Abstract
The densities of transposable elements (TEs) in the human genome display substantial variation both within individual chromosomes and among chromosome types (autosomes and the two sex chromosomes). Finding an explanation for this variability has been challenging, especially in light of genome landscapes unique to the sex chromosomes. Here, using a multiple regression framework, we investigate primate Alu and L1 densities shaped by regional genome features and location on a particular chromosome type. As a result of our analysis, first, we build statistical models explaining up to 79% and 44% of variation in Alu and L1 element density, respectively. Second, we analyze sex chromosome versus autosome TE densities corrected for regional genomic effects. We discover that sex-chromosome bias in Alu and L1 distributions not only persists after accounting for these effects, but even presents differences in patterns, confirming preferential Alu integration in the male germline, yet likely integration of L1s in both male and female germlines or in early embryogenesis. Additionally, our models reveal that local base composition (measured by GC content and density of L1 target sites) and natural selection (inferred via density of most conserved elements) are significant to predicting densities of L1s. Interestingly, measurements of local double-stranded breaks (a 13-mer associated with genome instability) strongly correlate with densities of Alu elements; little evidence was found for the role of recombination-driven deletion in driving TE distributions over evolutionary time. Thus, Alu and L1 densities have been influenced by the combination of distinct local genome landscapes and the unique evolutionary dynamics of sex chromosomes.
Transposable elements (TEs), once labeled as “junk” DNA, contribute to unprecedented levels of interspecific divergence (Lander et al. 2001; The Chimpanzee Sequencing and Analysis Consortium 2005; Han et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), intraspecific genome diversity (Batzer and Deininger 2002; Bennett et al. 2004; Seleme et al. 2006; Han et al. 2008), and human diseases such as hemophilias A and B, as well as common cancers (Batzer and Deininger 2002; Chen et al. 2005). Recently, the intra- and interchromosomal distributions of short and long interspersed nuclear elements (SINEs and LINEs, respectively), the most abundant classes of TEs in primate genomes (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), have received critical attention. For example, although SINEs are retrotransposed into the human genome via LINE (L1) reverse transcriptase (Jurka 1997; Cost et al. 2002), the distribution patterns of SINEs and LINEs within chromosomes are very different (Lander et al. 2001). Indeed, Alus (a primate-specific family of SINEs) and L1s both appear to insert into AT-rich sequences, yet Alus accumulate over time in GC-rich regions (Yang et al. 2004; Belle et al. 2005; Abrusan and Krambeck 2006).
TEs are also differentially distributed in the human genome between the sex chromosomes and autosomes. Young AluYs are present on chromosomes X, Y, and the autosomes at the ratios expected according to the relative time spent in the male germline; chromosome X has 2/3 the autosomal average Alu density, chromosome Y has twice the density of autosomes, and the Y:X ratio of Alu densities is 3:1 (Jurka et al. 2002). Furthermore, a larger proportion of Alus occurring in AT-rich regions (i.e., recently inserted) reside on the Y than on the X chromosome (Jurka et al. 2004). In contrast, old AluJs subfamily densities are reportedly highest on the X, intermediate on autosomes, and lowest on the Y, concurrent with the noted shift in preferred base compositions of young AluYs (GC-poor) versus old AluJs (GC-rich) (Jurka et al. 2004).
Interestingly, the sex-chromosome distribution of L1 densities appears to be more complex. Recent analyses suggest that primate-specific L1 subfamily densities (L1Ps) are significantly higher on both the Y and X as compared to autosomes (Boissinot et al. 2001; Abrusan et al. 2008). Sex-chromosome distributions of older L1 subfamilies have received less attention; thus, the evolutionary timing of establishing their genome-wide patterns remains unexplained.
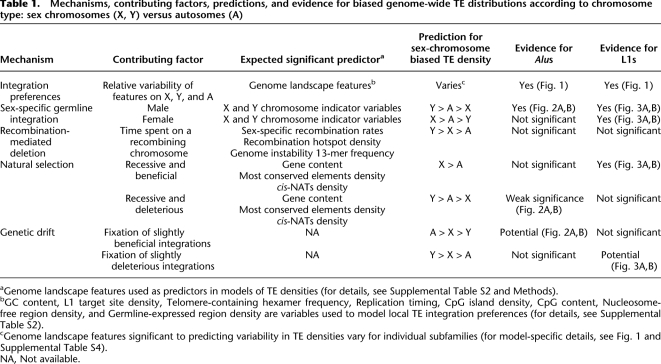
How is the sex-chromosome-biased genomic distribution of TEs achieved, and maintained, over evolutionary time? First, the male germline integration hypothesis predicts that densities of TEs on each of the chromosome types will correspond to the amount of time spent in the male germline (Table 1). Preferential integration in the male germline could explain the observed Y:A:X ratio of young Alu densities corrected for GC content (Jurka et al. 2002, 2004). The hypomethylation of Alu repeats during spermatogenesis (Rubin et al. 1994) is consistent with this hypothesis, since reduced levels of methylation lead to transcriptionally active regions, important to retrotransposition. Yet, L1s appear to have higher levels of methylation in the male than the female germline (Rubin et al. 1994), possibly impeding their activity in the former.
Table 1.
Mechanisms, contributing factors, predictions, and evidence for biased genome-wide TE distributions according to chromosome type: sex chromosomes (X, Y) versus autosomes (A)
aGenome landscape features used as predictors in models of TE densities (for details, see Supplemental Table S2 and Methods).
bGC content, L1 target site density, Telomere-containing hexamer frequency, Replication timing, CpG island density, CpG content, Nucleosome-free region density, and Germline-expressed region density are variables used to model local TE integration preferences (for details, see Supplemental Table S2).
cGenome landscape features significant to predicting variability in TE densities vary for individual subfamilies (for model-specific details, see Fig. 1 and Supplemental Table S4).
NA, Not available.
Second, recombination-driven deletion is presumed to facilitate the preferential loss of TEs on certain chromosomes. As the Y does not recombine outside of pseudo-autosomal regions and the X has lower recombination rates than do autosomes (Kong et al. 2002), this mechanism would result in higher TE densities on the Y, lower on the X, and the lowest on autosomes (Table 1). The importance of recombination in establishing TE distributions should also be reflected in the significance of recombination-related features to models of genome-wide TE densities. However, recombination-driven deletion cannot explain the observed change to lowest density of AluJs on the non-recombining Y chromosome (Jurka et al. 2002). Loss of L1 elements due to recombination is anticipated to result in the same sex-biased distributions noted for Alus; yet, comparisons of L1s among the chromosome types contradict this expectation (Boissinot et al. 2001; Abrusan and Krambeck 2006; Abrusan et al. 2008).
Third, chromosomal differences in genome characteristics such as base composition (Lander et al. 2001) could affect local retrotransposition rates and hence TE distributions (Table 1). Integration by target-primed reverse transcription (Cost et al. 2002) occurs concurrently with other cellular processes, most notably transcription, and possibly replication timing (transcription-rich regions tend to replicate early) (Woodfine et al. 2004). As the sex chromosomes are more GC-poor than autosomes (Skaletsky et al. 2003; Ross et al. 2005) and are known to replicate late in S phase (Woodfine et al. 2004), these differences might determine genome-wide TE densities if integration preferences are the primary source of variation.
Fourth, natural selection might play a role in influencing the interchromosomal distribution of TEs. On the one hand, natural selection operating against fixation of TEs capable of promoting deleterious ectopic recombination (Boissinot et al. 2001; Abrusan and Krambeck 2006; Sen et al. 2006; Han et al. 2008) would ultimately lead to similar genome-wide distributions as the male germline integration hypothesis: highest densities on the Y chromosome, due to its lack of recombination (and as a result inefficient selection) (Muller 1964; Felsenstein 1974), and lowest densities on the X, due to male hemizygosity and increased efficiency at purging deleterious recessive mutations (Table 1; Charlesworth et al. 1987).
On the other hand, natural selection could drive the fixation of TEs possessing a beneficial nature, particularly on the X (Charlesworth et al. 1987)—selection is thought to shape the distribution of L1 elements that are potential mediators of X-chromosome inactivation (XCI) (Table 1; Lyon 1998; Bailey et al. 2000; Carrel et al. 2006; Abrusan et al. 2008). As selection is difficult to measure directly, local gene content (as well as the content of other functional DNA classes) is often used as a proxy to reflect its impact (Abrusan et al. 2008). Interestingly, selection could also influence TE distributions because of their ability to regulate gene expression via cis-natural antisense transcripts (cis-NATs) (Conley et al. 2008) capable of RNA interference (Kloc and Martienssen 2008). L1 elements contain the largest proportion of TE-derived cis-NATs (Conley et al. 2008) and were recently suggested to accumulate on the X for this selectively advantageous purpose (Abrusan et al. 2008), although this hypothesis has not been formally tested.
