Abstract
We studied miRNA profiles in 4419 human samples (3312 neoplastic, 1107 nonmalignant), corresponding to 50 normal tissues and 51 cancer types. The complexity of our database enabled us to perform a detailed analysis of microRNA (miRNA) activities. We inferred genetic networks from miRNA expression in normal tissues and cancer. We also built, for the first time, specialized miRNA networks for solid tumors and leukemias. Nonmalignant tissues and cancer networks displayed a change in hubs, the most connected miRNAs. hsa-miR-103/106 were downgraded in cancer, whereas hsa-miR-30 became most prominent. Cancer networks appeared as built from disjointed subnetworks, as opposed to normal tissues. A comparison of these nets allowed us to identify key miRNA cliques in cancer. We also investigated miRNA copy number alterations in 744 cancer samples, at a resolution of 150 kb. Members of miRNA families should be similarly deleted or amplified, since they repress the same cellular targets and are thus expected to have similar impacts on oncogenesis. We correctly identified hsa-miR-17/92 family as amplified and the hsa-miR-143/145 cluster as deleted. Other miRNAs, such as hsa-miR-30 and hsa-miR-204, were found to be physically altered at the DNA copy number level as well. By combining differential expression, genetic networks, and DNA copy number alterations, we confirmed, or discovered, miRNAs with comprehensive roles in cancer. Finally, we experimentally validated the miRNA network with acute lymphocytic leukemia originated in Mir155 transgenic mice. Most of miRNAs deregulated in these transgenic mice were located close to hsa-miR-155 in the cancer network.
Characterization of genes that control the timing of larval development in Caenorhabditis elegans revealed two small regulatory RNAs, lin-4 and let-7 (Reinhart et al. 2000). Soon thereafter, lin-4 and let-7 were reported to represent a new class of small RNAs, named microRNAs (miRNAs) (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). miRNAs have since been found in plants, green algae, viruses, and animals (Griffiths-Jones et al. 2008). The number of mature miRNAs in the human genome has now surpassed 1000 (Ruby et al. 2006, 2007; Landgraf et al. 2007). Baek et al. (2008) used quantitative mass spectrometry to measure the proteome response as a function of miRNA activity. Although some targets were repressed without changes in mRNA levels, those translationally repressed by more than a third also displayed mRNA destabilization and, for the most highly repressed targets, mRNA destabilization usually was the major component of repression. In the same manner, another group (Selbach et al. 2008) showed that a single miRNA can repress the production of hundreds of proteins, typically in a mild fashion. They too demonstrated that miRNAs down-regulate target mRNA levels.
Evolutionarily conserved among distant organisms, miRNAs are involved in a variety of biological processes, including cell cycle regulation, differentiation, development, metabolism, neuronal patterning, and aging (Bartel 2009). Alterations in miRNA expression are also involved in the initiation, progression, and metastasis of human tumors (Spizzo et al. 2009). Germline mutations in the hsa-miR-15a and hsa-miR-16-1 cluster are associated with familial chronic lymphocytic leukemia (CLL), whereas a common SNP in pre-hsa-miR-146a decreases mature miRNA expression and predisposes to papillary thyroid carcinoma. Furthermore, Mir155 transgenic mice show proliferation of pre-B cells and develop lymphoblastic leukemia/high-grade lymphoma. Mice overexpressing mmu-miR-17/92 in lymphocytes develop lymphoproliferative disease and autoimmunity. hsa-miR-10b, hsa-miR-373, and hsa-miR-520c have been reported to promote tumor invasion and metastasis; whereas, research has shown hsa-miR-335, hsa-miR-206, and hsa-miR-126 to be suppressors of breast cancer metastasis. Moreover, reduced expression of Dicer and Drosha miRNA processors has been observed in various human cancers. Several transcription factors regulate the expression of miRNAs, e.g., the tumor suppressor protein TP53 regulates the expression of hsa-miR-34 family members; MYC is a negative regulator of miRNA expression; STAT3 regulates hsa-miR-21; and TWIST1 trans-activates hsa-miR-10b transcription. The consequences of altered patterns of miRNA expression are just starting to be understood (Spizzo et al. 2009). Insensitivity to anti-growth signals is achieved by inhibition of E2F transcription factors caused by overexpression of the hsa-miR-17/92 cluster on chromosome 13q31.1 and by overexpression of the hsa-miR-106b/25 cluster on chromosome 7q22.1. Modulation of apoptosis may occur by direct regulation of miRNAs that are: proapoptotic (TP53 and hsa-miR-34 family) or by targeting of anti-apoptotic proteins (BCL2, MCL1 proteins, and the hsa-miR-15/16 cluster).
Much of the current effort in miRNA studies is focused on the elucidation of their function. Typically miRNAs have been studied by using the gene profiling approach. Each miRNA has been studied for its single contribution to differential expression or to a compact predictive signature. However, the effect of miRNAs on cell pathology and physiology is likely to be complex for two reasons: (1) their activity is exerted in a one-to-many fashion, such that each miRNA can control translation of tens or even hundreds of different coding messengers and (2) a single messenger can be controlled by more than one miRNA. Thus, we propose a paradigm shift to the study of miRNAs in cancer by applying a systems biology approach. For this purpose we built miRNA gene networks by using our very large expression miRNA database.
Previously, Yoon and De Micheli (2005) and Tran et al. (2008) used computational methods to predict miRNA regulatory modules. Yoon based his approach solely on miRNA:mRNA interactions as predicted by TargetScan, without expression data. Tran and colleagues studied the correlation between the expression of 121 miRNAs and their 801 targets (as predicted by PicTar) in 89 human cancer samples.
