Abstract
X inactivation equalizes the dosage of gene expression between the sexes, but some genes escape silencing and are thus expressed from both alleles in females. To survey X inactivation and escape in mouse, we performed RNA sequencing in Mus musculus × Mus spretus cells with complete skewing of X inactivation, relying on expression of single nucleotide polymorphisms to discriminate allelic origin. Thirteen of 393 (3.3%) mouse genes had significant expression from the inactive X, including eight novel escape genes. We estimate that mice have significantly fewer escape genes compared with humans. Furthermore, escape genes did not cluster in mouse, unlike the large escape domains in human, suggesting that expression is controlled at the level of individual genes. Our findings are consistent with the striking differences in phenotypes between female mice and women with a single X chromosome—a near normal phenotype in mice versus Turner syndrome and multiple abnormalities in humans. We found that escape genes are marked by the absence of trimethylation at lysine 27 of histone H3, a chromatin modification associated with genes subject to X inactivation. Furthermore, this epigenetic mark is developmentally regulated for some mouse genes.
In mammalian females, one of two X chromosomes is randomly inactivated during early development to equalize dosage of X-linked genes between the sexes by a process called X inactivation (Lyon 1961). Although most genes are silenced on the inactive X, some remain expressed from both the active and inactive X chromosomes (Heard and Disteche 2006). In human, 15% of X-linked genes consistently escape X inactivation, and a further 10% escape in certain tissues or individuals (Prothero et al. 2009). In mouse, the proportion of genes that escape X inactivation is unknown; yet most advances in our understanding of the molecular mechanisms of X inactivation have been made in this model organism, where expression and regulation of genes can be followed and potentially manipulated during embryogenesis. A global survey of X inactivation and escape in mouse as done in the present study provides a basis for the molecular analysis of genes that remain expressed despite being embedded in silenced chromatin.
Deficiency in escape genes is thought to cause abnormal phenotypes observed in individuals with a single X chromosome (45,X) or with a deletion of part of the X (Zinn et al. 1993). Women with such chromosomal abnormalities have Turner syndrome, characterized by short stature, infertility, and multiple other physical anomalies. Interestingly, X0 mice have very few abnormal phenotypes (Burgoyne et al. 1983a,b), suggesting that humans and mice differ in the number and identity of escape genes (Disteche 1995). By determining the X-inactivation status of mouse genes, it should be possible to exclude genes that escape X inactivation in both species as candidates for the more severe Turner phenotypes, these being presumably caused by genes that escape X inactivation only in human. To date, only four mouse escape genes (Eif2s3x, Kdm5c, Kdm6a, Mid1) have been reported (Agulnik et al. 1994; Dal Zotto et al. 1998; Ehrmann et al. 1998; Greenfield et al. 1998). In addition, XIST/Xist (X-inactivation-specific transcript), a noncoding RNA essential for the onset of X inactivation, is exclusively expressed from the inactive X (Borsani et al. 1991; Brockdorff et al. 1991; Brown et al. 1991).
RNA sequencing in combination with single nucleotide polymorphism (SNP) identification has been used to follow allele-specific gene expression, for example, to discover novel mouse imprinted genes, which are expressed from a single parental allele (Babak et al. 2008). Since X inactivation is random (Lyon 1961), allele-specific expression can only be detected in single cells or in populations of cells with skewed X inactivation. We previously derived a mouse cell line that contains an active X chromosome from Mus spretus and an inactive X from Mus musculus (C57BL/6J) (Lingenfelter et al. 1998). In the present study, the frequent SNPs between mouse species were exploited to identify the allelic origin of X-linked transcripts. We found that mice, in contrast to humans, have few genes that escape X inactivation.
X inactivation is associated with ordered epigenetic changes established during embryogenesis. Silencing is initiated by the noncoding RNA Xist, which coats the future inactive X chromosome in cis (Clemson et al. 1996), followed by loss of histone modifications associated with active chromatin and gain of modifications associated with repressed chromatin (Heard and Disteche 2006). In particular, the Polycomb group repressive complex 2 (PRC2), responsible for the deposition of the repressive mark H3K27me3 (trimethylation at lysine 27 of histone H3), is recruited to the inactive X chromosome during early embryogenesis in a Xist-dependent process (Plath et al. 2003; Silva et al. 2003; Zhao et al. 2008). The global survey of X inactivation in mouse that we performed provides a baseline to examine molecular changes associated with X inactivation and escape. To explore enrichment of H3K27me3 at escape genes, H3K27me3 profiles were compared between female and male mouse tissues and embryos using chromatin immunoprecipitation with microarray hybridization (ChIP-chip). We determined that mouse genes that escape X inactivation are depleted in the repressive histone mark, a process that is apparently developmentally regulated for some genes.
Results
SNP identification
To identify novel genes that escape X inactivation in mouse, we performed high-throughput RNA sequencing (RNA-seq) on an interspecific female cell line (Patski) (Lingenfelter et al. 1998). This cell line, originally grown from embryonic kidney, from a cross between M. spretus and a C57BL/6J (BL6) mouse with an Hprt mutation, was subsequently grown in hypoxanthine aminopterin thymidine (HAT) medium to select for cells with inactivation of the BL6 X chromosome. Complete (100%) skewing of X inactivation was verified by allele-specific primer extension assays and by late replication analyses (Lingenfelter et al. 1998). Karyotyping confirmed that the Patski cell line is a female diploid cell line (Supplemental Fig. S1).