Finally, sampling effects due to random genetic drift can alter TE densities among the chromosome types, as the X has effectively 3/4, and the Y 1/4, the population size (Ne) of the autosomes. For instance, random allele fluctuations over time could lead to either the higher rate of fixation of slightly deleterious or, conversely, loss of slightly advantageous newly integrated TEs on the sex chromosomes versus autosomes, thus contributing to the observed sex-chromosome-biased distributions (Table 1).
Here, we aim to distinguish among the above mentioned explanations for sex chromosome biases in distributions of Alu and L1 subfamilies of different evolutionary ages. We employ multiple regression models to account for the variability in TE densities in 1-Mb windows and, here for the first time, considering multiple genomic landscape features simultaneously. Subsequently, we analyze corrected TE densities on the sex chromosomes and autosomes. By providing an explanation for the sex bias in Alu and L1 distributions, we shed light on the evolutionary forces shaping nearly a third of each primate genome (e.g., Lander et al. 2001; The Chimpanzee Sequencing and Analysis Consortium 2005) and illuminate significant processes in sex chromosome evolution.
Results
Identification of branch-specific transposable elements
To explain genome-wide variability in TE densities in primate genomes, we applied the following strategy. First, we compared TE annotations among all possible pairwise combinations of the four sequenced primate genomes: human (Lander et al. 2001), chimpanzee (The Chimpanzee Sequencing and Analysis Consortium 2005), orangutan (The Orangutan Genome Sequencing Consortium, in prep), and macaque (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). We classified TEs as lineage-specific, ancestral (integrated in the common ancestor of the studied species), or intermediate in integration timing (i.e., human–chimpanzee [HC] or human–chimpanzee–orangutan [HCO] branch-specific) (Supplemental Table S1; Methods). Consistent with a recent analysis (Walser et al. 2008), we observed orthologous repeats in human, orangutan, and rhesus that were not recovered in chimpanzee, highlighting differences in sequence and genome assembly quality among primates, and/or chimpanzee-specific deletions (e.g., van de Lagemaat et al. 2005; Sen et al. 2006; Han et al. 2008).
Multiple regression analyses
Next, we obtained transposable element densities (as measured by count per window) and genomic landscape features (Supplemental Table S2) in 1-Mb nonoverlapping windows for each of the studied primate genomes (for details, see Methods). TEs found in regions of segmental duplication (Cheng et al. 2005) were excluded, in order to focus our analyses on inferences of insertion rather than duplication dynamics (Methods). Windows of low sequence coverage and/or quality for each genome were also excluded to avoid potential biases in the detection of TEs and in the measurements of genome landscape predictors (Methods). This resulted in 2620, 146, and 21 windows on human autosomes, chromosome X, and chromosome Y, respectively (for final window numbers retained after pruning for each model, see Supplemental Table S3); genome-wide data for additional primates analyzed are provided in Supplemental Table S4.
Finally, for each species, we performed multiple regression analyses to model variability in each TE subfamily density (response) depending on genomic landscape features (predictors) that reflect particular mechanisms of TE integration preferences and accumulation over evolutionary time (see introduction and Methods; Table 1; Supplemental Table S2); however, we acknowledge the possibility that some of the predictors might be associated with multiple mechanisms. The relative contribution to variability explained (RCVE) (Kvikstad et al. 2007) was used to detect the relative predictive power of each predictor in each full model, when in the context of all other predictors (for details, see Methods). Initially we applied this framework to repeat densities in the human genome, not separating windows by location on Y, X, and autosomes. We compared the densities of TEs on sex chromosomes versus autosomes after adjustment for regional variation as measured by the residuals from the above genome regression models. Last, repeat densities in other primate genomes were analyzed, following a similar procedure.
Genomic features shaping the distribution of Alus
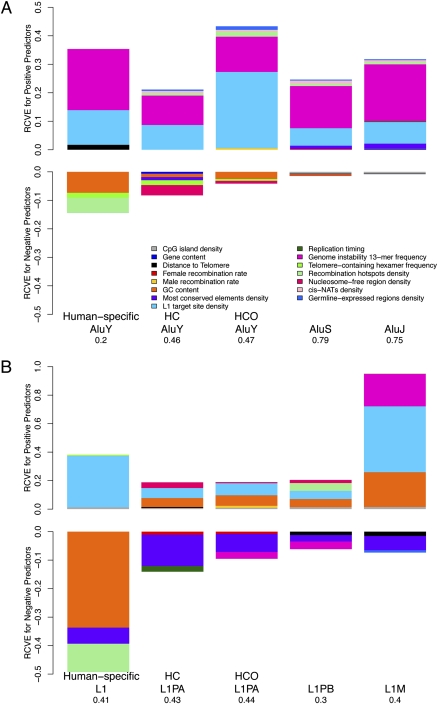
Our multiple regression models incorporating genomic landscape features (Fig. 1A) explain from 20% to 79% of the total genome-wide variability in Alu repeat densities, depending on the subfamily. Here, Alus were divided into human-specific AluYs; intermediate in integration timing, that is, human–chimpanzee (HC) or human–chimpanzee–orangutan (HCO) branch-specific AluYs; and ancestral AluS and AluJ subfamilies that integrated in the common ancestor of the studied species (Fig. 1A; Supplemental Tables S1, S3; see Methods). Models increase in explanatory power (measured by adjusted R2) with the estimated abundance of each Alu subfamily: the largest variability is explained for the most prolific (and oldest) AluS and AluJ subfamilies (Fig. 1A; Supplemental Table S1).
Figure 1.
Relative contribution to variability explained (RCVE) for each genome landscape feature significant to modeling variation in densities of Alus (A) and L1s (B) in 1-Mb windows across the human genome. Results of regressions with either the Alu or L1 density of various evolutionary integration timings as response and genome-wide features as predictors are depicted as bar plots (for details, see Supplemental Table S3). Color-coded areas correspond to the relative share (RCVE) that each predictor contributes to the total variability explained, in the presence of all other predictors (for details, see Methods). Bar plots are proportional to the sum of the RCVEs for each multiple regression model. (HC) Human–chimpanzee branch-specific elements; (HCO) human–chimpanzee–orangutan branch-specific elements.
The relative predictive power of each feature in each model corresponds to its RCVE value, depicted in Figure 1. The frequency (calculated as a ratio of the observed/expected frequency) of a 13-mer associated with genome instability (Supplemental Table S2; Myers et al. 2008) is among the most significant predictors that consistently explain variability in the densities of all Alu elements, independent of integration timing (Fig. 1A; regression details in Supplemental Table S3). Indeed, the 13-mer CCNCCNTNNCCNC, hypothesized to promote double strand breaks (DSBs) (Myers et al. 2008), is a significant positive predictor for the Alu densities of each analyzed subfamily, explaining ∼10%–22% of variability genome-wide (Fig. 1A; Supplemental Table S3). The frequency of this oligomer was calculated considering all nucleotides present in each genomic window, excluding oligomers located at TEs themselves, which did not alter our results. Consistent with the importance of L1 endonuclease target sites (TTTTAA) for Alu integration (Jurka 1997; Cost et al. 2002), all Alu densities strongly positively correlate with the density of these motifs: this predictor contributes 6.1%–26.7% of variation in Alu densities (Fig. 1A).
Whereas previous reports (e.g., Lander et al. 2001; Jurka et al. 2004; Abrusan and Krambeck 2006) observed a shift from AT to GC location preferences of young to old Alus, here, when simultaneously considering variability in other genomic features, GC content is negatively associated with the density of Alus at nearly all integration time points (except for ancestral AluJs) (Fig. 1A) and explains between 0.5% and 7% of overall Alu density variability (Supplemental Table S3). Interestingly, examination of pairwise correlations (i.e., not in the context of a multiple regression) between individual Alu subfamilies and GC content reveals that the negative Pearson correlations between GC content and young (human-specific, HC, HCO) AluYs decrease in their significance with time, whereas, conversely, the ancestral (AluS, and AluJ) subfamilies are weakly positively correlated with GC content (Supplemental Fig. S1). Thus, correlations between GC content considered alone and Alu densities recapitulate the previously noted Alu shift in base composition preferences (see introduction).