We built our miRNA networks exclusively from miRNA expression data. Here, we report the first miRNA network from normal tissues. In parallel, we built miRNA networks for coupled cancerous and noncancerous tissues. By comparing normal to cancer networks we attained a second goal: the identification of cancer variations in miRNA networks. Finally, we superimposed DNA variations onto expression data to generate a comprehensive miRNA alteration map in cancer.
Results and Discussion
The miRNA network in normal tissues
We assayed mature miRNAs in 17 groups of normal human tissues, from a total of 1107 chips. Tissue specificity was calculated by using the information content (IC) according to Landgraf et al. (2007) who measured expression levels by sequencing cloned miRNAs. The most tissue-specific miRNAs are the members of the hsa-miR-302 cluster, as shown in Supplemental Figure 1 and Supplemental Tables I and III. hsa-miR-302a/b/c were expressed in embryonic samples.
Complexity of genetic regulatory mechanisms in higher organisms is thought to also be achieved through controlled and coordinated networks of miRNAs. We exploited our microarray database to generate miRNA networks based exclusively on expression data. We applied Banjo (Smith et al. 2006) to infer the Bayesian network for normal tissues. miRNA relations were modeled as graphs where nodes represent the miRNAs and colored edges the relationships between them. The node degree distribution of the normal miRNA network is illustrated in Supplemental Figure 3A. The exponential decrease of both absolute frequency and inverse cumulative frequency curves shows that there were a lot more poorly connected nodes than highly connected (hubs). More than 40% of the nodes had degrees of 1 and almost 75% had degrees of ≤2. The normal tissues miRNA graph thus presented a scale free behavior. The highest degree hub was hsa-miR-16, followed by hsa-miR-215.
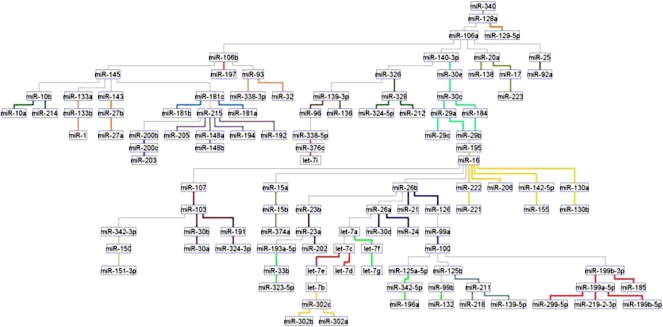
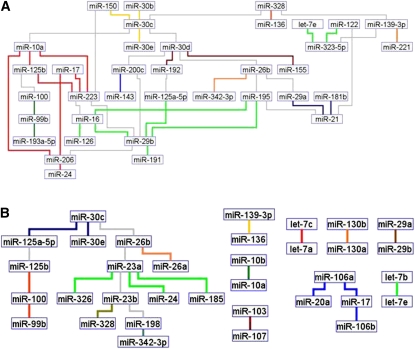
To discover miRNA groups with highly related expression patterns we extracted coherent groups of nodes by adopting clustering algorithms. Other researchers have developed algorithms to extract clusters (i.e., groups of densely connected nodes) from biological nets and we chose the MCL graph-based algorithm (Enright et al. 2002). This algorithm, which was implemented in Neat, (Brohee et al. 2008) has been shown to enable good performance in extracting coregulated genes from transcriptomes. Figure 1 displays the miRNA network of normal tissues, obtained from over 1000 samples and 50 cell types/tissues. We used all of the expressed miRNAs to build the Bayesian networks (rather than only the differentially expressed ones). The MCL clusters with high coexpression patterns throughout normal tissues are linked by specific colored edges. miRNAs are generally connected as expected from the published literature. For example, hsa-miR-133a/b was in a cluster with hsa-miR-1 (light orange) and all were involved in skeletal muscle proliferation and differentiation (Chen et al. 2006). A close cluster is hsa-miR-10a/b and hsa-miR-214 (green). hsa-miR-214 is expressed during early segmentation stages in somites and can modulate the expression of genes regulated by Hedgehog. Inhibition of hsa-miR-214 results in a reduction or loss of slow-muscle cell types (Flynt et al. 2007). Perhaps not surprisingly these muscle/differentiation clusters are linked to hsa-miR-143/145, a miRNA capable of pushing ES cells to differentiate (Xu et al. 2009). The top, right proliferation cluster hsa-miR-106a/b/93 (Petrocca et al. 2008) to hsa-miR-20a/17 and to hsa-miR-25/92a (MYC-associated) includes hsa-miR-223 (Spizzo et al. 2009) and is involved in cell cycle progression (Ivanovska et al. 2008). The hsa-miR-145 node links the proliferation clusters described above to the muscle differentiation clusters. This link might explain why the loss of hsa-miR-145 in cancer leads to an undifferentiated cellular state. The hsa-miR-29 family, that targets the anti-apoptotic protein MCL1 and plays a role in the TP53 pathway (Park et al. 2009) is linked to hsa-miR-30 and to hsa-miR-15/16, miRNAs that target the anti-apoptotic protein BCL2 (Spizzo et al. 2009). Overexpression of hsa-miR-29a leads to epithelial-to-mesenchymal transition (EMT) and metastasis, in cooperation with oncogenic Ras signaling (Gebeshuber et al. 2009). hsa-miR-221-222, regulators of the cell cycle, together with hsa-miR-206, hsa-miR-155 (pre-B cell proliferation), and hsa-miR-130a/b are in a yellow cluster (Spizzo et al. 2009). In the top center of the graph, hsa-miR-194 and hsa-miR-192 connect to hsa-miR-215, within a purple cluster associated with TP53 activation (Braun et al. 2008). In a close branch the hsa-miR-200 family, hsa-miR-203 and hsa-miR-205 are directly involved in TGF-beta-mediated EMT and differentiation (Spizzo et al. 2009). hsa-miR-181 family members are involved in B- and T-lineage and myoblast differentiation (Chen et al. 2004; Naguibneva et al. 2006; Li et al. 2007). hsa-miR-181, hsa-miR-200, hsa-miR-205, and hsa-miR-215 are all linked to hsa-miR-145. Thus, the loss of the hsa-miR-145 hub might also impact the TP53/EMT/differentiation branch. Another important hub appears to be hsa-miR-16, which is located in the other half of the net and coordinates hsa-miR-29 and hsa-miR-221/206/155/130 clusters. The hsa-miR-16 hub also feeds the hsa-miR-26/let-7/hsa-miR-302 branch. hsa-miR-26 is a key gene in hepatocellular carcinoma and its expression is associated with survival and response to adjuvant therapy with interferon alpha (Ji et al. 2009). Let-7 regulates Ras and hsa-miR-302 are expressed in ES cells and other early embryonic tissues.