Allele-specific expression of X-linked genes was determined based on RNA-seq reads that overlap SNPs between M. spretus and BL6. Two sources of SNPs were used to determine allelic expression. First, reads were aligned to M. spretus SNPs identified by genome sequencing (Wellcome Trust Sanger Institute, http://www.sanger.ac.uk). Second, SNPs were identified by aligning all reads to the BL6 genome, and a machine learning approach (http://svm.sdsc.edu/cgi-bin/nph-SVMsubmit.cgi) was used to discriminate SNPs from sequencing errors. This approach has the advantage of detecting novel SNPs only present in our cell line (Babak et al. 2008). However, it is biased toward regions preferentially expressed from the M. spretus allele (i.e., no SNPs would be detectable for genes with mono-allelic expression from BL6). Conversely, the first approach allowed us to detect transcripts solely expressed from the BL6 allele on the inactive X, for example, Xist. Using all identified SNPs, polymorphic regions on the mouse X chromosome were screened for bi-allelic expression by counting allele-specific reads that overlapped SNPs.
Based on the M. spretus genomic sequence available in the Sanger database, SNPs between M. spretus and BL6 cover 750 X-linked genes or ∼87% of the total number of identified X-linked genes based on a combination of UCSC mm9 RefSeq genes and Ensembl genes. Of the 15,792 SNPs located on the X chromosome, 5780 were detected by RNA-seq and thus expressed in the Patski cell line, representing 393/750 (or 52%) X-linked transcripts that have SNPs (Supplemental Table S1). Genes not detected in our analysis include those with no or low expression in the cell line, or else genes not included in the Sanger database, because of either incomplete coverage or high conservation between BL6 and M. spretus. A number of expressed SNPs were confirmed by conventional sequencing of genomic DNA and of cDNA (Figs. 1, 2). There was good agreement between SNPs present in the Sanger database and those discovered by the learning approach, which identified 187 (false discovery rate [FDR] = 0.03) novel expressed X-linked SNPs. It is possible that a subset of SNPs represented in only one set of data are false-positive calls. Alternatively, there may be differences between the genome of the M. spretus selected for sequencing and the M. spretus our cell line was derived from.
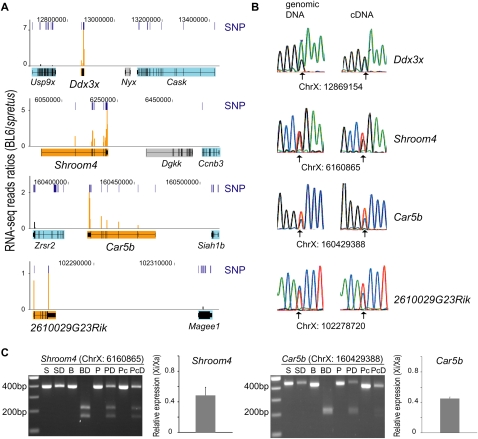
Figure 1.
RNA-seq identification of four novel escape genes (Ddx3x, Shroom4, Car5b, 2610029G23Rik). The escape genes are shown with six adjacent genes subject to X inactivation (Usp9x, Cask, Ccnb3, Zrsr2, Siah1b, Magee1). (A) RNA-seq data are graphed as the sequence read ratio (orange bars) between BL6 (inactive X) and M. spretus (active X) at each SNP (short, dark-blue bars) identified in the four escape genes (filled in orange). A few SNPs had ratios higher than those at the maximum scale (Supplemental Table S1). Flanking genes subject to X inactivation (filled in light blue) have very low BL6/spretus ratios. The inactivation status of the genes filled in gray is unknown due to the absence of expressed SNPs. Data were uploaded into the UCSC Genome Browser (Mouse July 2007 [mm9] assembly). (Top) Chromosome X coordinates. (B) Validation of SNPs and of bi-allelic expression of the escape genes by conventional sequencing of PCR and RT-PCR products. Chromatograms are shown for genomic DNA and cDNA with SNP coordinate underneath. (Arrows) Heterozygous bases. (C) Quantification of escape level of Shroom4 and Car5b. Genomic DNA and cDNA were amplified by PCR, followed by restriction endonuclease digestion. Products were separated by gel electrophoresis as shown in the gel pictures (SNP coordinate on top). (S) M. spretus DNA; (SD) M. spretus DNA digested; (B) BL6 DNA; (BD) BL6 DNA digested; (P) Patski genomic DNA; (PD) Patski genomic DNA digested; (Pc) Patski cDNA; (PcD) Patski cDNA digested. Graphs at right of the gels show relative expression levels from the inactive X (Xi) versus the active X (Xa) after real-time qPCR analyses.
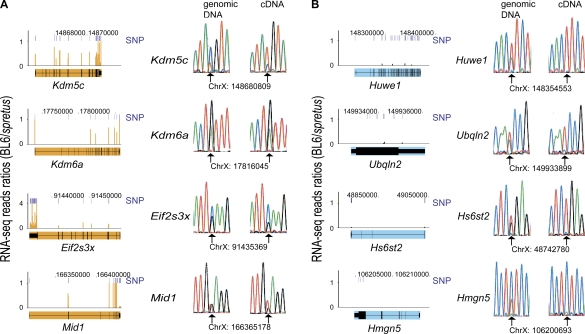
Figure 2.
