Abstract
Pre-mRNA 5′ spliced-leader (SL) trans-splicing occurs in some metazoan groups but not in others. Genome-wide characterization of the trans-spliced mRNA subpopulation has not yet been reported for any metazoan. We carried out a high-throughput analysis of the SL trans-spliced mRNA population of the ascidian tunicate Ciona intestinalis by 454 Life Sciences (Roche) pyrosequencing of SL-PCR-amplified random-primed reverse transcripts of tailbud embryo RNA. We obtained ∼250,000 high-quality reads corresponding to 8790 genes, ∼58% of the Ciona total gene number. The great depth of this data revealed new aspects of trans-splicing, including the existence of a significant class of “infrequently trans-spliced” genes, accounting for ∼28% of represented genes, that generate largely non-trans-spliced mRNAs, but also produce trans-spliced mRNAs, in part through alternative promoter use. Thus, the conventional qualitative dichotomy of trans-spliced versus non-trans-spliced genes should be supplanted by a more accurate quantitative view recognizing frequently and infrequently trans-spliced gene categories. Our data include reads representing ∼80% of Ciona frequently trans-spliced genes. Our analysis also revealed significant use of closely spaced alternative trans-splice acceptor sites which further underscores the mechanistic similarity of cis- and trans-splicing and indicates that the prevalence of ±3-nt alternative splicing events at tandem acceptor sites, NAGNAG, is driven by spliceosomal mechanisms, and not nonsense-mediated decay, or selection at the protein level. The breadth of gene representation data enabled us to find new correlations between trans-splicing status and gene function, namely the overrepresentation in the frequently trans-spliced gene class of genes associated with plasma/endomembrane system, Ca2+ homeostasis, and actin cytoskeleton.
A striking evolutionary variation in eukaryotic gene expression mechanisms is the presence or absence in diverse organismal groups of a form of RNA splicing—pre-mRNA spliced leader (SL) trans-splicing (Davis 1996; Nilsen 2001; Hastings 2005). SL trans-splicing is closely related to conventional RNA splicing, or cis-splicing. The same nucleotide sequence features define donor and acceptor sites, and both processes occur in spliceosomes and involve formation of a 5′,3′,2′ branchpoint upstream of the acceptor site (Agabian 1990; Nilsen 1993). Whereas cis-splicing joins paired donor and acceptor sites within a single RNA molecule, in SL trans-splicing the donor exon is the 5′-segment of a specialized small 5′-capped RNA molecule—the SL RNA—and the target is an unpaired acceptor site near the 5′-end of a pre-mRNA molecule. Transfer of the SL RNA 5′-segment, the SL sequence, to the pre-mRNA molecule occurs with loss of the pre-mRNA's initial 5′-segment upstream of the acceptor site—a segment termed the “outron” (Conrad et al. 1991). In organisms that carry out SL trans-splicing, many different pre-mRNAs are trans-spliced with the same SL RNA species, with the result that many mature mRNA species share a common 5′-end sequence. Apart from being short, from 16 to ∼50 nucleotides (nt), the SL sequences of diverse organisms are not similar (Nilsen 2001).
The best understood function of SL trans-splicing is to resolve polycistronic operon transcripts into individual 5′-capped mRNAs by trans-splicing to unpaired acceptor sites adjacent to downstream cistron open reading frames (Clayton 2002; Blumenthal and Gleason 2003). However, it is likely that SL trans-splicing has additional unknown functions because in trans-splicing metazoa the majority of trans-spliced genes are not present in operons. A variety of possible functions for SL trans-splicing in monocistronic genes have been hypothesized but not firmly established, including direct effects of the leader itself on mRNA stability or translation, and an indirect effect, i.e., the removal of potentially deleterious elements within the outron (5′ untranslated region [UTR] “sanitization”) (Hastings 2005).
Due to its sporadic phylogenetic distribution, it is not clear whether SL trans-splicing is an ancestral eukaryotic feature that has been lost in several lineages or whether it was absent from the ancestor and arose independently in several lineages (Nilsen 2001; Stover and Steele 2001). In the deuterostome division of the metazoa, we discovered SL trans-splicing in the chordate tunicate ascidian Ciona intestinalis (Vandenberghe et al. 2001) and it has since been found in several other tunicate species (Yuasa et al. 2002; Ganot et al. 2004). Tunicates are of particular evolutionary interest because, as chordates, they are related to vertebrate ancestors (Dehal et al. 2002). Moreover, because vertebrates are among the groups that almost certainly do not carry out SL trans-splicing (Nilsen 2001; Hastings 2005), it follows that either SL trans-splicing has been lost in the vertebrate lineage or was invented in the tunicate lineage after the tunicate/vertebrate divergence. Thus, in-depth knowledge of trans-splicing in the tunicates may provide valuable insight into genome evolution in the chordates and the evolutionary dynamics of RNA splicing.