Figure 1.
The miRNA network in normal tissues (1107 samples, 50 tissues, 115 miRNAs). The network was inferred for all expressed and varying miRNAs, without preselecting for differential expression. Standard Banjo parameters were adopted with a q6 discretization policy. The consensus graph depicted here was obtained from the best 100 nets (after searching through 8.3 × 109 networks). MCL graph-based clustering algorithm was applied to clusters extraction (miRNAs with highly related expression pattern have edges of the same color); thus, different clusters in the network are linked by different color edges. yEd graph editor (yFiles software) was employed for graphs visualization. See text for further discussion.
The miRNA networks in solid cancers are reprogrammed
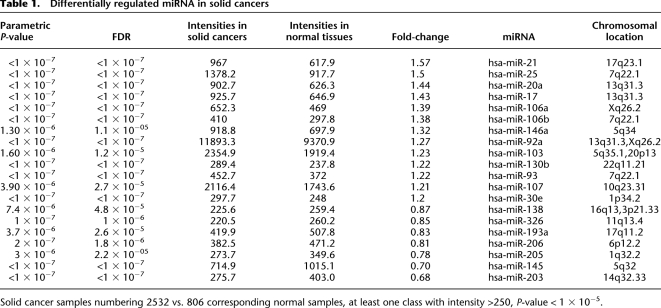
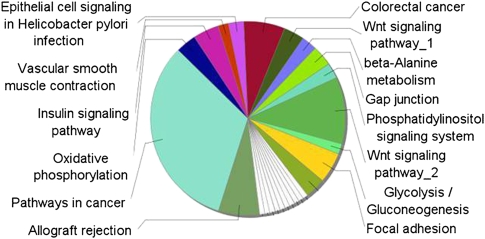
In previous research, we studied six types of solid cancer in about 500 different samples (Volinia et al. 2006). Here, we expanded our study to 51 types of cancer in 3312 samples (2532 solid cancers and 780 leukemia samples). The results of miRNA differential expression in solid cancers are listed in Table 1 and in Supplemental Tables IV and V. They confirm our earlier report and the established miRNA profiling in cancer. For example, hsa-miR-21 and hsa-miR-17/20/25/92/103/106/146a were overexpressed, whereas hsa-miR-203/205 and hsa-miR-143/145 were down-regulated. The KEGG pie chart in Figure 2 and the corresponding network graphs (Supplemental Figs. 5, 6) show the functional effect of differentially expressed miRNAs on cellular pathways. The large majority of the affected pathways is related to cancer or signal transduction (i.e., Wnt, VEGF, TGF-beta, insulin, phosphatidylinositol signaling, focal adhesion, and colorectal cancer). We also applied the IC measure to identify cancer-specific miRNAs. The ICs were almost as high as those measured for the normal tissues, indicating that there were miRNAs with high cancer-type specificity (Supplemental Fig. 4; Supplemental Table VI).
Table 1.
Differentially regulated miRNA in solid cancers
Solid cancer samples numbering 2532 vs. 806 corresponding normal samples, at least one class with intensity >250, P-value < 1 × 10−5.
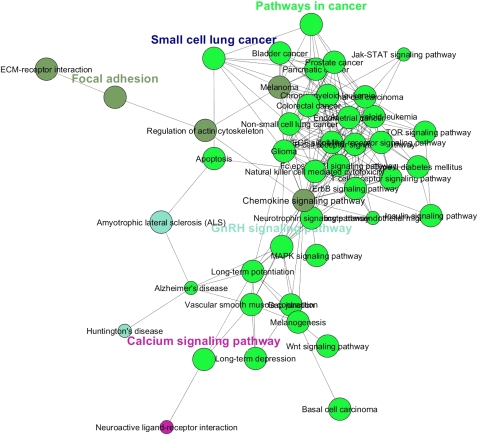
Figure 2.
Cellular pathways regulated by differentially expressed miRNAs in cancer. KEGG analysis by ClueGO (Bindea et al. 2009) of pathways (score = 3, P-value < 1 × 10−3) simultaneously targeted by both up-regulated and down-regulated miRNAs (listed in Table 1; Supplemental Tables IV, V). The KEGG pie-chart shows the functional effect of differentially expressed miRNAs on cellular pathways in cancer. The large majority of the affected pathways is related to cancer or signal transduction (i.e., Wnt, VEGF, TGF-beta, insulin, and phosphatidylinositol signaling, focal adhesion, and colorectal cancer). Target genes selection was performed with DIANA-miRpath, microT-V4.0 (Papadopoulos et al. 2009). The union of the target mRNAs with a score above 3 was used as an input to ClueGO. Right-sided hypergeometric test yielded the enrichment for GO terms. Benjamini-Hochberg correction for multiple testing controlled the P-values. GO term fusion was applied for redundancy reduction.