RNA-seq identification and conventional sequencing validation of the X-inactivation status of mouse genes. (A) Four genes previously known to escape X inactivation (Kdm5c, Kdm6a, Eif2s3x, Mid1; filled in orange) correctly identified by RNA-seq; (B) four examples of genes subject to X inactivation (Huwe1, Ubqln2, Hs6st2, Hmgn5; filled in light blue) identified by RNA-seq. (Left) RNA-seq data are graphed as the sequence read ratio (orange bars) between BL6 (inactive X) and M. spretus (active X) at each SNP (short, dark-blue bars). A few SNPs had ratios higher than those at the maximum scale (Supplemental Table S1). Data were uploaded into UCSC Genome Browser (Mouse July 2007 [mm9] assembly). (Top) Chromosome X coordinates. (Right) Validation of SNPs and of bi-allelic (A) or mono-allelic (B) gene expression by conventional sequencing of PCR and RT-PCR products. Chromatograms are shown for both genomic DNA and cDNA with SNP coordinate underneath. (Arrows) Heterozygous bases.
Identification of escape genes and of genes subject to X inactivation in mouse
To quantify allelic expression of X-linked genes, the ratio of BL6 (inactive X) to M. spretus (active X) sequence reads was calculated for each SNP. The majority of transcripts (260/393, or 66%) identified as polymorphic showed no evidence of BL6 sequences, indicating that these genes are subject to X inactivation (Fig. 2B; Supplemental Tables S1, S2). The rest of the transcripts (133/393, or 34%) had at least one sequence read derived from BL6, consistent with some escape from X inactivation (Supplemental Tables S1, S3). However, for most of these transcripts (120/393), expression from the inactive X was very low, suggesting low or leaky expression from the silenced allele or overlap with a false-positive SNP (Fig. 3; Supplemental Table S3). Consequently, these genes were classified as subject to X inactivation. Forty-four genes were also classified as subject to X inactivation based on a single SNP; three of these genes (Tsx, Rps4x, Efhc2) are known to be inactivated. A total of 380 genes were ultimately classified as subject to X inactivation (Supplemental Tables S2, S3), including many well-known genes (e.g., Iqsec2, G6pdx, Pgk1, Rbmx, Smc1a, Tspyl2, Zfx).
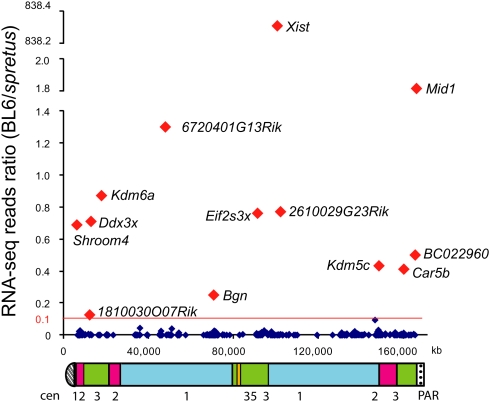
Figure 3.
Distribution of escape genes on the mouse X chromosome. The ratio between RNA-seq reads from BL6 (inactive X) and M. spretus (active X) is graphed for each gene against its location (UCSC Genome Browser, Mouse July 2007 [mm9] assembly). (Red) Genes with an average expression level from the inactive X higher than 10% of the active X; (dark blue) other genes. Underneath, an ideogram of the mouse X chromosome indicates regions that correspond to evolutionary strata (1, 2, 3, and 5) defined in human (Ross et al. 2005). Genes in strata 4 are apparently deleted in mouse. (Cen) Centromere; (PAR) pseudoautosomal region.
The total number of mouse genes that escape X inactivation based on at least 10% expression from the inactive X versus the active X, which was the cutoff previously used to identify human genes that escape X inactivation (Carrel and Willard 2005), was only 13, representing 3.3% of mouse X-linked transcripts assayed (Table 1; Fig. 3; Supplemental Fig. S3). One of the escape transcripts we identified is a noncoding RNA, 6720401G13Rik. Our survey correctly identified four previously known mouse escape genes—Kdm5c, Eif2s3x, Kdm6a, and Mid1 (Fig. 2A; Table 1). Bi-allelic expression of four additional transcripts with robust expression from the inactive X—Ddx3x, Shroom4, Car5b, and 2610029G23Rik—was confirmed by conventional sequencing of PCR and RT-PCR products (Fig. 1B; Table1). Note that tracings obtained by conventional sequencing do not necessarily reflect the allele-specific copy number, that is, heterozygous peaks do not always show a 50/50 ratio for genomic DNA. This is due to the fact that peak height is highly dependent on neighboring DNA sequence (Parker et al. 1995). For Shroom4 and Car5b the relative expression levels from the inactive X versus the active X were confirmed using RT-PCR products digested with a restriction endonuclease that distinguishes alleles based on the SNP. Bi-allelic expression was quantified by real-time qPCR, which showed 48% and 45% expression from the inactive X, compared to 42% and 44% BL6 reads, for Shroom4 and Car5b, respectively (Fig. 1C; Supplemental Table S1); thus, the RNA-seq data and the qPCR results were in agreement.
