-value = 10−41).
Estrogen binding alone identified regulated genes 1.4-fold more often than did nonregulated genes. In contrast, the ER-bound genes showing colocalization of ER and cohesin (in the absence of CTCF) were more than twice as likely to be E2-regulated (P-value < 0.0001). Importantly, this effect was similar for both up- and down-regulated genes as identical calculations on the independent sets afforded similar results to the combined sets. These results demonstrate that in breast cancer cells, the genes bound by ER and cohesin together, compared with ER binding only, are more likely to be regulated in response to estrogen treatment.
This observation indicates that cohesin marks functionally active target genes, and further suggests that it may play a role in mediating estrogen-dependent transcriptional responses.
Cohesin is required for estrogen-mediated re-entry into the cell cycle
Estrogen-driven proliferation in breast cancer cells proceeds via activation of the ER-regulated gene expression program that mediates re-entry into cell cycle. The functional effect of RNAi-mediated removal of candidate regulators on estrogen-induced proliferation can be used to determine whether cofactors are required for ER-driven transcription (Prall et al. 1997). Under normal circumstances, estrogen-deprived MCF-7 cells arrest in G0/G1 phase, and their re-entry into cell cycle upon estrogen treatment can be quantitated (Fig. 6, mock RNAi; Prall et al. 1997). In such an experiment, a small number of cycling cells are still observed in hormone-depleted cultures, which has been attributed to the presence of the residual estrogen, as well as small amounts of endogenously produced E2.
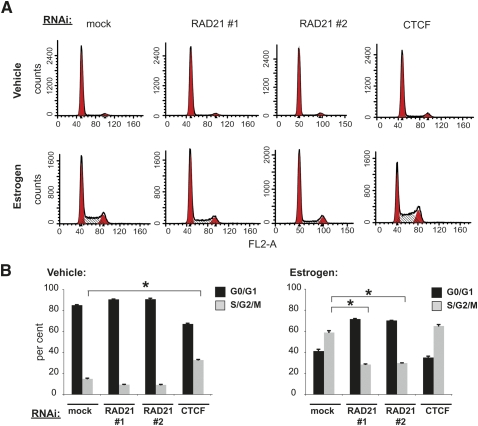
Figure 6.
Cohesin is functionally required for correct estrogen-induced cell cycle progression. (A) Following RNAi-mediated knockdown of RAD21 and CTCF, cell cycle distributions after vehicle or estrogen treatment were assessed by propidium iodide staining followed by flow cytometry. (B) The percentage of cells in G0/G1 (black bars) and S/G2/M (gray bars) phases were quantitated after 24 h of vehicle or estrogen treatment (mean of n = 2; error bars, ±SD; *, t-test P-value ≤ 0.001). In vehicle-treated cells, suppression of CTCF caused a doubling of the cells in S/G2/M phases, compared with mock treatment. After 24 h of estrogen treatment, the removal of the cohesin subunit RAD21 caused a decrease in the number of cells that have transitioned to S/G2/M, indicating reduced cell-cycle progression. Removal of CTCF again enhanced entry into S/G2/M, but not with statistical significance.
We used this approach to assess whether cohesin is required for ER-mediated re-entry into the cell cycle by silencing the cohesin subunit RAD21 using two different siRNA molecules, treating with estrogen, and then using flow cytometry to identify the fraction of cells that have exited G0/G1. Western blotting confirmed effective RNAi silencing (Supplemental Fig. S3). We chose to evaluate the fraction of MCF-7 cells re-entering the cell cycle after only 24 h of hormone treatment, as temporally longer experiments may result in biases due to cohesin's known role in chromosome cohesion, which is established in S phase (Ghiselli 2006).
Cohesin-depleted cells showed a strong reduction in estrogen-induced cell cycle re-entry compared with mock-treated cells (P-value < 0.001, t-test) (Fig. 6B). This result indicates that cohesin is functionally required for efficient estrogen-dependent G0/G1–S phase transition in breast cancer cells.
To determine whether the observed effects are independent of cohesin's role in the CTCF insulator complex, we suppressed the expression of CTCF using RNAi and characterized the effect on the G0/G1–S transition. In principle, if the observed inhibition were caused by a genome-wide decrease in cohesin–CTCF binding, then removal of CTCF should produce a similar decrease in cell cycle re-entry, as seen when cohesin is suppressed. Notably, however, removal of CTCF had the opposite effect on cell cycle progression. The number of cells in S/G2/M phase increased both before and after estrogen stimulation when CTCF was depleted (Fig. 6B).