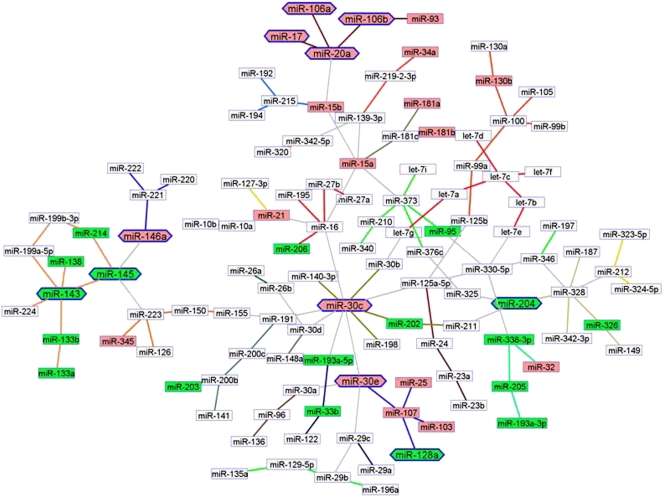
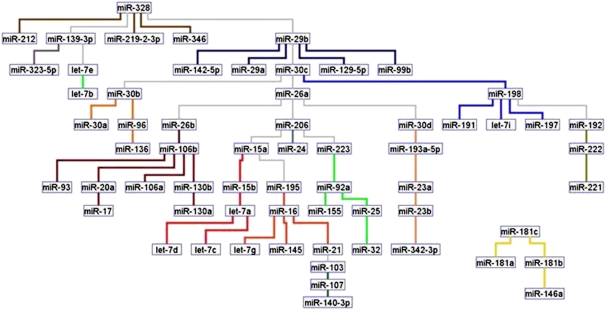
We generated a global miRNA expression network for solid cancers (Fig. 3). It is important to note that, to build the Bayesian net, we used all the expressed and varying miRNAs as input, rather than only using the differentially expressed ones (miRNAs with low variation were excluded from the analysis). The node degree distribution of the solid cancer miRNA network is illustrated in Supplemental Figure 3B. Like the normal tissues miRNA graph, the solid cancer net also presented a scale-free behavior. In cancer, the most connected hub was hsa-miR-30c (degree 10), followed by hsa-miR-16 (degree 6). Whereas, in nonmalignant tissues, hsa-miR-16 was the most connected node (degree 8) and hsa-miR-30c had only a low degree of 3. Opposite behavior had TP53 regulated hsa-miR-215 (degree 6 in normal tissues and degree 3 in cancer) and hsa-miR-103/106a (degree 5 in normal tissues and only degree 1 in cancer). The exchanges of hubs between nonmalignant and cancer tissues were the first notable sign of divergences in their respective miRNA programs. The MCL clustering algorithm was employed to map the subnetworks with high coexpression patterns (these MCL clusters, or cliques, are linked by specific colored edges). Additionally, we color-coded the miRNA nodes according to their differential expression in tumors (red, overexpressed; green, down-regulated). Neighbors preferentially appeared with the same trend, such that clustered miRNAs were either overexpressed or down-regulated. For example, hsa-miR-17/20a (chr 13q31.3), hsa-miR-106a/b (chr Xq26.2 and chr 7q22.1), and hsa-miR-93 (chr 7q22.1) were all up-regulated in cancers. Conversely, hsa-miR-143/145 (chr 5q32), hsa-miR-133a/b (chr 18q11.2, chr 20q13.3 and chr 6p12.2), hsa-miR-214 (chr 1q24.3), and hsa-miR-138 (chr 3p21.33 and chr 16q13), all in the same coexpression clique, were down-regulated.
Figure 3.
The miRNA network in solid cancers (2532 samples, 31 cancer types, 120 miRNAs). The network was inferred for all expressed and varying miRNAs, without preselecting for differential expression. Standard Banjo parameters were adopted with a q6 discretization policy. The consensus graph depicted here was obtained from the best 100 nets (after searching through 8.4 × 109 networks). MCL expression clusters are linked by different color edges. yEd graph editor (yFiles software) was employed for graph visualization. The miRNAs expressed differentially in the tumors are color-coded and in the graph miRNA neighbors are of the same color code: Closely clustered miRNAs are either pink (overexpressed) or green (down-regulated). The node labels, for which expression and physical alteration (CGH, see “miRNA copy number variations in cancer and leukemia” section) were concordant (i.e., overexpression and amplification), were emboldened and visually reinforced with a hexagonally shaped border.