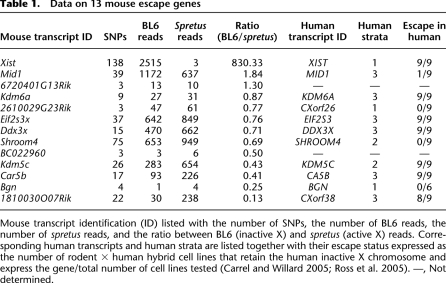
Table 1.
Data on 13 mouse escape genes
Mouse transcript identification (ID) listed with the number of SNPs, the number of BL6 reads, the number of spretus reads, and the ratio between BL6 (inactive X) and spretus (active X) reads. Corresponding human transcripts and human strata are listed together with their escape status expressed as the number of rodent × human hybrid cell lines that retain the human inactive X chromosome and express the gene/total number of cell lines tested (Carrel and Willard 2005; Ross et al. 2005). —, Not determined.
Xist was correctly identified as a transcript expressed uniquely from the inactive X (Fig. 3; Table 1; Borsani et al. 1991). Surprisingly, Mid1 also had an apparently higher expression from the inactive X compared to the active X, as indicated by the large number of BL6 reads for SNPs located at its 3′ end. However, SNPs located in exon 2 showed 46% expression from the inactive X (Fig. 2; Supplemental Table S1). The high number of BL6 reads for the 3′-end region of Mid1 is probably due to amplification of the pseudoautosomal portion of this gene in M. musculus (Dal Zotto et al. 1998). No other gene showed multiple SNPs exclusively from BL6. However, some genes showed discrepant results between SNPs: six genes showed a single SNP with a substantial number of BL6 reads but still lower than spretus counts, while 29 other genes had BL6 reads higher than spretus reads for one or two SNPs. In theory, these discrepant SNPs could represent either alternative transcripts or polymorphisms/mutations in the M. spretus used to derive the Patski cell line. However, since conventional sequencing of several of these SNPs (Mmgt1, Atp6ap1, Idh3g, Huwe1, Hmgn5, Ubqln2, Hs6st2, Slc6a8, and B230358A15Rik) did not show heterozygosity in genomic DNA, we conclude that they are not real SNPs (Supplemental Fig. S2). SNPs that were inconsistent with the majority of SNPs within the same gene were not further considered.
Human and mouse differ in the number and identity of escape genes
Significant differences were observed between mouse and human in terms of X inactivation and escape (Table 1; Supplemental Tables S2, S3). Eight of 11 genes that we found to escape X inactivation in mouse also escape in human (i.e., are expressed from at least one out of nine hybrids that retain the human inactive X) (Carrel and Willard 2005), suggesting that escape from X inactivation is partially conserved (Table 1). Of the 380 genes subject to X inactivation in mouse (with no or few BL6 sequence reads), 133 escape X inactivation in human (expressed in at least one out of nine hybrid lines); 184 are subject to X inactivation, and 63 do not have orthologs or have not been studied in human (Supplemental Tables S2, S3).
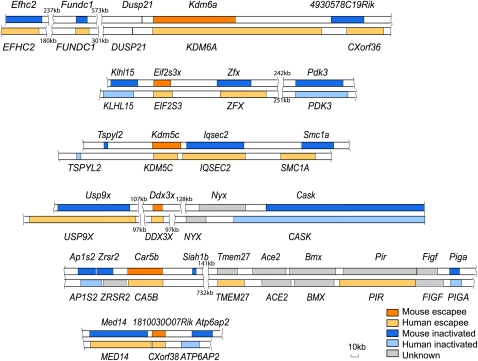
There was no clustering of escape genes in mouse, unlike the large domains of escape in human (Fig. 4). Thus, in mouse, escape from X inactivation is apparently controlled at the level of individual genes and not of chromatin domains. Furthermore, escape genes appeared to be randomly distributed along the mouse X chromosome, with no preference for a region corresponding to a specific evolutionary strata as defined in human (Fig. 3; Table 1; Ross et al. 2005).
Figure 4.
Mouse escape genes are not clustered in domains. Six mouse escape genes with surrounding regions are each aligned with the corresponding human region underneath. CXorf38 is the corresponding human gene to 1810030O07Rik. 4930578C19Rik was previously found to be subject to X inactivation, while CXorf36 escapes X inactivation (DK Nguyen and CM Disteche, unpubl.). The size of regions with no known genes is indicated.