Thus, cohesin appears to be functionally required for estrogen-induced cell cycle re-entry, independent of cohesin's role in CTCF insulator pathways.
Cohesin is enriched at ER binding events involved in chromatin interactions
Cohesin has been shown to be required for chromosomal interactions mediated by CTCF (Hadjur et al. 2009), and ER itself is known to mediate chromatin interactions in breast cancer cells (Carroll et al. 2005; Pan et al. 2008). Long-range chromosomal interactions have recently been reported for ER genome-wide in breast cancer cells (Fig. 7A; Fullwood et al. 2009) using chromatin interaction ligations followed by paired-end sequencing. From the correspondence between FOXA1 binding and ER long-range chromosomal interactions, it was suggested that FOXA1 participates in tethering chromatin interactions near key ER target genes.
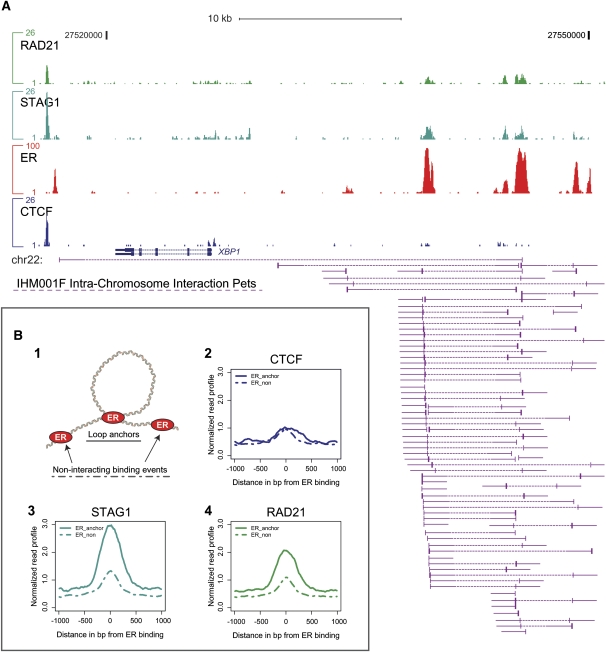
Figure 7.
Cohesin preferentially associates with estrogen receptor–mediated through-space chromatin interactions. (A) Genomic binding of cohesin (RAD21 and STAG1), estrogen receptor alpha, and CTCF are shown as genomic enrichment tracks above the genome annotation of the known ER target gene XBP1, and ChIA-PET interaction data (Fullwood et al. 2009) are shown as purple sequencing tracks beneath the genome annotation. (B, 1) ER binding events were divided into loop anchors and noninteracting binding events. Association of CTCF (2), STAG1 (3), and RAD21 (4) with ER-interacting binding events (loop anchors, solid lines) and ER noninteracting binding events (dashed lines) are shown.
To determine whether cohesin may play a similar role in tethering chromatin interactions, we examined CTCF and cohesin occupancy near ER binding events involved in chromatin loops (loop anchors) as well as ER noninteracting binding events. Cohesin was significantly more enriched at ER binding events involved in chromatin interactions than ER binding events not interacting in long-range looping (Fig. 7B). In contrast, CTCF binding was not observed to vary based on whether ER binding events were participating in long-range looping events.
Although correlative, this result suggests that cohesin, but not CTCF, binding predicts long-range interactions at ER binding events, and that cohesin can tether chromatin interactions independently of CTCF.
Discussion
Our understanding of cohesin's molecular function has expanded beyond its central role in chromatid cohesion to novel functions such as mediating long-range chromosomal interactions, transcriptional insulation, and transcriptional coregulation in model organisms. Previous reports have indicated that the large majority of cohesin binding occurred across the genome with CTCF (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008), suggesting that cohesin binding may also be cell-type independent. Our data confirm that cohesin–CTCF complexes are indeed largely cell-type invariant, yet also reveal a novel set of binding events that are highly tissue-specific. We found that cohesin colocalizes with ER in breast cancer cells; similarly, cohesin cobinds with HNF4A and CEBPA in liver cells. The similar cohesin behavior in two different cell types, where it colocalizes with the respective master regulators, suggests that cohesin might contribute to tissue-specific transcription. Indeed, we showed that tissue-specific cohesin binding events are associated with ER-regulated genes independently of CTCF, and that cohesin is required for the correct ER-mediated cell cycle re-entry.