The density of germline-expressed genes, a feature related to potential integration preferences and likely representing transcriptionally active DNA, is a weak positive predictor for all but the human-specific Alus (RCVEs less than 1% to 1.2%) (Fig. 1A; Supplemental Table S3). Our models further demonstrate that the intermediate (HC, HCO) AluYs and AluS elements show a negative relationship with nucleosome-free regions (e.g., HC RCVE is 3.6%) (Fig. 1A; Supplemental Table S3).
In addition, our models tested features related to recombination. Human-specific AluYs strongly negatively correlate with computationally predicted recombination hotspots (Myers et al. 2005), explaining ∼5.4% of variability (Supplemental Table S3); the remaining Alu subfamilies, however, show relatively weak (RCVEs ranging from less than 1% to 1.8%) positive associations with such hotspots. The frequency of telomere-containing hexamer TTAGGG, a hallmark of telomerase-dependent RNA retrotranscription and repair of DSBs (Nergadze et al. 2007), is also negatively associated with particularly young (human-specific, HC and HCO) AluY integrations with RCVEs = 0.6%–1.7%, but is not significant for ancestral AluS and AluJ integrations. Other measures of recombination, namely, male- and female-specific recombination rates (Kong et al. 2002), explain <1% of variability in Alu densities for two subfamilies only (Fig. 1A; Supplemental Table S3). Interestingly, the distance to the telomere, an important factor in determining variability in substitution and indel rates (Kvikstad et al. 2007; Tyekucheva et al. 2008), is a positive predictor for human-specific AluY densities (RCVE 1.7%), but explains <1% of AluS and AluJ variability (Fig. 1A; Supplemental Table S3).
Several genomic features likely to reflect local selection pressure also display (usually weak) associations with local Alu densities (Fig. 1A). The density of most conserved elements is negatively associated with the density of HC AluYs, while positively correlated with the density of ancestral AluSs and AluJs, and contributes up to ∼1.6% in the RCVE. Gene content, on the other hand, explains <1% of the total variability in Alu density (Supplemental Table S3). Additionally, the local density of cis-NATs thought to regulate gene expression via RNA interference (Conley et al. 2008) is a relatively weak but significant positive predictor for all Alus except for human-specific ones (RCVEs ranging up to 1.2%) (Supplemental Table S3). The remaining features together explain only a small fraction of the overall variability in Alu densities genome-wide.
Genomic landscape features important to L1 subfamily densities
Depending on repeat integration timing, our multiple regression models capture 30%–44% of total genome-wide variability in L1 densities. Here, separate models were built for human lineage-specific L1s, ancestral L1s—L1PB and L1M repeats, and L1s intermediate in integration timing—HC and HCO L1PA repeats (Fig. 1B; Supplemental Tables S1, S3). In contrast to Alus, our explanatory power for individual models of L1 subfamily densities correlates with neither evolutionary timing of integration nor repeat quantity: for example, L1Ms are the most abundant and the oldest L1 subfamily studied here (Supplemental Table S1), yet models capture a similar proportion of their variability in repeat density as for the rarer human-specific L1s (Fig. 1B).
Several predictors, including L1 target sites, most conserved elements, and GC content, are significant for all (or almost all) regression models of L1 densities, independent of L1 evolutionary age. Predictably, L1 target sites (Berry et al. 2006) positively associate with the density of all L1 elements (Fig. 1B) and can alone explain the major portion of total L1 density variability (Supplemental Table S3), reflecting target-primed reverse transcription (Jurka 1997; Cost et al. 2002).
When simultaneously considering variability in other cofactors, GC content is a strong positive predictor for almost all (but the human-specific) L1 integrations and contributes up to an additional 34% to determining variation in L1 densities genome-wide (Fig. 1B; Supplemental Table S3). This is observed despite the known AT preferences of L1 endonuclease cleavage (Jurka 1997; Lander et al. 2001; Cost et al. 2002). Due to their AT-rich nature, L1 target sites display a strong negative correlation with GC content (data not shown), requiring an interaction term when both predictors are selected in the full regression model (Methods). The negative coefficient for the interaction term in the human-specific L1 model (Supplemental Table S3), for instance, indicates that despite the strong positive correlation between L1 density and L1 target site density, the magnitude of this relationship decreases in regions with increasing GC content. Nevertheless, although human-specific L1s insert into GC-poor regions, all other L1s increase in density with increasing GC content, as reported in previous pairwise analyses of L1s and GC content (e.g., Lander et al. 2001; Jurka et al. 2004).
Our models reveal that the density of most conserved elements is a strong negative predictor for all L1 densities, explaining ∼2%–11% of L1 density variation genome-wide (Fig. 1B; Supplemental Table S3). This implies that L1 densities are sparse in regions of the genome rich in most conserved elements. In contrast, other features used as proxies of natural selection, namely, gene content and cis-NATs, are not significant for any L1 model (Supplemental Table S3).
As observed for Alus, human-specific L1 densities strongly negatively correlate with the density of recombination hotspots: this variable explains ∼9.9% of total human-specific L1 density variability genome-wide (Fig. 1B; Supplemental Table S3). Recombination hotspots display a positive association with L1PB densities (RCVE ∼ 5.5%), yet, the sex-specific recombination rates (as for Alus) contribute only small fractions (RCVE ≤ 1.5%) of the overall variability in L1 densities (Fig. 1B; Supplemental Table S3). Interestingly, the telomerase-associated hexamer is a weak positive predictor for human-specific L1 densities (with RCVE ∼ 1%), possibly reflecting an alternative L1 endonuclease-independent mechanism for integration (Morrish et al. 2007; Sen et al. 2007) and/or their recruitment during DSB repair (Sen et al. 2007). The frequency of the genome instability 13-mer, a strong positive predictor for all Alu densities, displays a negative correlation with intermediate HCO L1PAs and L1PBs (RCVEs ∼ 2.5%) and a strong positive correlation with ancestral L1Ms explaining an additional 23% of their variability (Fig. 1B; Supplemental Table S3).
Similar to the results for variation in Alu repeat densities, some genomic features predict genome-wide variability of L1s of a particular evolutionary age only. For instance, the densities of intermediate L1s (HC and HCO L1PAs) and ancestral L1PBs positively associate with density of nucleosome-free regions that explain ∼1%–4% of L1 variability; human-specific L1s and ancestral L1Ms are not significantly associated with nucleosome-free regions (Supplemental Table S3).
In contrast to Alus, however, CpG islands are a weak positive predictor for all L1 elements (RCVEs ∼ 1%–1.5%; Fig. 1B; Supplemental Table S3). The density of germline-expressed genes is a significant but weak negative factor in determining variability in ancestral L1M densities (RCVE < 1%).
Distribution of Alus and L1s on sex chromosomes versus autosomes
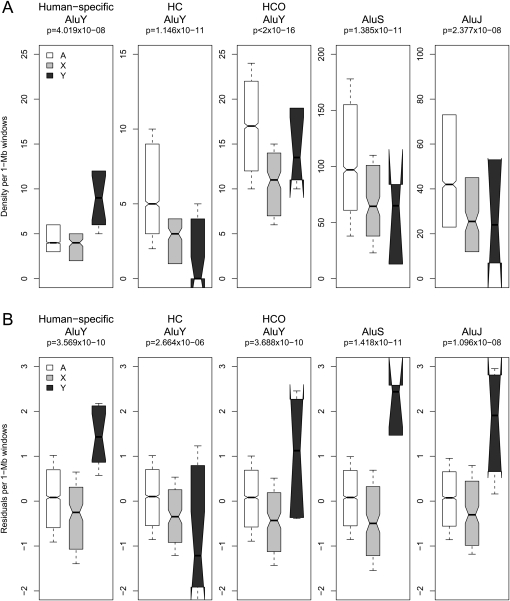
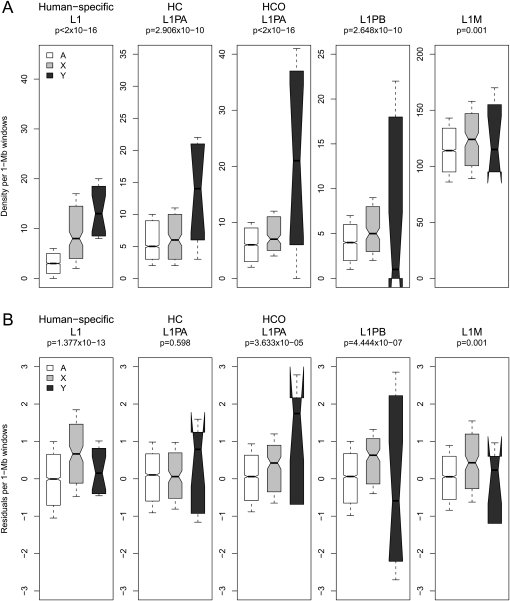
Next, we compared Alu and L1 densities on sex chromosomes with their corresponding densities on autosomes, separately for each integration time point. We performed regressions of the observed TE densities using categorical variables indicating location of a window on an autosome (A) or sex chromosome (X or Y) (Methods). Consistent with previous analyses (see introduction), we detect significant differences in Alu and L1 densities both among the various chromosome types and the individual subfamilies (Figs. 2, 3). In particular, human-specific AluY densities measured in 1-Mb windows are significantly higher on Y, followed by autosomes and lowest on X (categorical regression P-value = 4.02 × 10−8); all other Alu element densities are significantly higher on the autosomes with respect to both sex chromosomes (P-values ≤ 2.38 × 10−8; for details, see Fig. 2A). In contrast, observed L1 densities are significantly higher on the Y for all relatively recent L1 integrations analyzed here (human-specific, HC, and HCO; P-values ≤ 2.91 × 10−10; Fig. 3A), but highest on X for L1PB (P-value = 2.65 × 10−10) and L1M elements (P-value = 0.001; Fig. 3A).