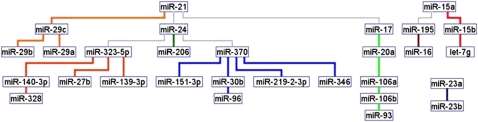
To individually investigate cancer types (lung, colon, breast, prostate) we built graphs for normal and cancer tissues and applied MCL to extract miRNA coexpression clusters. Normal lung miRNome was represented by a single complete miRNA network (Fig. 4A), while adenocarcinomas were by one major and eight unconnected subnetworks (Fig. 4B). KEGG functional analysis of the miRNAs in the eight minor subnetworks unconnected in cancer showed that they target genes involved in cancer-related pathways (Fig. 5). Strikingly, a similar situation of disjointed miRNA cliques was also present in the other cancers we studied, which included: colon, breast, and prostate cancers (Supplemental Figs. 8–10, respectively). In particular, we identified a number of notable, and often common, miRNAs in the unconnected cancer clusters. hsa-miR-10a/b was identified in lung, colon, and breast cancers (Ma et al. 2007), miR 26a/b in colon and prostate cancers (Kota et al. 2009), hsa-miR-29a/b in breast, colon, and lung cancers (Mott et al. 2007; Gebeshuber et al. 2009), hsa-miR-181 family members in colon and breast cancers (Spizzo et al. 2009), and hsa-miR-107/ hsa-miR-103 in breast, prostate, lung, and colon cancers (Volinia et al. 2006). The let-7c/a miRNAs were prominent in colon, lung, and prostate cancers (Johnson et al. 2005) and hsa-miR-106a/b (Petrocca et al. 2008), linked to hsa-miR-17 and hsa-miR-20, in colon and lung cancers. Other miRNA cliques included hsa-miR-200c (Korpal and Kang 2008), linked to TP53-associated hsa-miR-192/215 (Braun et al. 2008; Georges et al. 2008), in a colon subnetwork. Thus, miRNA networks were reprogrammed in solid cancer, and the expression of a few notable miRNAs was independent from the major network. The single graph in the overall solid cancer net can be explained only by the same miRNAs having variable roles in a range of cancers, such that a miRNA regulates different targets in different cell types. The cell type and context specific action of miRNAs has been described often (Spizzo et al. 2009). An extreme example for such a double play is the apparently controversial role of hsa-miR-29, which acts as a tumor suppressor in leukemia and an oncogene in solid tumors (Gebeshuber et al. 2009).
Figure 4.
Comparison of miRNA networks in normal lung and adenocarcinoma. (A) Normal lung (71 samples). A single complete miRNA network is shown. (B) Lung adenocarcinoma (125 samples). In this graph, one major and eight minor subnetworks were detected. For example, hsa-miR-10a/b, hsa-miR-29a/b, hsa-miR-107, and hsa-miR-103 are in minor independent subnetworks disjoint from the main one.
Figure 5.
The KEGG functional analysis of eight disjointed minor miRNA networks in lung adenocarcinoma. The miRNA present in the unconnected cliques target genes are involved in many cancer-related terms, such as focal adhesion, small cell lung cancer, and calcium signaling. The detailed list of significant GO terms is shown in Supplemental Figure 7.
Leukemias confirm that miRNA networks are aberrant in neoplasia
In addition to looking at miRNA networks in solid cancers, we generated the networks for two hematological cancers, acute myeloid leukemia (AML) and chronic lymphocytic leukemia (CLL). Both of these leukemias have been well characterized in terms of miRNA profiles and their relations to prognosis. The miRNA network in AML also had disjointed cliques (Fig. 6). The most prominent finding here was that hsa-miR-155 and hsa-miR-181, two miRNAs with clinical relevance (Debernardi et al. 2007; Marcucci et al. 2008a,b), were positioned in two separated subnetworks, as expected from their prognostic independence. In fact, hsa-miR-181 was associated to hsa-miR-146a in a detached yellow miniclique (Taganov et al. 2006; Labbaye et al. 2008), while hsa-miR-155 (Thai et al. 2007; O'Connell et al. 2009) belonged to the main subnetwork, in the same red MCL clique as hsa-miR-223, hsa-miR-92a, hsa-miR-25, and hsa-miR-32. Finally, hsa-miR-29b has a key role in AML (Garzon et al. 2008) and, in accordance, it acts as a hub in the AML net.
Figure 6.
The miRNA network in acute myeloid leukemia (589 samples, two subnetworks). Standard Banjo parameters were adopted with a q6 discretization policy. The consensus graph depicted here was obtained from the best 100 nets (after searching through 8.5 × 109 networks). The miRNA network in AML has disjointed cliques. hsa-miR-155 and hsa-miR-181, two miRNAs with clinical relevance are in two separated subnetworks, as expected from their prognostic independence. hsa-miR-181 is associated with hsa-miR-146a in a detached yellow miniclique. mir-155 belongs to the main subnetwork, in the same red MCL clique of hsa-miR-223, hsa-miR-92a, hsa-miR-25, and hsa-miR-32. Finally, hsa-miR-29b has a key role in AML and acts as a hub in the AML net.
In chronic lymphocytic leukemia (CLL) two small cliques were separated from the main net (Fig. 7): hsa-miR-23a/b (Gao et al. 2009) and a second one embracing the hsa-miR-15/16 pair. hsa-miR-15 and hsa-miR-16, two miRNAs frequently deleted in CLL, have been showed to regulate apoptosis via BCL2 (Calin et al. 2005; Cimmino et al. 2005). Thus, the network topologies for these two leukemias could recapitulate their respective molecular pathology, with the key AML hsa-miR-29b acting as a hub in AML, but only a branch in CLL. AML prognostic hsa-miR-181 was disjointed in AML, but not in CLL, with the reverse being true for the CLL prognostic hsa-miR-15/16 pair.
Figure 7.