Recently, techniques have been reported that reveal how whole mammalian genomes are structured within the nucleus in complex three-dimensional patterns (Fullwood et al. 2009; Lieberman-Aiden et al. 2009). ER-regulated enhancers have been shown to bridge to proximal promoter regions at the TFF1 and NRIP1 loci in breast cancer cells (Carroll et al. 2005; Pan et al. 2008), and indeed, recent genome-wide work has shown that ER binding directs three-dimensional chromatin structure (Fullwood et al. 2009). Our analyses suggest that cohesin can help stabilize higher-order complexes, including direct looping of distant regulatory regions. Because cohesin has no known DNA binding domains, it could mediate enhancer-promoter looping initially established and maintained by the binding of tissue-specific transcriptional master regulators like ER and CEBPA.
CdLS is caused by mutations in cohesin loading factors as well as proteins of the cohesin complex and characterized by severe developmental defects. The relative contributions of sister chromatid cohesion and CTCF-dependent cohesin binding to CdLS have been actively debated. Our finding that cohesin has CTCF-independent functional roles, mediated by colocalization with tissue-specific master regulators, suggests a third possible contributor to the observed developmental defects.
Methods
Cell culture
MCF-7 human breast cancer cells were grown as previously described (Neve et al. 2006). Unless otherwise stated, MCF-7 cells were grown in phenol red-free DMEM supplemented with 5% charcoal-dextran-treated serum for at least 3 d, and during all experiments 17β-estradiol (estrogen, E2) was added at a final concentration of 100 nM for 45 min (ChIP) or 24 h (cell cycle analysis) or not (RNAi–ChIP-seq, see under siRNA). HepG2 cells were grown in DMEM supplemented with 10% FBS.
siRNA
Cells were grown in phenol red-free DMEM supplemented with 5% charcoal-dextran-treated serum for at least 3 d (cell cycle analysis) or in DMEM supplemented with 10% FBS (RNAi–ChIP-seq). Cells were transfected with siRNA for 48 h using Lipofectamine2000 (Invitrogen). AllStars Negative Control siRNA (Qiagen) was used as negative (mock) controls. CTCF RNAi (Invitrogen Stealth, sense: GCGCUCUAAGAAAGAAGAUUCCUCU; antisense: AGAGGAAUCUUCUUUCUUAGAGCGC); RAD21#1 (Invitrogen Stealth, sense: CAGCUUGAAUCAGAGUAGAGUGGAA; antisense: UUCCACUCUACUCUGAUUCAAGCUG); RAD21#2 (ON-TARGETplus).
Western blot analysis
Nuclear extracts were harvested. Antibodies were the same as for ChIP and beta-actin (Abcam, ab6276).
Cell cycle analysis
Cells were plated at equal confluence, deprived of hormones, and transfected using siRNA as described above. Total cells were harvested and stained with propidium iodide for flow cytometry analysis.
ChIP sequencing
ChIP experiments were performed with well-characterized antibodies against CTCF (Millipore, 07-729), STAG1 (Abcam, ab4457), RAD21 (Abcam, ab992), ER (Santa Cruz, sc-543), CEBPA (Santa Cruz, sc-9314), and HNF4A (Aviva Systems Biology, ARP31946) (Carroll et al. 2006; Kim et al. 2007; Lefterova et al. 2008; Wendt et al. 2008), as recently described (Schmidt et al. 2009). Briefly, the immunoprecipitated material was end-repaired, A-tailed, ligated to the sequencing adapters, amplified by 18 cycles of PCR, and size selected (200–300 bp) followed by single end sequencing on an Illumina Genome Analyzer according to the manufacturer's recommendations.
Heatmaps of ChIP-seq data
To generate the heatmaps of the raw ChIP-seq data, the appropriate binding regions were used as targets to center each window. Each window was divided into 100 bins of 100 bp in size. An enrichment value was assigned to each bin by counting the number of sequencing reads in that bin and subtracting the number of reads in the same bin of an input library. Each data set was normalized to 10 million reads. Data were visualized with Treeview (Saldanha 2004).