Escape genes are depleted of H3K27me3
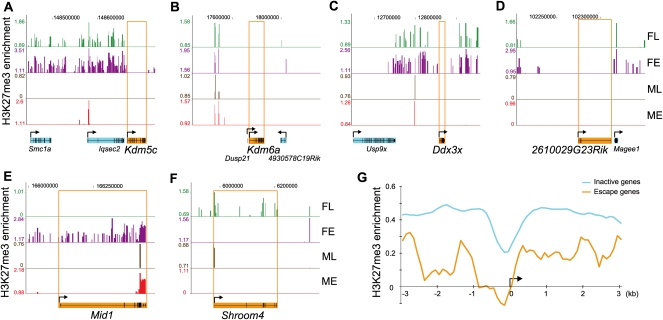
To investigate the role of H3K27me3 in escape from X inactivation, enrichment profiles were generated from female and male mouse adult liver and 12.5 days postcoitum (dpc) embryos by ChIP-chip. The majority of escape genes (Kdm5c, Eif2s3x, Kdm6a, Ddx3x, 6720401G13Rik, 2610029G23Rik) were completely depleted in H3K27me3 in both female and male liver and embryos, while adjacent genes subject to X inactivation (e.g., Iqsec2, Magee1, Usp9x) were specifically enriched in female but not male samples (Fig. 5A–D; Supplemental Fig. S3). The escape genes Car5b, Bgn, and 1810030O07Rik, which had the lowest levels of escape, had some enrichment in H3K27me3 in female liver and embryos. Conversely, some genes were not enriched in H3K27me3 even though they are subject to X inactivation, for example, Klhl15 (Supplemental Fig. S3). Nonetheless, a majority of genes subject to X inactivation (251/366, or 68%) displayed enrichment in H3K27me3, and the average enrichment was higher at the 5′-end of these genes compared to escape genes (Fig. 5G).
Figure 5.
H3K27me3 is usually depleted at escape genes in female tissues. (A–D) Escape genes Kdm5c, Kdm6a, Ddx3x, and 2610029G23Rik are completely devoid of H3K27me3 in female tissues, while adjacent genes subject to X inactivation (e.g., Iqsec2, Magee1, Usp9x) are enriched. (E–F) Depletion in H3K27me3 at escape genes Mid1 and Shroom4 differs between tissues/developmental stages. (FL) Female liver; (FE) female embryos; (ML) male liver; (ME) male embryos. ChIP-chip peak files were uploaded in the UCSC Genome Browser (Mouse July 2007 [mm9] assembly). (Top) X chromosome coordinates. Escape genes are filled in orange, and inactive genes in light blue. (Arrow) Transcription direction. (G) H3K27me3 enrichment at the 5′ end of genes subject to X inactivation is higher than at escape genes in female liver. Average H3K27me3 enrichment for genes subject to X inactivation (366 genes; light blue curve) is compared to average enrichment for escape genes (10 genes; orange curve). Data are shown as log2 of the signal ratio between ChIP and input fractions for 3 kb upstream and 3 kb downstream of the transcription start site (black arrow).
For some genes H3K27me3 profiles differ in relation to the developmental stage. For example, Mid1 was enriched in H3K27me3 only in female embryos but not in liver (Fig. 5E). Thus, Mid1 may be initially silenced and only escape X inactivation later in development or in some tissues, which would explain previous conflicting results (Dal Zotto et al. 1998; Li and Carrel 2008). In contrast, Shroom4 was enriched in H3K27me3 only in female liver but not in embryos, suggesting a progressive and possibly tissue-specific onset of silencing (Fig. 5F). The RNA-sequencing data were generated using a cell line and thus would not address tissue-specific differences in escape.
Discussion
Our study demonstrates the usefulness of high-throughput RNA sequencing to identify genes that escape X inactivation and genes subject to X inactivation. Few mouse genes with robust expression from the inactive X chromosome were identified. Mouse escape genes are embedded within inactive chromatin as single genes, unlike the large domains of escape in human, as shown by our current comparisons of six regions, which significantly add to a previous comparison of one domain around Kdm5c (Tsuchiya and Willard 2000; Tsuchiya et al. 2004). We conclude that the mouse has very few escape genes, as previously suggested (Disteche 1995). The significant difference in the number of genes that escape X inactivation between human and mouse is consistent with the more severe phenotypes of 45,X women compared to 39,X mice. As many as 99% of human conceptions with a single X chromosome result in death in utero (Hook and Warburton 1983). Those who survive have Turner syndrome, including infertility, short stature, and other physical anomalies, but no mental impairment except for difficulty with spatial recognition (Bondy 2009). Candidate genes deficient in Turner individuals are located either in the pseudoautosomal region—for example, the SHOX gene, clearly implicated in short stature—or in the short arm of the X chromosome based on deletion analysis (Clement-Jones et al. 2000; Zinn and Ross 2001). It should be noted that the survey of X inactivation and escape in human was mainly done using rodent × human hybrid lines that retained an inactive human X chromosome. The different systems used in that survey and the present survey could explain some species-specific differences. However, the survey of human genes was validated at least in part using SNPs in human fibroblast cell lines (Carrel and Willard 2005).
A subset of escape genes conserved between human and mouse must be under selection to retain expression from the inactive X. Whether these genes are implicated in sex-specific phenotypes remains to be determined. Re-examination of previously published expression array data to compare expression levels between female and male mouse tissues (Raefski and O'Neill 2005; Yang et al. 2006; Isensee et al. 2008) and between female and X0 mouse brains (Raefski and O'Neill 2005) indicates higher female-specific expression of mouse escape genes, which is tissue-dependent (Supplemental Table S4). However, expression is not doubled in females, consistent with our RNA-seq data, in which ratios between BL6 and spretus reads were lower than 1, except for Xist, Mid1, and 6720401G13Rik. Similarly, human escape genes show a relatively modest increase in expression in female versus male cell lines, suggesting lower expression from the inactive X (Carrel and Willard 2005; Johnston et al. 2008).