The miRNA network in chronic lymphocytic leukemia (254 samples, three subnetworks). Standard Banjo parameters were adopted with a q6 discretization policy. The consensus graph depicted here was obtained from the best 100 nets (after searching through >1 × 1010 networks). The network graph shows a major net and two separated minicliques: hsa-miR-23a/b and the hsa-miR-15/16 cluster. hsa-miR-15 and hsa-miR-16, two miRNAs frequently deleted in CLL, have been shown to regulate apoptosis via BCL2. The key hsa-miR-29b, acting as a hub in AML, is only a branch in CLL. AML prognostic hsa-miR-181 is disjointed in AML but not in CLL, while the reverse happens in CLL for prognostic hsa-miR-15/16 genes.
miRNA copy number variations in cancer and leukemia
miRNAs are differentially expressed in human cancer (Spizzo et al. 2009), but little is known about their chromosomal alterations, such as amplifications (hsa-miR-17/92) and deletions (hsa-miR-15a/16-1). To systematically study miRNA copy number alterations in cancer, we investigated 744 samples (solid cancers and leukemia), at medium resolution (150 kb). We used data from array comparative genomic hybridization (aCGH) and calculated, for each of 20,000 different chromosomal locations, two P-values, one for deletion and one for amplification. To measure miRNA copy number alterations we used their respective host genes or, when unavailable, their two flanking genes. In addition, to focus on the functional role of miRNAs, to increase the statistical power of our approach and to possibly dilute the contribution of the host/associated genes, we considered miRNAs as families (Grimson et al. 2007). miRNA families have similar targeting properties and thus their members are expected to have similar impacts on oncogenesis.
We worked on aCGH samples from lung, pancreas, breast, colon, and nasopharyngeal carcinomas, glioblastoma, melanoma, Ewing sarcoma, osteosarcoma, T-cell acute lymphoblastic leukemia (T-ALL), AML, CLL, myelodysplasia, various lymphomas, and mucosa-associated lymphoid tissue MALT (Supplemental Table VII). We used aCGH from the NCBI Gene Expression Omnibus (GEO) and Stanford Microarray Database (SMD). CDKN2A and CDKN2B were identified as the most deleted genes in human cancers, followed by other tumor suppressors PTEN, ATM, and TP53. Oncogenes, like EGFR, MYC, LYN, MET, and MOS, were amplified. Supplemental Tables VIII and IX list amplified and deleted miRNA families. The detection of an amplified hsa-miR-17-5p/20/93/106 family was a successful validation of our approach. It is also noteworthy that the MIR17HG host gene for the hsa-miR-17/92 cluster was not present in the arrays, but its flanking genes successfully compensated for its absence. The top deleted miRNA family was hsa-miR-204/211, followed by other families including hsa-miR-200b/c/429, hsa-miR-141/200a, hsa-miR-125/351, and hsa-miR-218. Down-regulation of hsa-miR-200a/b/c/429 and 141 have been linked to breast cancer stem cells by targeting BMI1, a stem cell self-renewal regulator (Shimono et al. 2009). Likewise hsa-miR-211 is involved in stem cells as it shows the highest Information content in an ES cell differentiation series (Supplemental Fig. 2; Supplemental Table III). Therefore, we suggest that loss of hsa-miR-211 might be involved in regulation of cancer differentiation. We suggest the same possibility for hsa-miR-218, which is deleted in cancer and highly expressed in spontaneously differentiated monolayers. The results from aCGH were overlaid on the expression network in solid cancers (Fig. 3). The node labels, for which expression and physical alteration were concordant (i.e., overexpression and amplification), were emboldened and visually reinforced with a hexagonally shaped border.
Deregulated miRNAs in a Mir155-induced leukemia are preferentially located around hsa-miR-155 in the miRNA network
We generated two cases of leukemias in an Eμ/VH Mir155 transgenic mice. These leukemias were positive for CD43 and T-cell markers (CD3, CD8) and negative for B220. Both cases exhibited VDJ and TCR oligoclonal rearrangement. This T-cell immunophenotype might be caused by the proliferation of lymphoid progenitors that atypically differentiated into T cells. The disease started early, at 2 and 4 mo of age, respectively, and had a rapid course with the mice dying 2 wk later. Their autopsy revealed a widespread leukemic infiltration, with organomegaly and lymphadenopathy, histologically diagnosed as an aggressive malignant lymphoproliferation similar to Burkitt lymphoma (data not shown). The injection of single sick splenocytes into 30 syngeneic mice was sufficient to reproduce the full blown malignancy.
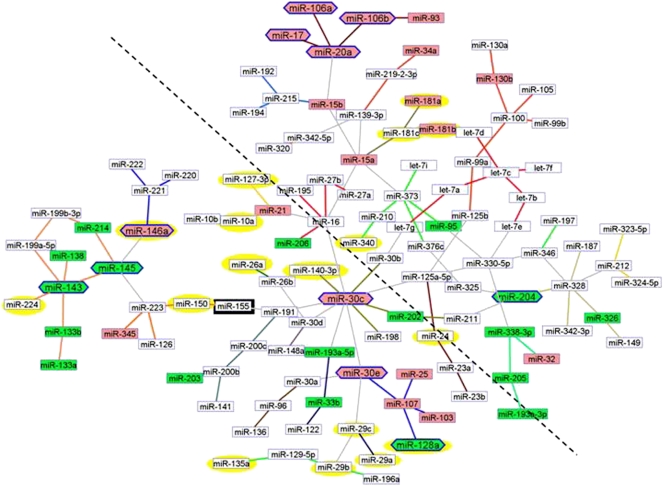
We compared the miRNA profiles of three leukemia samples from these Mir155 trangenes to controls from wild-type mice. Then we located the positions in the network for the miRNAs regulated in the transgene's leukemias (Supplemental Table X). We did not have an acute lymphocytic leukemia miRNA network as reference, therefore we mapped the deregulated miRNAs onto the generic cancer network and highlighted the nodes in yellow (Fig. 8). The yellow nodes appeared concentrated around the hsa-miR-155 node (black). When a diagonal, separating the hsa-miR-155 half from the other one, was drawn and the two sides compared, the difference in yellow node concentrations was significant (14 vs. 43, 4 vs. 57, Fisher's exact test, two-tail P-value < 0.009). The topological distribution was even more skewed if hsa-miR-29s and hsa-miR-181s were not considered as hsa-miR-155 regulated. In fact, hsa-miR-181 overexpression and hsa-miR-29 down-regulation are hallmarks miRNAs in leukemia; thus, they are likely to be independent events in cellular transformation and not directly related to the Mir155 transgene.