Read mapping
All sequencing reads were aligned using MAQ (Li et al. 2008) with default parameters in a replicate specific fashion to the human NCBI36 genome assembly. HepG2 reads were mapped against a male genome, while a female genome was used in MCF-7 cells.
Peak calling
We merged the data from the MAQ aligned replicates, filtering out nonunique reads as well as reads with a MAQ quality score below 10. Positive binding events were discovered using a dynamic programming algorithm (SWEmbl), which incorporates a scoring function that is increased by alignments of ChIP reads and decreased by alignments of input reads by a general decay function after read observation (S Wilder, D Thybert, D Sobral, B Ballester, P Flicek, in prep.). We explored several SWEmbl parameters (−R 0.01, −R 0.007, and −R 0.005), obtaining different peak numbers, but with peaks showing the same behavior in terms of general characteristics and overlap proportions. We noted that −R 0.005 outputs much shorter regions, so that two such peaks often correspond to one −R 0.01 determined peak. We report our results obtained with −R 0.01 for all factors, except for the cohesin non-CTCF sites, which are described below.
Factor overlaps and cohesin–non-CTCF subsets
We define overlaps according to a 1-bp proximity criterion; nonoverlaps, as the remaining sites. However, this does not ensure that some residual binding (reads just under the peak-calling threshold) is not still present in the “nonoverlap” sets. For obtaining a high-quality cohesin–non-CTCF binding site set, we filtered the cohesin, respectively cohesin overlapping ER, CEBPA or HNF4A peaks (called with SWEmbl 0.005, in order to have shorter regions, like described above) for CTCF reads. Our final sets, which we refer to as cohesin–non-CTCF, respectively, cohesin–ER (or CEBPA/HNF4A)–non-CTCF, fulfill the following criterion: log(normalized_ctcf/normalized_input) < 1.2. The significance of the difference between ER–cohesin–non-CTCF sites and cohesin–CTCF sites was assessed by comparing the CTCF binding profile 2000 bp around cohesin binding summits with a Mann-Whitney U test. For both sets, the number of CTCF reads in 100-bp bins were summed over all analyzed sites, and the two distributions were compared.
Genomic distribution
We determined localization of different peak sets with respect to Ensembl version 54 genes. For each peak region, all genes overlapping or in proximity of a peak were considered. We defined the following categories: “5′ and 3′” if a peak was located 3 to 1 kb of the annotated Ensembl gene start or end, “start” and “end” if they were within 1 kb of annotated gene starts or ends, intronic, exonic, and intergenic. Finally, we compared the obtained proportions with the genome background, obtaining the enrichments shown in Figure 1C.
Read profiles
For both the chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) data association and the CTCF knockdown analysis, we calculated read profiles around the sites of interest by counting reads in 10-bp bins 10 kb upstream and downstream of the peak summits and normalizing these over input reads, total read yield, and peak numbers. For the ChiA-PET data analysis, ER anchor-loop and noninteracting site coordinates were available from Fullwood et al. 2009. Read profiles for RAD21, STAG1, and CTCF at these two binding site categories were compared. When looking at binding changes after CTCF knockdown, we profiled STAG1 and RAD21 reads at both ER–cohesin–non-CTCF and cohesin–CTCF binding sites.
Cohesin binding change upon ER stimulation
Cohesin–CTCF and CTCF-independent cohesin bound sites were split into 10-bp bins, centered on the peak summits. For each binding site category, we calculated the difference between normalized STAG1, respectively, RAD21 reads upon estrogen treatment. The significance of the difference between the observed profiles was assessed with the Kolmogorov-Smirnov test.
Motif analysis
De novo motif discovery was conducted with MEME (Bailey and Elkan 1994) using the settings “-maxw 25 -nmotifs 5 -revcomp -dna” and with NestedMica (Down and Hubbard 2005) using the parameters “-numMotifs 5 -minLength 5 -maxLength 20 –revComp.” We used 800–1000 50-bp-long regions centered on the SWEmbl summit as input for the motif discovery programs, obtaining highly similar motifs for different peak score categories and the two analysis programs.
We searched for the discovered motif in all of the high-confidence bound regions using the PWM score. The scan was performed with the TFBS Perl module (Lenhard and Wasserman 2002), with two different thresholds for the CTCF PWM match (0.75 and 0.80) as well as with Patser (van Helden et al. 2000), using the parameters “-A a:t 3 g:c 2 -R 1000 -M 10” and a cutoff of 10. We obtained very similar relative motif occurrence numbers with all three methods. The ER motif enrichment analysis was performed with the CEAS program (http://ceas.cbi.pku.edu.cn/).