The novel mouse escape genes we identified are candidates for the phenotypes of 39,X mice (Lynn and Davies 2007). Abnormal phenotypes in X0 mice include mild behavioral deficiencies, slightly reduced fertility, and postnatal growth retardation. In particular, Shroom4, which does not escape X inactivation in human, may play a role in behavioral deficits, which include anxiety and abnormal reaction to fear. The SHROOM protein family interacts with actin filaments to generate thickened epithelial sheets and is involved in neurulation (Hagens et al. 2006; Lee et al. 2009). Mutations of SHROOM4 are associated with X-linked mental retardation in humans (Hagens et al. 2006). Ddx3x encodes a helicase and has been implicated in dosage-dependent immune responses against viral invasion (Schroder et al. 2008). Although immune responses are disrupted in Turner individuals, no such effect has been reported in X0 mice. Car5b, which encodes a carbonic anhydrase localized in mitochondria, is more highly expressed in female, versus male, mouse hearts, consistent with escape from X inactivation (Supplemental Table S4; Isensee et al. 2008).
Comparisons of H3K27me3 enrichment in female and male liver and embryos showed that the repressive histone mark is depleted at escape genes. H3K27me3 profiles in mouse female ES cells showed a similar depletion at escape genes (J Berletch, unpubl.), confirming a previous study of a limited region of the mouse X chromosome (Marks et al. 2009). Our current findings of abrupt changes in chromatin modifications at transitions between genes subject to X inactivation and escape genes support our previous findings that suggest a role for the chromatin insulator protein CTCF in protecting chromatin domains (Filippova et al. 2005). Depletion of H3K27me3 at escape genes is consistent with the recent findings that Xist RNA, which recruits the PRC2 complex to deposit H3K27me3 on the inactive X (Zhao et al. 2008), is not present at escape genes such as Kdm5c and Kdm6a (Murakami et al. 2009). Furthermore, depletion of EED, a component of the PRC2, leads to reactivation of the inactive X chromosome (Kalantry et al. 2006), suggesting that H3K27me3 is important for the maintenance of the inactive X status. Based on their H3K27me3 profiles, Mid1 and Shroom4 may escape from X inactivation only in specific tissues and/or developmental stages. Previous studies have indicated that another escape gene, Kdm5c, also shows variation in escape in different tissues and developmental stages (Carrel et al. 1996; Sheardown et al. 1996; Lingenfelter et al. 1998). In this study, we did not observe H3K27me3 enrichment at Kdm5c in 12.5-dpc embryos, suggesting that its transient silencing (Lingenfelter et al. 1998; Chaumeil et al. 2006) either may be largely over at that stage, or may be mediated by another epigenetic mechanism.
Together, our global survey of genes that escape X inactivation in mouse provides a baseline for developmental and tissue-specific studies of molecular mechanisms of escape. In addition, we identified 133 genes subject to X inactivation in mouse, which are known to escape X inactivation in human (Carrel and Willard 2005). The significant differences between species will help identify genes important for the phenotypes associated with Turner syndrome.
Methods
Mouse tissues and cell lines
Male and female adult liver and embryos were collected from BL6 mice. The Patski cell line derived from a cross between M. spretus and BL6 (Hprt−), was previously selected in HAT so that the X chromosome from BL6 is always inactive (Lingenfelter et al. 1998). Patski cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% FBS and 1% penicillin/streptomycin.
Library construction and RNA sequencing
RNA was prepared using an mRNA-Seq sample preparation kit (Illumina). mRNA was purified from 10 μg of total RNA using magnetic oligo(dT) beads, followed by fragmentation and synthesis of first-strand and second-strand cDNA. Double-stranded cDNA was end-repaired, and additional A bases were added to the 3′ end of the DNA fragments. After ligation of adaptors, size selection of the library was done by gel electrophoresis followed by excision and purification of DNA (using the QIAGEN gel extraction kit) in the 200 ± 25-bp range. PCR was performed to enrich the purified DNA templates. Sequencing of the RNA-seq shotgun library was done on an Illumina Genome Analyzer, yielding 36-bp single-end reads.
SNP identification and assessment of escape from X inactivation
Of 39 million reads, 32.5 million mapped uniquely to the reference mouse genome (BL6). As a primary source of polymorphisms between BL6 and spretus, we used X chromosome SNPs identified by the Mouse Genome Project (Wellcome Trust Sanger Institute, http://www.sanger.ac.uk) that intersected with our transcriptome data. To identify additional SNPs (e.g., novel to our cell line or missed by large-scale genomic sequencing), Novoalign (Novocraft) was used to align sequence reads against NCBI mouse genome release 36 (UCSC Feb. 2006 release [mm8]) and a collection of splice junctions generated from RefSeq genes, Ensembl genes, and UCSC Known Genes (Pruitt et al. 2007; Hubbard et al. 2009; Kuhn et al. 2009). Predicted splice junctions from ESTs, Genscan, and N-scan (Kuhn et al. 2009) predictions were also considered. All possible splice sites that would generate transcripts resulting from skipping events spanning up to two exons were represented. A minimum of 5 nt overlap per flanking junction sequence was required for alignment to be considered, based on maximizing overall alignment sensitivity (data not shown). All reads that aligned uniquely to the genome or splice sites were retained for further analysis. Reads that contained mismatches to the reference BL6 genome were used to identify SNPs as described previously (Babak et al. 2008) with several modifications. Briefly, at all genomic positions where at least one mismatch existed, the number of all represented bases and the sum of the phred qualities supporting those base calls were tallied. The Support Vector Machine (SVM) (Pavlidis et al. 2004) was used to quantify the likelihood that mismatches at these positions arose from sequencing errors. The SVM was trained on previously published RNA-seq data from BL6/CAST (Babak et al. 2008) using publicly known SNPs (Frazer et al. 2007) and was run using default settings (http://svm.sdsc.edu/cgi-bin/nph-SVMsubmit.cgi). At a score threshold corresponding to a FDR of 3%, 169,031 SNPs were discovered, of which 3571 (2%) were on the X chromosome. SNPs could be missed by this approach because the BL6 X chromosome in this case is inactivated and the SVM was trained to identify heterozygous SNPs (i.e., both alleles represented).