Figure 8.
Deregulated miRNAs in leukemia from Mir155 transgenic mice are preferentially located close to hsa-miR-155 in the cancer network. We compared miRNA profiles of three leukemia samples from Mir155 transgenes to controls from wild-type mice. The deregulated miRNAs (Supplemental Table X) were mapped onto the cancer network and highlighted in yellow. Most of the other miRNAs are concentrated around hsa-miR-155 node (black). When a diagonal is drawn and the two sides compared the difference between yellow nodes is significant (Fisher's exact test, two-tail P-value < 0.009).
Conclusion
We have presented a thorough analysis of miRNA tissue specificity in 50 different normal tissues grouped by 17 systems, corresponding to 1107 human samples. A small set of miRNAs were tissue-specific, while many others were broadly expressed. We also studied 51 oncologic or hemato-oncologic disorders and identified cancer-type-specific miRNAs. Then we inferred genetic networks for miRNAs in normal tissues and in their pathological counterparts. Normal tissues were represented by single complete miRNA networks. Cancers instead were portrayed by separate and unlinked miRNA subnets. Intriguingly, miRNAs independent from the general transcriptional program were often known as cancer-related. This “egocentric” behavior of cancer miRNAs could be the result of positive selection during cancer establishment and progression, as supported by aCGH. Leukemias were also rewired, but to a much lower extent. Nevertheless, miRNAs related to AML and CLL pathogenesis, such as hsa-miR-155, hsa-miR-181, and hsa-miR-15/16, were still removed from coordinated control. The dissimilar behavior of solid cancers and leukemia might be due to the diverging pathogenetic mechanisms, which include differing oncogenic miRNA networks. In the former, complex chromosomal aberrations are frequent, whereas in the latter, translocations often represent the major driving force.
Overall, miRNA networks in cancer cells defined independently regulated miRNAs. The target genes of these uncoordinated miRNA were involved in specific cancer-related pathways.
Methods
miRNA expression arrays
Microarray analysis was performed as previously described (Volinia et al. 2006). Briefly, 5 μg of total RNA were used for hybridization of miRNA microarray chips. These chips contain gene-specific oligonucleotide probes, spotted by contacting technologies and covalently attached to a polymeric matrix. The microarrays were hybridized in 6× SSPE (0.9 M NaCl, 60 mM NaH2PO4 · H2O, 8 mM EDTA at pH 7.4), 30% formamide at 25°C for 18 h, washed in 0.75× TNT (Tris-HCl, NaCl, Tween 20) at 37°C for 40 min, and processed by using a method of detection of the biotin-containing transcripts by streptavidin-Alexa647 conjugate. Processed slides were scanned using a microarray scanner (Axon), with the laser set to 635 nm, at a fixed PMT setting, and a scan resolution of 10 mm. Microarray images were analyzed by using GenePix Pro and post-processing was performed essentially as described earlier (Volinia et al. 2006) . Briefly, average values of the replicate spots of each miRNA were background-subtracted and subject to further analysis. miRNAs were retained, when present, in at least 20% of samples and when at least 20% of the miRNA had a fold change of more than 1.5 from the gene median. Absent calls were thresholded prior to normalization and statistical analysis. Normalization was performed by using the quantiles method. MiRNA nomenclature was according to the miRNA database at Sanger Center (Griffiths-Jones et al. 2008).
Data analysis
An SQL miRNA internal database was built with the data retrieved from a large number of different experiments performed in our laboratory. The description of the procedure and statistics for the database will be reported elsewhere (L Tagliavini, G Gamberoni, S Rossi, M Galasso, J Palatini, CM Croce, and S Volinia, in prep.). Briefly, the F635-background values were used. Bad spots were removed. Nonexpressed spots were averaged for each gpr files (chip). For each mature miRNA, we computed the geometric mean of its multiple reporters in the chip. A NaN value was assigned to miRNAs with more than 50% of corrupted spots, as reported by the GenePix image analysis software. All the results were log2-transformed. The normalization was performed by using the quantiles normalization, as implemented in the Bioconductor “affy” package (Bolstad et al. 2003). BRB Arraytools was used to perform t-test over two classes' experiments or F-tests over multiple classes (i.e., different normal tissues) (Zhao and Simon 2008). Target genes selection was performed by DIANA-miRpath, microT-V4.0 (Papadopoulos et al. 2009). The union of the target mRNAs with a score >3 was used as an input to ClueGO (Bindea et al. 2009). ClueGO was used to relate differential expression in cancer to functional pathways (KEGG). ClueGO visualizes the selected terms in a functionally grouped annotation network that reflects the relationships between the terms based on the similarity of their associated genes. The size of the nodes reflects the statistical significance of the terms. The degree of connectivity between terms (edges) is calculated using kappa statistics. The calculated kappa score is also used for defining functional groups. A term can be included in several groups. The reoccurrence of the term is shown by adding “n.” The not grouped terms are shown in white color. The group leading term is the most significant term of the group. The network integrates only the positive kappa score term associations and is automatically laid out using the Organic layout algorithm supported by Cytoscape. A right-sided hypergeometric test yielded the enrichment for GO-terms. Benjamini-Hochberg correction for multiple testing controlled the P-values. (Please refer to the Supplemental material for the complete list of data sets analyzed in this study.)