Expression analysis
The raw bead-level data was preprocessed and normalized by log2 transformation and quantile normalization using beadarray, a Bioconductor package developed at the Cambridge Research Institute. The Bioconductor limma package was used for the statistical analysis. Only genes that obtained the quality scores “perfect” and “good” after this processing were selected for the analysis. The regulated set was defined as having a logFC > 0.2 or < −0.2 and an adjusted P-value < 0.01 measured at least one time point, while the nonregulated set had a logFC between −0.2 and 0.2 and a P-value > 0.01. Genes in the regulated and nonregulated categories were scanned for the presence of the binding sites of interest 20 kb of the Ensembl annotated gene starts and significance of the association determined by the Fisher's exact test.
All computational analyses were performed with in-house Perl and Python scripts; statistical analysis and visualization was done in R and Bioconductor. ChIP-seq data are displayed using the UCSC Genome Browser (Kent et al. 2002) and in Figure 7A and Supplemental Figure S5 normalized to 10 million total aligned sequencing reads.
Acknowledgments
We thank the CRI Genomics and Bioinformatics Cores especially Nick Matthews, Claire Fielding, James Hadfield, and Roslin Russell. Supported by the European Research Council, EMBO Young Investigator Program, and Hutchinson Whampoa (D.T.O.), University of Cambridge (D.S., P.C.S., C.R., J.S.C., D.T.O.), Cancer Research UK (D.S., C.S.R., A.H., G.D.B., J.S.C., D.T.O.), the Wellcome Trust (P.F.) and EMBL (P.C.S., P.F.), Commonwealth Fund (C.R.).
Author Contributions: D.S., J.S.C., and D.T.O. conceived and designed the experiments; D.S., C.S.R., and A.H. performed experiments; D.S., P.C.S., C.R., and G.B. analyzed the data; D.S., P.C.S., and D.T.O. wrote the manuscript; and J.S.C., P.F., and D.T.O oversaw the work.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray and sequencing data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae) under accession nos. E-MTAB-158 and E-TABM-828, respectively.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100479.109.
References
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Brown AM, Jeltsch JM, Roberts M, Chambon P 1984. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci 81: 6344–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2008. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Hubbard TJ 2005. NestedMICA: Sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res 33: 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-alpha–bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli G 2006. SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol Cancer 5: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K 2003. Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K 2008. The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS 2007. Cohesin-dependent regulation of Runx genes. Development 134: 2639–2649 [DOI] [PubMed] [Google Scholar]
- Kaur M, DeScipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID 2005. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A 138: 27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. 2004. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36: 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi E, Pezzi N, Prieto I, Barthelemy I, Carreiro C, Martínez A, Maldonado-Rodríguez A, López-Cabrera M, Barbero JL 2004. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem 279: 6553–6559 [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, et al. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes & Dev 22: 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Wasserman WW 2002. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics 18: 1135–1136 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L 2006. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 38: 528–530 [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E 2008. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem 283: 32977–32988 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Prall OW, Sarcevic B, Musgrove EA, Watts CK, Sutherland RL 1997. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E–Cdk2. J Biol Chem 272: 10882–10894 [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D 2004. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24: 3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Stark R, Wilson MD, Brown GD, Odom DT 2008. Genome-scale validation of deep-sequencing libraries. PLoS One 3: e3713. doi: 10.1371/journal.pone.0003713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT 2009. ChIP-seq: Using high-throughput sequencing to discover protein–DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T 2005. Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr Opin Genet Dev 15: 258–264 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36: 636–641 [DOI] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J 2000. A web site for the computational analysis of yeast regulatory sequences. Yeast 16: 177–187 [DOI] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, et al. 2005. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37: 468–470 [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, et al. 2007. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: Evidence for impaired recombinational repair. Hum Mol Genet 16: 1478–1487 [DOI] [PubMed] [Google Scholar]
- Watrin E, Peters JM 2006. Cohesin and DNA damage repair. Exp Cell Res 312: 2687–2693 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J 2007. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134: 3191–3201 [DOI] [PubMed] [Google Scholar]