A combined X chromosome SNP list was generated by merging the SNPs identified by genomic sequencing (Wellcome Trust Sanger Institute) with the SNPs identified by the machine learning approach. All reads were remapped with MAQ (mapping and assembly with quality) (Li et al. 2008) to a version of the reference mouse genome (mm9) that excluded the Y chromosome, as this was an XX cell line. Except for Eif2s3x and Pgk1, reads with a MAQ mapping score of zero were excluded from consideration. A MAQ score of zero means that two regions or more have an equal mapping score on the reference genome, so that these reads will be assigned to one of the locations at random. For two genes, Eif2s3x and Pgk1, the MAQ score was zero for most SNPs due to high sequence similarity between Eif2s3x and another region on the X chromosome with no annotated transcript and between Pgk1 and the testis-specific pseudogene Pgk2. The number of reads was tabulated for the BL6 versus spretus alleles at the coordinates of anticipated or predicted X chromosome SNPs covered by 1+ reads. Three-hundred -thirty-six SNPs with BL6 reads but no spretus reads, which were inconsistent with other SNPs in the same gene, were not considered further.
SNP validation by conventional sequencing and quantification of level of escape from X inactivation
Genomic DNA and RNA were extracted from Patski cells, followed by PCR or RT-PCR using a first-strand cDNA synthesis kit (Invitrogen) and primers designed around candidate SNPs (Supplemental Table S5). After purification of PCR products, conventional sequencing was performed to validate the SNPs. To quantify allelic expression of Shroom4 and Car5b, PCR was done to amplify genomic DNA or cDNA from Patski cells and parental M. spretus and BL6, using primers designed around SNPs (Supplemental Table S5). After purification (QIAGEN kit) amplification products were split into two aliquots; one served as undigested control, while the other was digested with a restriction endonuclease (StyI for Shroom4 and ApaI for Car5b) (New England BioLabs). Products were examined by gel electrophoresis. For real-time qPCR analysis, initial PCR reactions were done for 20 cycles to get linear amplification from Patski genomic DNA or cDNA before restriction endonuclease digestion. Real-time qPCR was done in triplicate using a 7900 PCR system (Applied Biosystems) to compare digested and undigested samples using primers that flank the restriction site (Supplemental Table S5).
ChIP-chip analyses
ChIP was performed according to a fast chromatin immunoprecipitation method (Nelson et al. 2006). Briefly, after cross-linking with 1% formaldehyde, chromatin was sonicated to ∼500-bp fragments and immunoprecipitated with a histone H3K27me3 antibody (Upstate) followed by pull-down using protein A–Sepharose beads (Amersham). Controls included ChIP carried out without antibody (no antibody fraction) and the input fraction. After incubation with the antibody overnight, protein A–Sepharose beads were added to pull down the chromatin. DNA was purified from the ChIP fractions using a PCR purification kit (QIAGEN). ChIP samples and corresponding input controls were amplified using a whole-genome amplification kit (Sigma), and labeled with Cy5 and Cy3, respectively, according to Roche's protocol. Labeled DNA was hybridized to high-density tiling arrays that contained X chromosome probes every 100 bp (2.1M format, Array10; Roche). Nimblescan software (Roche) was used to search for peaks of enrichment using a 500-bp sliding window. A FDR score was assigned to each enriched region using a stringent cutoff of <0.05% FDR.
Computational analyses of enrichment of H3K27me3 at the 5′-end of genes
Average H3K27me3 enrichment at the 5′-end of genes was calculated using a platform generated by J. Henikoff and S. Henikoff (Fred Hutchinson Cancer Research Center, Seattle) (Mito et al. 2005). Briefly, NimbleGen ratio files were uploaded into the platform to identify 100-bp tiling intervals and search for overlapping 50-mers in 3-kb intervals on either side of the 5′-end of X-linked transcripts prior to calculating average enrichment at each interval. To avoid interference from overlapping or neighboring genes, the search stopped at the overlapping or end of the current gene on either side.
Acknowledgments
This work was supported by NIH grants GM046883 and GM079537 (C.M.D.). We thank S. Henikoff and J. Henikoff (Fred Hutchinson Cancer Research Center, Seattle) for access to the computational analysis platform; E. Turner and C. Lee for assistance with library construction protocols and Illumina sequencing; and D.K. Nguyen and J. Berletch (University of Washington, Seattle) for access to unpublished data.
Footnotes
[Supplemental material is available online at http://www.genome.org. The ChIP-chip data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE20617. The sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA010053.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103200.109.