Figure 2.
Human sex chromosome (X, Y) versus autosome (A) distribution of observed (A) and corrected (B) densities of Alus in 1-Mb windows genome-wide. (A) Observed densities are plotted for Alu elements of various evolutionary ages including human-specific AluYs; human–chimpanzee branch-specific (HC) AluYs; and human–chimpanzee–orangutan branch-specific (HCO) AluYs; AluS and AluJ subfamilies, on the autosomes (white), X (light gray), and Y (dark gray) chromosomes. (B) Residuals from genome-wide multiple regression models represent densities corrected for local variation in genome landscape features. Notches on boxplots indicate the 95% confidence interval of the median.
Figure 3.
Human sex chromosome (X, Y) versus autosome (A) distribution of observed (A) and corrected (B) densities of L1 elements in 1-Mb windows genome-wide. (A) Observed densities are plotted for L1s of various evolutionary ages including human-specific L1s; human–chimpanzee branch-specific (HC) L1PAs; and human–chimpanzee–orangutan branch-specific (HCO) L1PAs, L1PB, and L1M subfamilies, on the autosomes (white), X (light gray), and Y (dark gray) chromosomes. (B) Residuals from genome-wide multiple regression models represent densities corrected for local variation in genome landscape features. Notches on boxplots indicate the 95% confidence interval of the median.
Since observed TE densities on the various chromosome types reflect the combined influences of local genome landscape features and location on X, Y, or autosomes, we next analyzed the densities of TEs on sex chromosomes versus autosomes after accounting for variation due to inherent chromosomal differences in genomic properties. In other words, we sought to determine whether the sex-chromosome-biased distribution observed above is preserved after we correct for the unique genomic landscapes on sex chromosomes. For each Alu and L1 subfamily, the residuals from the above genome-wide models (resulting in “corrected” TE densities) were regressed on categorical variables indicating window location on a particular chromosome type, similar to above.
Despite substantial variation in many genomic characteristics, almost all of the TEs analyzed here continue to exhibit sex-chromosome-biased distributions after applying the correction. Additionally, we frequently note a stark contrast between observed versus corrected densities in 1-Mb windows, on the autosomes, X, and Y (Figs. 2, 3)—suggesting that the significance of location on a particular chromosome type is not merely a reflection of the differences in landscape features. For instance, the corrected Alu densities are significantly higher on the Y than on the X chromosome for all subfamilies except for HC AluY (P-values ≤ 1.10 × 10−8; Fig. 2B; regression model details in Supplemental Table S3); at the same time, the corrected Alu densities are still significantly higher on autosomes than on X for all subfamilies (Fig. 2B), consistent with the observed densities (Fig. 2A). The explanatory power for location of a window on the X, the Y, or an autosome explains an additional ∼1%–2% variability for corrected Alu densities (Supplemental Table S3).
The corrected densities of human-specific L1 integrations are significantly higher on the X, intermediate on the Y, and lowest on autosomes, in contrast with the pattern for observed densities (P-value = 1.38 × 10−13; Fig. 3B). The pattern of highest corrected densities on the X is also noted for the ancestral L1PB (P-value = 4.44 × 10−7) and L1M (P-value = 0.001) subfamilies (Fig. 3B; Supplemental Table S3). The corrected densities of HC L1PAs are not significantly different among the various chromosome types (regression model P-value not significant) (Fig. 3B; Supplemental Table S3), whereas corrected HCO L1PAs display the same distribution pattern as the observed densities. Similar to Alus, the sex-chromosome indicator variables explain relatively small but significant portions of variability in corrected L1 densities: location of a window on chromosomes X, Y, or autosomes contributes from <1% to at most 2% to the variability in corrected densities of the analyzed human L1s (Supplemental Table S3).
Genome landscape features driving chromosome-specific effects
We investigated which of the above genome landscape features could be important to establishing the noted change in observed versus corrected distribution patterns for Alu and L1 densities, suggesting that a feature exhibits chromosome-specific effects leading to the observed relative TE densities on the autosomes, X, and Y. To accomplish this, we tested each significant predictor in genome-wide regressions (Supplemental Table S3) for each TE model as follows. We removed each predictor of interest sequentially, performed multiple regression on the subset of genome landscape features (reduced model, i.e., excluding the terms related to the predictor of interest; see Methods), and regressed residuals of each reduced model on categorical variables indicating location of a window on the X, Y, or autosomes, as above. If the resulting pattern (the relative densities among the chromosome types) was similar to that for the corrected TE densities, we inferred that the predictor of interest was not important to determining chromosome-specific effects, since other predictors remaining in the reduced model contribute to the distribution pattern. However, a resulting pattern similar to observed densities implied that the predictor removed from the model was, indeed, driving the change in pattern, and responsible for chromosome-specific effects.
Of the Alus analyzed here, HCO AluY, AluS, and AluJ densities noticeably change in pattern between observed versus corrected densities (Fig. 2). Interestingly, the reduced models for each of these Alus, regardless of which predictor of interest is excluded, display patterns similar to the corrected density distributions on A, X, and Y (data not shown).
Several L1 subfamilies also exhibit a stark contrast in their distribution patterns on the chromosome types, before versus after correction for genome landscape features (Fig. 3). Reduced models for the human-specific L1s, when excluding most conserved elements density and recombination hotspot density, separately, each display distribution patterns similar to observed densities (Supplemental Fig. S2) and thus likely drive the observed sex-chromosome-biased distributions. Similarly, for the HC L1s, the only TE model analyzed here that shows no significant difference in corrected densities among the chromosome types (Fig. 3B), the reduced model omitting most conserved elements shows a pattern similar to observed densities (Supplemental Fig. S2).
Distributions of Alu and L1 subfamilies in primate genomes
Following the strategy described above, we analyzed lineage-specific TE densities in chimpanzee and orangutan, separately, using landscape features available for each of these genomes (Supplemental Table S4). Despite fewer annotated features, our models explain considerable fractions of variability in primate lineage-specific Alu and L1 densities (from ∼20% to 64% of variability) (Supplemental Table S4). Shared model characteristics across primates reveal diagnostic patterns predictive of Alus and L1s: Alus negatively correlate with GC content; most conserved elements, rather than genes, provide substantial explanatory power in predicting TE densities genome-wide; and, the 13-mer associated with DSBs is a positive predictor for Alus in all analyzed primates (Supplemental Table S4).
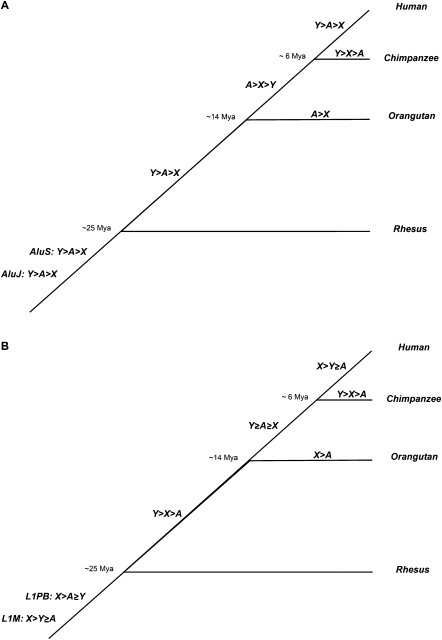
We observe similar sex-chromosome-biased distributions of corrected lineage-specific L1 and Alu densities in both the orangutan and the human genomes (Fig. 4; Supplemental Fig. S3). Orangutan (for which Y chromosome sequence is not yet available), like human, exhibits higher corrected densities of lineage-specific Alus on autosomes than on the X, but densities of lineage-specific L1s are highest on X relative to autosomes.