Network generation and clustering
Banjo was used to infer the Bayesian network for the different tissues and diseases. For each tissue or disease all the mature expressed and varying miRNAs were used as input to Banjo. The expression values were preprocessed with Gene Pattern to only filter out nonvarying miRNAs, according to the following parameters: #filter.flag = filter (variation filter and thresholding flag); #preprocessing.flag = no disk or norm (discretization and normalization flag); #minchange = 10 (minimum fold change for filter); #mindelta = 512 (minimum delta for filter); #threshold = 64 (value for threshold); #ceiling = 20,000 (value for ceiling); #max.sigma.binning = 1 (maximum sigma for binning); #prob.thres = 1 (value for uniform probability threshold filter); #num.excl = 2% of total chips (number of experiments to exclude (max and min) before applying variation filter); #log.base.two = no (whether to take the log base two after thresholding); #number.of.columns.above.threshold = 1% of total chips (remove row if n columns not ≥ than given threshold above.threshold); and #column.threshold = 512 (threshold for removing rows). We then performed a quality control step to remove chips with abnormal expression distribution across miRNAs: Chips were retained only if less than 25% of miRNAs were absent (expression value < 64). Similarly, miRNAs were retained only when less than 25% of samples had absent expression (value < 64). The static Bayesian network inference algorithm was run on the miRNA expression matrix by using standard parameters, with a discretization policy of q6. Consensus graphs, based on the top 100 networks, were obtained from at least 8 × 109 searched networks. We applied the MCL graph-based clustering algorithm to extraction of clusters (i.e., groups of densely connected nodes) from miRNA networks. MCL (Neat) has been shown to enable good performances in extracting coregulated genes from transcriptome networks. yEd graph editor (yFiles software, Tubingen, Germany) was employed for graphs visualization.
miRNA family array CGH
Seven hundred forty-four comparative genomic hybridization arrays were studied (537 samples from GEO and 207 from SMD). All platforms were two-channel based, data were downloaded as normalized values, and genes were annotated according to the gene symbol. All normalized log ratios were converted to log2 ratios, with the cancer value at the numerator and the control value at the denominator. Bootstrap analysis was used (10,000 random swaps of cancer and control channels) to obtain P-values and confidence limits for deletion and amplifications. We investigated 306 miRNA loci; 168 miRNA loci were associated to a host gene, and 138 miRNA loci to flanking genes. miRNA families were defined according to TargetScan. The threshold P-value for a miRNA family was set at 0.05 to the number of family members, n (0.05n). To control for multiple testing, we performed 100 bootstrapping cycles and used the results to calculate the false discovery rate (FDR). The resampling analysis was executed by randomly assigning the original P-values to the miRNA loci, while all family structures and chromosomal locations were kept unchanged. The FDR was defined as the percentage of families in the simulation evaluating better (lower P-values) than in the original test. Since the number of family member was variable (from a minimum of 2 to 7), FDRs were computed for each family according to its size (n, number of miRNA members).
Acknowledgments
S.V. is supported by AIRC (IG 8588), PRIN MIUR 2008, and Regione Emilia Romagna PRRIITT BioPharmaNet grants; A.V. is supported by AIRC (IG 5573). Microarray analyses were performed using BRB-ArrayTools developed by Richard Simon and the BRB-ArrayTools Development Team, GenePattern (Broad Institute), BioConductor and R.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae) under accession nos. E-TABM-969–E-TABM-975.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.098046.109.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J 2009. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M 2008. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68: 10094–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohee S, Faust K, Lima-Mendez G, Vanderstocken G, van Helden J 2008. Network Analysis Tools: From biological networks to clusters and pathways. Nat Protoc 3: 1616–1629 [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. 2005. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353: 1793–1801 [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. 2005. hsa-miR-15 and hsa-miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci 102: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD 2007. MicroRNA hsa-miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 21: 912–916 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG 2007. Zebrafish hsa-miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of hsa-miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, et al. 2008. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci 105: 3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeshuber CA, Zatloukal K, Martinez J 2009. hsa-miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 10: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, et al. 2008. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, hsa-miR-192 and hsa-miR-215. Cancer Res 68: 10105–10112 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. 2008. MicroRNAs in the hsa-miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. 2009. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med 361: 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Korpal M, Kang Y 2008. The emerging role of hsa-miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol 5: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, et al. 2008. A three-step pathway comprising PLZF/hsa-miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol 10: 788–801 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. 2007. hsa-miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129: 147–161 [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, et al. 2008a. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A cancer and leukemia group B study. J Clin Oncol 26: 5078–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, et al. 2008b. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ 2007. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26: 6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A 2006. The microRNA hsa-miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8: 278–284 [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D 2009. Inositol phosphatase SHIP1 is a primary target of hsa-miR-155. Proc Natl Acad Sci 106: 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG 2009. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics 25: 1991–1993 [DOI] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN 2009. hsa-miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol 16: 23–29 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. 2008. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VA, Yu J, Smulders TV, Hartemink AJ, Jarvis ED 2006. Computational inference of neural information flow networks. PLoS Comput Biol 2: e161 doi: 10.1371/journal.pcbi.0020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Croce CM, Calin GA 2009. SnapShot: MicroRNAs in cancer. Cell 137: 586–586.e1 [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D 2006. NF-κB-dependent induction of microRNA hsa-miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. 2007. Regulation of the germinal center response by microRNA-155. Science 316: 604–608 [DOI] [PubMed] [Google Scholar]
- Tran DH, Satou K, Ho TB 2008. Finding microRNA regulatory modules in human genome using rule induction. BMC Bioinformatics 9: S5 doi: 10.1186/1471-2105-9-S12-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- Yoon S, De Micheli G 2005. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics 21: ii93–ii100 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Simon R 2008. BRB-ArrayTools data archive for human cancer gene expression: A unique and efficient data sharing resource. Cancer Inform 6: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]