References
- Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE 1994. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet 3: 879–884 [DOI] [PubMed] [Google Scholar]
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr Biol 18: 1735–1741 [DOI] [PubMed] [Google Scholar]
- Bondy CA 2009. Turner syndrome 2008. Horm Res 71 (Suppl 1): 52–56 [DOI] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329 [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44 [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Evans EP, Holland K 1983a. XO monosomy is associated with reduced birthweight and lowered weight gain in the mouse. J Reprod Fertil 68: 381–385 [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Tam PP, Evans EP 1983b. Retarded development of XO conceptuses during early pregnancy in the mouse. J Reprod Fertil 68: 387–393 [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Carrel L, Hunt PA, Willard HF 1996. Tissue and lineage-specific variation in inactive X chromosome expression of the murine Smcx gene. Hum Mol Genet 5: 1361–1366 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes & Dev 20: 2223–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement-Jones M, Schiller S, Rao E, Blaschke RJ, Zuniga A, Zeller R, Robson SC, Binder G, Glass I, Strachan T, et al. 2000. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Genet 9: 695–702 [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB 1996. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen CH, Chapman V, Kalcheva I, et al. 1998. The mouse Mid1 gene: Implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet 7: 489–499 [DOI] [PubMed] [Google Scholar]
- Disteche CM 1995. Escape from X inactivation in human and mouse. Trends Genet 11: 17–22 [DOI] [PubMed] [Google Scholar]
- Ehrmann IE, Ellis PS, Mazeyrat S, Duthie S, Brockdorff N, Mattei MG, Gavin MA, Affara NA, Brown GM, Simpson E, et al. 1998. Characterization of genes encoding translation initiation factor eIF-2γ in mouse and human: Sex chromosome localization, escape from X-inactivation and evolution. Hum Mol Genet 7: 1725–1737 [DOI] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8: 31–42 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Hagens O, Dubos A, Abidi F, Barbi G, Van Zutven L, Hoeltzenbein M, Tommerup N, Moraine C, Fryns JP, Chelly J, et al. 2006. Disruptions of the novel KIAA1202 gene are associated with X-linked mental retardation. Hum Genet 118: 578–590 [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes & Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hook EB, Warburton D 1983. The distribution of chromosomal genotypes associated with Turner's syndrome: Livebirth prevalence rates and evidence for diminished fetal mortality and severity in genotypes associated with structural X abnormalities or mosaicism. Hum Genet 64: 24–27 [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. 2009. Ensembl 2009. Nucleic Acids Res 37: D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Noppinger PR 2008. Sexually dimorphic gene expression in the heart of mice and men. J Mol Med 86: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT 2008. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 4: e9 doi: 10.1371/journal.pgen.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T 2006. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol 8: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Le MP, Wallingford JB 2009. The shroom family proteins play broad roles in the morphogenesis of thickened epithelial sheets. Dev Dyn 238: 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Carrel L 2008. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci 105: 17055–17060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, Chapman VM, Disteche CM 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet 18: 212–213 [DOI] [PubMed] [Google Scholar]
- Lynn PM, Davies W 2007. The 39,XO mouse as a model for the neurobiology of Turner syndrome and sex-biased neuropsychiatric disorders. Behav Brain Res 179: 173–182 [DOI] [PubMed] [Google Scholar]
- Lyon MF 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG 2009. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 19: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 37: 1090–1097 [DOI] [PubMed] [Google Scholar]
- Murakami K, Ohhira T, Oshiro E, Qi D, Oshimura M, Kugoh H 2009. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet Genome Res 125: 19–25 [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185 [DOI] [PubMed] [Google Scholar]
- Parker LT, Deng Q, Zakeri H, Carlson C, Nickerson DA, Kwok PY 1995. Peak height variations in automated sequencing of PCR products using Taq dye-terminator chemistry. Biotechniques 19: 116–121 [PubMed] [Google Scholar]
- Pavlidis P, Wapinski I, Noble WS 2004. Support vector machine classification on the web. Bioinformatics 20: 586–587 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Prothero KE, Stahl JM, Carrel L 2009. Dosage compensation and gene expression on the mammalian X chromosome: One plus one does not always equal two. Chromosome Res 17: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2007. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raefski AS, O'Neill MJ 2005. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet 37: 620–624 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Baran M, Bowie AG 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKε-mediated IRF activation. EMBO J 27: 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown S, Norris D, Fisher A, Brockdorff N 1996. The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum Mol Genet 5: 1355–1360 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 Polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Tsuchiya KD, Willard HF 2000. Chromosomal domains and escape from X inactivation: Comparative X inactivation analysis in mouse and human. Mamm Genome 11: 849–854 [DOI] [PubMed] [Google Scholar]
- Tsuchiya KD, Greally JM, Yi Y, Noel KP, Truong JP, Disteche CM 2004. Comparative sequence and X-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res 14: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn AR, Ross JL 2001. Molecular analysis of genes on Xp controlling Turner syndrome and premature ovarian failure (POF). Semin Reprod Med 19: 141–146 [DOI] [PubMed] [Google Scholar]
- Zinn AR, Page DC, Fisher EM 1993. Turner syndrome: The case of the missing sex chromosome. Trends Genet 9: 90–93 [DOI] [PubMed] [Google Scholar]