Figure 4.
Sex-chromosome biased patterns of corrected TE densities in primates. Relative densities of Alus (A) and L1s (B), after accounting for regional variation in local genome landscape features, are compared among the X, Y, and autosomes (A) for each branch in the primate phylogeny. Statistically significant differences are indicated with inequality symbols (>), whereas insignificant differences are indicated with equality symbols (≥).
However, corrected lineage-specific TE distribution trends in chimpanzee, the only non-human primate species for which Y-chromosome sequence is available (The Chimpanzee Sequencing and Analysis Consortium 2005; Hughes et al. 2005; Kuroki et al. 2006), exhibit differences to those in human (Fig. 4; Supplemental Fig. S4). Interestingly, the distributions by chromosome type for both observed and corrected densities of chimpanzee-specific AluYs and L1s share the same patterns: densities are highest on Y, intermediate on X, and lowest on the autosomes (Fig. 4; Supplemental Fig. S4). To test whether differences between human- and chimpanzee-specific sex chromosome distributions are due to the limited subset of genomic features available for chimpanzee, we reanalyzed human-specific TEs using the subset of genomic features common for all primates (Supplemental Table S2). Despite a moderate decrease in the variability explained, the sex-chromosome patterns observed for human-specific TE densities hold (Supplemental Table S4).
Discussion
Here, we demonstrate that local genome landscape features and location on sex chromosomes versus autosomes contribute significantly to shaping Alu and L1 densities over evolutionary time (Fig. 4). To our knowledge, this is the first comprehensive study combining local variation modeling and sex-chromosome versus autosome analysis of TE densities. The multiple regression models of TE densities presented here explain substantial proportions of variability in Alu and L1 genome-wide densities and illuminate the mechanisms shaping TE integration and fixation preferences over evolutionary time.
Mechanisms contributing to the genome-wide distribution of Alu densities
What mechanisms could be shaping the genome-wide distribution of Alus? Here (and also below for L1s), we highlight the most likely mechanisms associated with each significant genomic factor (Table 1; Table S2); we cannot exclude the possibility that other processes might also be involved. Our analysis confirms the role of previously identified target site sequences for Alu integration (Supplemental Tables S3, S4; Jurka 1997; Cost et al. 2002), and the AT-rich local base composition preferences of young (here, human-specific) Alus (Fig. 1; Lander et al. 2001; Hardison et al. 2003; Yang et al. 2004). As previously noted (e.g., Lander et al. 2001; Hardison et al. 2003; Yang et al. 2004; see introduction), both Alu and L1 densities strongly associate with GC content. However, here we observe that, when simultaneously considering other genome features, GC content is a negative predictor for almost all Alus analyzed but decreases in its explanatory power over time (Fig. 1; Supplemental Table S3). This distinction between pairwise and multiple regression analyses could be due to several factors. For example, our observations might emphasize the importance of scale for genomic analysis (Berry et al. 2006); previous analyses of Alus and the dependence on local GC content have focused on scales of 50 kb (Jurka et al. 2004; Yang et al. 2004) and 5 Mb (Hardison et al. 2003), but not 1 Mb, as done here. Additionally, the subtle changes noted for Alus over evolutionary time could be masked when all Alus (or all SINEs) are analyzed together. Alternatively, differences could be attributed to the ability of multiple regression models to tease apart the underlying correlations between GC content and additional genome characteristics that are aggregated when GC content is modeled individually (e.g., Lander et al. 2001; Hardison et al. 2003; Yang et al. 2004). Remaining features related to integration preferences, such as measurements of transcriptional activity (CpG island density, nucleosome-free region density, and germline-expressed gene density), each contribute comparatively small fractions to explaining variation in Alu densities (Supplemental Table S3).
We demonstrate that simple sequence degradation (via accumulation of substitutions and/or small indels) is not a likely explanation for establishing Alu densities over evolutionary time because these predictors did not contribute to any of the Alu models (Supplemental Table S3; Belle et al. 2005). Nor do the genome regression models indicate a significant role for recombination-mediated deletion in shaping Alu densities. Male- and female-specific recombination rates display very weak correlations with older Alu elements (Supplemental Table S3), whereas computationally predicted recombination hotspots most strongly negatively associate with human-specific Alu densities (Fig. 1A), suggesting that Alus insert into regions of the genome experiencing low levels of recombination. The corresponding contributions of recombination hotspots to models of Alu densities decrease with the age of TE integration (Supplemental Table S3), consistent with the rapid evolutionary movement of hotspots (for review, see Coop and Przeworski 2007). In addition, the high explanatory power of Alu models (37%–51%) for each primate species analyzed here, including species that yet lack recombination data, suggest that features other than recombination contribute more to shaping Alu densities over evolutionary time scales (Table 1; Supplemental Table S4).
In contrast, the frequency of a 13-mer defining genome instability (Myers et al. 2008) is positively associated with all human Alu subfamilies (Fig. 1A) and with lineage-specific Alus in all primate species analyzed here (Supplemental Table S4). We calculate motif's frequency as the ratio of observed to expected frequencies; thus, the observed positive correlation implies that Alus increase in density with regions enriched for this motif, and thus double-stranded breaks, after correction for sequence similarity. Alu densities also strongly negatively correlate with GC content, consistent with elevated inter-Alu recombination in GC-rich regions (Sen et al. 2006). Together, these results imply that Alus contribute to instability themselves by providing sequences that promote nonallelic homologous recombination (NAHR) (Batzer and Deininger 2002; Sen et al. 2006). Conversely, Alu elements might contribute to the evolution of the recombination landscape; in light of recent findings that revealed a lack of correspondence between the 13-mer and current recombination hotspot activity in chimpanzee (Myers et al. 2010), a potential role for Alus in this process awaits further investigation.
Interestingly, NAHR could potentially explain the contradictory pattern for observed versus corrected densities of Alus on the Y chromosome: over time, we note a decline in observed densities of Alus on Y, while the relative corrected densities are highest compared to autosomes and X (Fig. 2). Gene conversion is known to occur frequently on the Y (Rozen et al. 2003; Skaletsky et al. 2003).
Our results suggest that selection could be at most a weak determinant in shaping the distribution of Alus. Gene content explains only small fractions in models of genome-wide Alu densities (Supplemental Table S3), and although most conserved elements and cis-NATs increase in their explanatory power for ancestral integrations, together they explain only a small proportion of genome-wide variability of Alu densities (Fig. 1A; Supplemental Table S3).
Mechanisms contributing to the genome-wide distribution of L1 densities
Which mechanisms act to establish the distribution of L1s? First, several features related to potential integration preferences were tested in our analysis. Similar to Alus, we confirm that previously identified target site sequences (Jurka 1997; Cost et al. 2002) are a major determinant for L1 integration (Fig. 1B). In fact, the density of L1 target sites is consistently among the most significant predictors explaining variability in all TEs analyzed here, particularly for the L1s (Supplemental Tables S3, S4; Berry et al. 2006). However, in contrast to Alus, GC content positively correlates with densities of all but human-specific L1s and remains a highly significant factor at all integration time points (Fig. 1; Supplemental Table S3). Associations with replication timing and transcriptionally active DNA, measured by CpG island density, contribute only small fractions to the genome-wide variability in L1 densities; whereas, in contrast to Alus, germline-expressed gene density is significant to L1Ms only (Supplemental Table S3). Thus, L1 genome-wide densities are predominantly determined by the local DNA sequence composition.
Second, natural selection could be a significant factor in shaping genome-wide L1 densities (Table 1). Interestingly, the density of most conserved elements rather than gene content is a significant predictor for TE densities in all primates analyzed, confirming previous observations (Simons et al. 2006; Sironi et al. 2006). Indeed, most conserved elements explain up to ∼11% of genome-wide variability in L1 densities (Fig. 1); young L1s are not tolerated in regions rich in these elements, and older L1s maintain this strongly negative association throughout the evolutionary time studied here (Fig. 1B). These results indicate a possible role for natural selection, since most conserved elements are likely to contain functional DNA (Siepel et al. 2005). Third, neither substitution rates nor small indel rates were significant in our genome-wide models of L1 densities (Supplemental Table S3); thus, simple degradation over evolutionary time is an unlikely explanation (Belle et al. 2005). Fourth, like the case for Alus, meiotic recombination is not sufficient to explain the densities of L1s because of the minimal contributions of recombination-related features in our models. Human-specific L1s strongly negatively correlate with the density of recombination hotspots (Fig. 1; Supplemental Table S3), yet such hotspots explain smaller fractions of variability for the ancestral L1PBs only (Fig. 1; Supplemental Table S3). Moreover, male and female sex-specific recombination rates contribute only ∼1% to variability in HC and HCO L1 models, again diminishing the potential role of recombination in determining L1 densities over time (Supplemental Table S3).
Genome landscape features and mechanisms accounting for sex-chromosome-biased distributions in Alu and L1 densities
Our results indicate a drastic change, hence “revolution,” in pattern of sex-chromosome-specific densities for many Alu and L1 subfamilies after correcting for variation in genome landscape features. Which genomic variables might account for such a substantial difference? None of the features alone was significant in shaping the observed density patterns of AluSs, AluJs, and HCO AluYs, suggesting that either a combination of several variables accounts for Alu chromosome-specific effects, and/or that chromosomal location produces a true effect that cannot as yet be captured by the features in our models. For human-specific L1s, the densities of most conserved elements and recombination hotspots, separately, reproduced the observed density pattern—meaning that these features themselves have significantly different densities between the sex chromosomes and autosomes that assist in establishing the sex-chromosome-biased distribution. Interestingly, only most conserved elements' density was a significant predictor in determining the observed distribution pattern of HC L1s. Together, these results reinforce the potential for natural selection to have shaped the genome-wide densities of L1s throughout the evolutionary time frame investigated here.
Mechanisms explaining sex-chromosome-biased TE distributions, after accounting for local genomic landscapes
The persistence of the sex chromosome bias in many analyzed Alu and L1 subfamilies, after applying a correction for local genomic factors, implies that location on X, Y, or autosome is important to establishing TE density variation in addition to the genomic landscape differences among chromosomes. What could potentially explain this?
First, patterns for Alus are consistent with elevated integration in the male germline, whereas L1s appear to integrate both in the male and female germlines (Table 1). Indeed, after correcting for genome landscape features (Supplemental Table S2), the densities for all Alus become highest on Y, intermediate on autosomes, and lowest on X (Fig. 2B). This pattern is consistent with the amount of time each chromosome type spends in the male germline (Jurka et al. 2002).
The situation is different for L1s: distribution patterns for corrected human-specific L1 densities (Fig. 3) are not consistent with either exclusively male- or female-specific germline integration, nor are they consistent with equal integration in both sexes (Table 1). Nevertheless, male germline integration of L1 elements is suggested by the higher densities (both observed and corrected) of human-specific L1s on the Y chromosome relative to autosomes, after accounting for many genomic characteristic differences between the sex chromosomes and autosomes. Furthermore, the X chromosome exhibits higher observed and corrected densities of human-specific L1s than do autosomes (Fig. 3), suggesting that L1 integration is likely to occur prior to meiotic sex chromosome inactivation (MSCI) (for review, see Ellis and Affara 2006), when the X is inactivated during spermatogenesis. Importantly, the sex chromosomes, not the autosomes, remain mostly unpaired during prophase I, thus accommodating the integration of TEs on the sex chromosomes in male germ cells (Morelli and Cohen 2005). L1 integration is likely to occur in the female germline as well, since the X exhibits higher corrected densities of human-specific L1s than either autosomes or Y (Fig. 3); thus, our results agree with the integration of L1s in early embryogenesis (Kano et al. 2009). Consistent with recent evidence suggesting that telomerase might facilitate noncanonical L1-endonuclease-independent integrations in the rodent female germline (Morrish et al. 2007), we observe a positive correlation between the human-specific L1s and the frequency of telomerase hexamer motif (Supplemental Table S3). L1s are hypothesized to act as genomic “bandages” that repair DSBs (Sen et al. 2007), and female meioses are known to be more error-prone than male meioses during recombination, DSB formation, and repair pathways (Morelli and Cohen 2005).
Second, in addition to the germline integration of L1s, our results suggest that a potential involvement of L1s in XCI (Lyon 1998) might also contribute to determining their sex-chromosome-biased densities. Indeed, regulation of gene expression is critical on the X, where dosage compensation between males and females is achieved via XCI (Carrel and Willard 2005; Chow et al. 2005). Interestingly, a recent study suggested that L1M elements, rather than recent L1PA integrations, are more likely to play a role in XCI (Abrusan et al. 2008). Our models of sex-chromosome-biased distributions of L1s indicate a stronger significant difference for corrected densities of the L1P elements on the X, in contrast to this hypothesis and consistent with another study (Carrel et al. 2006).
Third, comparisons with other primate species highlight additional peculiarities of the sex-chromosome-biased distributions of Alu and L1 densities over evolutionary time (Figs. 3, 4). Indeed, sex-chromosome L1 densities in human and chimpanzee are likely more than mere reflections of original integration preferences. For instance, although models of human- and chimpanzee-specific L1s show parallel associations with genomic features (Supplemental Tables S3, S4) and similar observed TE distributions (Fig. 3A; Supplemental Fig. S4A), the patterns of corrected distributions are strikingly different: chimpanzee densities are highest on Y, intermediate on X, and lowest on autosomes (Supplemental Fig. S4B), whereas human-specific densities are highest on X, intermediate on Y, and also lowest on autosomes (Fig. 3B). This implies that L1s experienced a relative increase in density on the human X, or conversely, decreased density on the human Y since the time of human–chimpanzee divergence.
We hypothesize that genetic drift could potentially account for the higher corrected Alu densities on the chimpanzee X versus autosomes (Fig. 4; Supplemental Fig. S4), since chimpanzee effective population size is estimated to be relatively large, about 35,000 individuals compared to about 11,000 for human (Kaessmann et al. 1999) and about 10,000 for orangutan (Goossens et al. 2006). An alternative would be preferential integration of Alus on chimpanzee X rather than autosomes; however, since models of lineage-specific Alus in primates share significant predictors (and thus integration preferences), we are unaware of a mechanism that would lead to such a pattern.
Methods
Classification of primate transposable elements
Repeat densities for various L1 (L1PA, L1PB, L1M) and Alu (AluY, AluS, AluJ) subfamilies were obtained from RepeatMasker (AFA Smit, R Hubley, P Green. 1996–2004. RepeatMasker Open-3.0, http://www.repeatmasker.org) annotations for each genome analyzed here (hg18 for human, panTro2 for chimpanzee, ponAbe2 for orangutan [The Oranguntan Genome Sequencing Consortium, in prep.], and rheMac2 for macaque), available at the UCSC Genome Browser (Karolchik et al. 2008). Given the well-established phylogeny of these species (Fig. 4; Glazko and Nei 2003; Burgess and Yang 2008), we considered the following evolutionary scenarios for integration of transposable elements: lineage- or species-specific (along external branches), intermediate (along internal branches), and ancestral (in the common ancestor of the studied species).
To determine TE integration timing, we modified previous methods (Walser et al. 2008). Namely, we determined the coordinates of orthologous TEs in target genomes (e.g., chimpanzee) by converting the coordinates of species-specific TEs from the query genome (e.g., human), using the lift-Over utility implemented in Galaxy (minimum mapping ratio of 0.9 bases) (Blankenberg et al. 2007). Each studied primate genome was sequentially considered as a query. This was performed for recently active subfamilies of L1PA (active ∼3–100 million years ago [Mya]) (Khan et al. 2006) and AluY (0∼30 Mya) (Batzer and Deininger 2002) for each target genome. “Lineage-specific TE integrations” were inferred as repeat annotations unique to the target lineage, that is, not aligning (or aligning at most by 10% coverage of repetitive sequence) with orthologous repeats from any other studied genome. In addition, annotated species-specific TEs, for example, L1HSs for human (Smit et al. 1995), and L1Pts for chimpanzee (The Chimpanzee Sequencing and Analysis Consortium 2005; Mills et al. 2006), were added to our lineage-specific data set (Supplemental Table S1).
“TE integrations corresponding to intermediate timing” were defined as repeats of the same subfamily annotation present in the same genomic location: in human and chimpanzee (HC) and absent from orangutan and rhesus; or in human, chimpanzee, and orangutan (HCO), and absent from rhesus. We allowed for up to 10% variability in the lengths of orthologous repeat copies, consistent with allowed indel divergence between repeat and consensus sequences during the annotation process (AFA Smit, R Hubley, P Green. 1996–2004. RepeatMasker Open-3.0. http://www.repeatmasker.org). The Y chromosome was an exception to this strategy, as the orangutan genome was sequenced from a female and thus currently lacks Y sequence. Since sequence for chimpanzee Y is of finished quality (Hughes et al. 2005; Kuroki et al. 2006), we analyzed the distribution of human–chimpanzee coverage for TEs on Y, in comparison to the coverage on X, and autosomes (data not shown). We confirmed that a strict filtering criterion was appropriate to distinguish HC shared integration from HCO integration on Y (100% length match was inferred as HC integration, otherwise we allowed up to 10% variability to infer an HCO integration); however, altering these thresholds did not influence our final results.
“Ancestral integrations” were defined as those elements inserted into the common ancestor of the species analyzed: events of the same subfamily annotation, present in the same genomic location, and overlapping at >90% of their lengths in each of human, chimpanzee, orangutan, and rhesus genomes. We investigated various thresholds (100%, 95%, 90%) of orthologous repeat coverage for classification of all repeat integrations, to confirm the robustness of general regression model trends (data not shown). Ancestral integrations were represented by TE subfamilies that ceased active retrotransposition prior to the divergence of the analyzed primates, namely, L1PB (active ∼46–96 Mya), L1M (61–102 Mya), AluS (∼35 Mya), AluJ (55–65 Mya) (Supplemental Table S1; Batzer and Deininger 2002; Khan et al. 2006). A limited number of repeats from AluYs and L1PAs inserted in the common ancestor of the primates analyzed (data not shown) and were not studied here.
To focus on the forces shaping TE insertions, we excluded all TEs that intersected with segmental duplications, which could be the result of duplication rather than retrotransposition. We obtained coordinates of duplicated regions for both human and chimpanzee (Cheng et al. 2005). Next, using Galaxy, we used the lift-Over utility to convert to hg18, and intersected human TEs at all integration time points with resulting coordinates of both human-specific and human–chimpanzee-shared duplication (Blankenberg et al. 2007); these events were removed from analysis. Although we applied a similar strategy intersecting chimpanzee-specific TEs with chimpanzee-specific and human–chimpanzee-shared segmental duplications, together, the resulting numbers were sufficiently small (178 AluYs and 183 L1s, <1.5% of each subfamily) (Supplemental Table S1) that filtering was not deemed necessary. Since human–chimpanzee-shared segmental duplications could have occurred prior to orangutan divergence, coordinates of these segmental duplications were determined in ponAbe2 and intersected with orangutan-specific TEs; again, small numbers (74 AluYs and 85 L1PAs, <1% of events) precluded filtering.
Genome landscape features
Sequence features available for hg18 were obtained from the UCSC Genome Browser (Karolchik et al. 2008) and placed in 1-Mb windows (Supplemental Table S2). Several window features were computed directly for hg18 sequences. We defined three statistics to measure features in 1-Mb windows: density (counts per window), content (fraction of a window in base pairs [bp]), and frequency (ratio observed/expected frequency). The ratio of observed/expected frequency for a particular K-mer motif was calculated as the number of K-mers per site of length K divided by the total of all possible K-mers. Given that K is the motif length and L is the total window sequence length, then (L − K + 1) is the number of sites of length K, and 1/(4K) is the number of all possible sequences of length K (under the null assumption of equal base frequencies).
Several predictors likely affect TE integration, namely: GC content to reflect the local base composition; density of TTTTAA L1 target site sequence characteristic of target-primed reverse transcription (Jurka 1997; Cost et al. 2002); frequency of a telomerase hexamer TTAGGG characteristic of telomerase-dependent RNA retrotranscription and of noncanonical L1 integrations (Morrish et al. 2007; Nergadze et al. 2007); average replication timing in the S phase of the cell cycle, calculated as a ratio of S:G1-phase DNA (Woodfine et al. 2004); CpG island density as a measure of local transcription activity (Jones 1999; Hellmann et al. 2005); density of nucleosome-free regions, predicted from MNase cleavage (Ozsolak et al. 2007); and density of germline expressed genes to model transcriptional activity, corresponding to the number of genes per window expressed (defined as average difference >200) (Su et al. 2002) in either testis germ cell or ovary tissue (Su et al. 2004).
Genome sequence features contributing to mechanisms of simple sequence degradation were also incorporated. Lineage-specific nucleotide substitution rates in the primate phylogenetic tree (Fig. 4, also including marmoset) were estimated in ancestral repeats (ARs) (Hardison et al. 2003) in 1-Mb windows according to the REV model (Rodriguez et al. 1990) implemented in HyPHY using the Galaxy interface (Kosakovsky Pond et al. 2005; Blankenberg et al. 2007). Human-specific (since divergence from chimpanzee) insertion and deletion rates were calculated using previous methods (Kvikstad et al. 2007).
Several predictors were included to model the effects of recombination: sex-specific recombination rates (Kong et al. 2002), density of computationally predicted recombination hotspots estimated from linkage disequilibrium among SNPs (Myers et al. 2005), and the frequency of a 13-mer CCNCCNTNNCCNC associated with genome instability (Myers et al. 2008). To reflect the influence of selection we included gene content, density of most conserved elements that likely contain regulatory sequences (Siepel et al. 2005), and density of cis natural antisense transcripts (cis-NATs) capable of regulating gene expression (Conley et al. 2008). cis-NATs were determined following previously established methods (Conley et al. 2008): briefly, transcription start sites (TSSs) determined by cap analysis of gene expression (cage) from Japan's National Institute of Genetics (http://genomenetwork.nig.ac.jp/public/download/cage_Database_e.html; release 2007.3.28) were mapped to hg18 using Galaxy (Blankenberg et al. 2007). TSSs were intersected with human known genes (Karolchik et al. 2008) using the longest annotated transcription start and end coordinate (to score a putative TSS only once); TSSs were defined as “sense” if mapping to the same orientation as the transcribed gene, or “antisense” if mapping to the opposite orientation. Our numbers, although slightly different, were consistent with those reported in Conley et al. (2008). Last, we scored densities of cis-NATs per 1-Mb window genome-wide. A subset of sequence features in common to all primates analyzed here were obtained for each lineage, following the above methods (Supplemental Table S2).
Regression analysis
Prior to regression analyses, we applied filters to account for the lower sequence quality and coverage of draft genomes. Windows of low coverage (containing >50% Ns) and/or low sequence quality (>50% bases with phred scores < 20) were excluded. Additionally, we excluded X- and Y-chromosome windows corresponding to pseudo-autosomal regions (PAR; defined as ancestral to human–rhesus last common ancestor) (Ross et al. 2005; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), and human-specific X-transposed regions (Skaletsky et al. 2003; Hughes et al. 2005; Kuroki et al. 2006).
Multiple regression analyses, implemented in the R statistical package (R Development Core Team 2005), were performed for each repeat subfamily's density in 1-Mb windows as the response, using the various sequence features as predictors (Supplemental Table S2). First, orthogonal polynomials of second degree were included for each predictor to test for nonlinear relationships with the response, and to reduce the impact of multicollinearity (Kutner et al. 2005). Subfamily densities were log10-transformed (after addition of a constant of 1) to produce normally distributed errors and assure constant error variance; the power transformation was determined by the Box-Cox test (Kutner et al. 2005). Second, the best subsets selection procedure was used to determine the best model (smallest Mallow's Cp) (Kutner et al. 2005) from among this set of terms. Third, standard regression diagnostics were performed to identify outliers (e.g., according to Cook's distance and residuals >3 standard deviations), and assess model performance—for example, additive variable plots, residual analysis, interaction terms, variance inflation factor (VIF), and spatial autocorrelation (Kutner et al. 2005). Last, the best model was refined by excluding any non-significant predictors (P-values of the t-tests) determined by Bonferroni multiple hypothesis testing. The relative contribution to variability explained (RCVE) (see Kvikstad et al. 2007) was calculated to summarize each predictor's contribution to the full model in the context of all other predictors. Briefly, this statistic is similar to the commonly used measure partial R2 (share of variability explained); RCVE compares the regression sum of squares (SSR) of the full model (including all significant terms) to that of the reduced model (i.e., the full model excluding terms related to the predictor of interest), to quantify the improved reduction in error of the regression model (i.e., “increased fit”) attained by inclusion of the predictor of interest.
Next, the residuals of above regressions were used as “corrected” subfamily densities (i.e., accounting for inherent variability in the genome features), to determine chromosomal biases due to location of a 1-Mb window on an autosome (A), or sex chromosome (X and, where available, Y). We performed a one-way analysis of variance (ANOVA) with three categorical variables (one for each chromosome type) to determine the significance (F-test) of the regression model, applied Tukey HSD test to correct for multiple comparisons and determine significant differences among variables, and assessed the explanatory power of each model using adjusted R2.
During calculation of RCVE (above), we used the reduced regression models to determine each significant predictor's contribution to chromosome-specific effects for each TE model. We performed regressions of the residuals from each reduced model (minus predictor of interest) using the sex-chromosome categorical variables. Resulting patterns of residuals on the X, Y, and autosomes were compared to observed and corrected densities to infer the action of the excluded predictor of interest in shaping chromosomal biases.
Acknowledgments
We thank Francesca Chiaromonte for insightful discussions, Melissa Wilson for comments on the manuscript, the Galaxy support team for helpful assistance, the Genome Sequencing Center at WUSTL for providing and allowing us to use the orangutan sequence data, and Jeffrey Sorley for graphic design. This study was supported in part by NIH grant R01-GM072264 (K.D.M.) and The Pennsylvania State University Jeanette Ritter Mohnkern Graduate Student Scholarship in Biology (E.M.K.).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099044.109.
References
- Abrusan G, Krambeck H-J 2006. The distribution of L1 and Alu retroelements in relation to GC content on human sex chromosomes is consistent with the ectopic recombination model. J Mol Evol 63: 484–492 [DOI] [PubMed] [Google Scholar]
- Abrusan G, Giordano J, Warburton PE 2008. Analysis of transposon interruptions suggests selection for L1 elements on the X chromosome. PLoS Genet 4: e1000172. doi: 10.1371/journal.pgen.1000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci 97: 6634–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–380 [DOI] [PubMed] [Google Scholar]
- Belle EMS, Webster MT, Eyre-Walker A 2005. Why are young and old repetitive elements distributed differently in the human genome? J Mol Evol 60: 290–296 [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE 2004. Natural genetic variation caused by transposable elements in humans. Genetics 168: 933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2: e157. doi: 10.1371/journal.pcbi.0020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova K, et al. 2007. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res 17: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano A 2001. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18: 926–935 [DOI] [PubMed] [Google Scholar]
- Burgess R, Yang Z 2008. Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol Biol Evol 25: 1979–1994 [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, Makova KD 2006. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet 2: e151. doi: 10.1371/journal.pgen.0020151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat 130: 113–146 [Google Scholar]
- Chen J-M, Stenson PD, Cooper DN, Ferec C 2005. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet 117: 411–427 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, de Jong P, Wilson RK, Paabo S, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93 [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Chow JC, Yen Z, Ziesche SM, Brown CJ 2005. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet 6: 69–92 [DOI] [PubMed] [Google Scholar]
- Conley AB, Miller WJ, Jordan IK 2008. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet 24: 53–56 [DOI] [PubMed] [Google Scholar]
- Coop G, Przeworski M 2007. An evolutionary view of human recombination. Nature 8: 23–34 [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD 2002. Human L1 target-primed reverse transcription in vitro. EMBO J 21: 5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis PJI, Affara NA 2006. Spermatogenesis and sex chromosome gene content: An evolutionary perspective. Hum Fertil 9: 1–7 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 1974. The evolutionary advantage of recombination. Genetics 78: 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G, Nei M 2003. Estimation of divergence times for major lineages of primate species. Mol Biol Evol 20: 424–434 [DOI] [PubMed] [Google Scholar]
- Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW 2006. Genetic signature of anthropogenic population collapse in orang-utans. PLoS Biol 4: e25. doi: 10.1371/journal.pbio.0040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, Huang CT, Sandifer E, Hebert K, Barnes EW, et al. 2007. Mobile DNA in Old World monkeys: A glimpse through the rhesus macaque genome. Science 316: 238–240 [DOI] [PubMed] [Google Scholar]
- Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA 2008. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci 105: 19366–19371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Roskin KM, Yang S, Diekhans M, Kent WJ, Weber R, Elnitski L, Li J, O'Connor M, Kolbe D, et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res 13: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE 2005. Why do human diversity levels vary at a megabase scale? Genome Res 15: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC 2005. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature 437: 101–104 [DOI] [PubMed] [Google Scholar]
- Jones P 1999. The DNA methylation paradox. Trends Genet 15: 34–37 [DOI] [PubMed] [Google Scholar]
- Jurka J 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci 94: 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Krnjajic M, Kapitonov V, Stenger JE, Kokhanyy O 2002. Active Alu elements are passed primarily through paternal germlines. Theor Popul Biol 61: 519–530 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kohany O, Pavlicek A, Kapitonov VV, Jurka MV 2004. Duplication, coclustering, and selection of human Alu retrotransposons. Proc Natl Acad Sci 101: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Paabo S 1999. Extensive nuclear DNA sequence diversity among chimpanzees. Science 286: 1159–1162 [DOI] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH Jr 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & Dev 23: 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res 36: D773–D779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Martienssen R 2008. RNAi, heterochromatin, and the cell cycle. Trends Genet 24: 511–517 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet 31: 241–247 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S, Frost S, Muse S 2005. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21: 676–679 [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Toyoda A, Noguchi H, Taylor TD, Itoh T, Kim D-S, Kim D-W, Choi S-H, Kim I-C, Choi HH, et al. 2006. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat Genet 38: 158–167 [DOI] [PubMed] [Google Scholar]
- Kutner MH, Nachtsheim CJ, Neter J, Li W 2005. Applied linear statistical models McGraw-Hill, New York [Google Scholar]
- Kvikstad E, Tyekucheva S, Chiaromonte F, Makova K 2007. A macaque's-eye view of human insertions and deletions: Differences in mechanisms. PLoS Comput Biol. 3: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lyon M 1998. X-chromosome inactivation: A repeat hypothesis. Cytogenet Cell Genet 80: 133–137 [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE 2006. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet 78: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli MA, Cohen PE 2005. Not all germ cells are created equal: Aspects of sexual dimorphism in mammalian meiosis. Reproduction 130: 761–781 [DOI] [PubMed] [Google Scholar]
- Morrish T, Garcia-Perez J, Stamato T, Taccioli G, Sekiguchi J, Moran JV 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446: 208–212 [DOI] [PubMed] [Google Scholar]
- Muller HJ 1964. The relation of recombination to mutational advance. Mutat Res 1: 2–9 [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324 [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet 40: 1124–1129 [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Santagostino M, Salzano A, Mondello C, Giulotto E 2007. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol 8: R260. doi: 10.1186/gb-2007-8-12-r260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Song J, Liu X, Fisher D 2007. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25: 244–248 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2005. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 13: 222–234 [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Oliver JL, Marin A, Medina J 1990. The general stochastic model of nucleotide substitution. J Theor Biol 142: 485–501 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423: 873–876 [DOI] [PubMed] [Google Scholar]
- Rubin C, VandeVoort C, Teplitz R, Schmid C 1994. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res 22: 5121–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH Jr 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci 103: 6611–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA 2006. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet 79: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Huang CT, Han K, Batzer M 2007. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res 35: 3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Pheasant M, Makunin IV, Mattick JS 2006. Transposon-free regions in mammalian genomes. Genome Res 16: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Comi G, Cereda M, Cagliani R, Bresolin N, Pozzoli U 2006. Gene function and expression level influence the insertion/fixation dynamics of distinct transposon families in mammalian introns. Genome Biol 7: R120. doi: 10.1186/gb-2006-7-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837 [DOI] [PubMed] [Google Scholar]
- Smit A, Toth G, Riggs A, Jurka J 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246: 401–417 [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci 99: 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyekucheva S, Makova KD, Karro JE, Hardison RC, Miller W, Chiaromonte F 2008. Human–macaque comparisons illuminate variation in neutral substitution rates. Genome Biol 9: R76. doi: 10.1186/gb-2008-9-4-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat L, Gagnier L, Medstrand P, Mager D 2005. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res 15: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser J-C, Ponger L, Furano A 2008. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res 18: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP 2004. Replication timing of the human genome. Hum Mol Genet 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Yang S, Smit AF, Schwartz S, Chiaromonte F, Roskin KM, Haussler D, Miller W, Hardison RC 2004. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res 14: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]