Among the metazoan organisms that carry out SL trans-splicing, a significant fraction of genes are conventionally expressed, i.e., are not trans-spliced (e.g., in Ciona, ∼50% of genes are apparently not trans-spliced; Satou et al. 2006). It is not known why some monocistronic genes are trans-spliced and others are not. Genome-wide knowledge of the trans-splicing status of individual genes would provide a basis for elucidating those aspects of gene structure or function that may affect the “choice” of SL trans-splicing versus conventional gene expression. Although there have been several studies of individual genes or of small samples of the trans-spliced or non-trans-spliced gene sets (Satou et al. 2006; Sierro et al. 2009), there has not yet been a genome-wide identification of trans-spliced and non-trans-spliced gene populations in any organism. Because the trans-spliced and non-trans-spliced gene subpopulations represent a significant fraction of the genome, generating a comprehensive overview has been beyond the reach of conventional sequencing methodologies. However, recently developed high-throughput methods have made it feasible to study genome-wide processes through DNA sequencing. We have developed and employed an approach to high-throughput characterization of the trans-spliced mRNA population of the ascidan Ciona intestinalis based on 454 Life Sciences (Roche) pyrosequencing.
In order to sample a wide range of expressed genes we used the whole organism at a stage of active development and differentiation, the tailbud embryo. Our results identify the majority of trans-spliced genes in Ciona and precisely localize their trans-splice acceptor sites. Analysis of this extensive data set provides new insight into the splicing mechanism and into the nature of the trans-spliced and non-trans-spliced gene sets. In addition to improved understanding of trans-splicing, our results also provide a wealth of specific genetic information on Ciona, a key model organism for genomic and developmental genetics studies relevant to vertebrate early development and evolution (Satou et al. 2005; Imai et al. 2006; Munro et al. 2006).
Results
Spliced-leader-PCR (SL-PCR) and massively parallel DNA sequencing approach
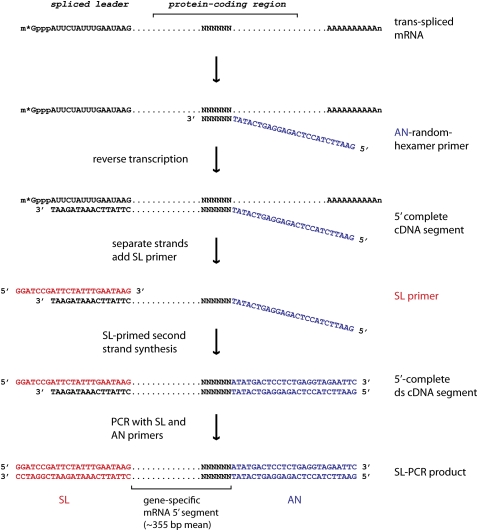
In order to globally identify the set of trans-spliced genes, we developed a method to selectively PCR-amplify 5′-segments of all trans-spliced mRNAs. This method, random-primed SL-PCR, consists of reverse transcription primed by random-hexamer sequences linked 3′ to an arbitrary 25-nt anchor sequence (AN), followed by PCR using the AN sequence for leftward priming and a 23-mer primer terminating in the 16-nt SL sequence, which is present at the 5′-end of trans-spliced mRNAs, as the rightward primer (SL primer) (see Fig. 1). We applied random-primed SL-PCR to total RNA isolated from tailbud stage embryos of Ciona intestinalis and analyzed the products (average length ∼400 bp) using 454 Life Sciences (Roche) high-throughput DNA sequencing technology (Margulies et al. 2005). We obtained 249,239 high-quality sequence reads from two runs on the GS20 system. The average read length was 190 nt (see Supplemental Fig. S1) and 24% of reads covered the full lengths of their SL-PCR products, the remainder terminating within the SL-PCR product before reaching the far end (see Supplemental Figs. S1, S2, and Supplemental material, section 5).
Figure 1.
SL-PCR amplification of 5′ segments of trans-spliced mRNAs. Starting from a capped (m*Gppp) and polyadenylated (AAAAAAAAAAn) trans-spliced mRNA (top line) with the 16-nt SL sequence at its 5′ terminus, the diagram illustrates the basis of AN-linked random-hexamer-primed reverse transcription, SL-primed second strand cDNA synthesis, and PCR amplification with SL and AN primers.
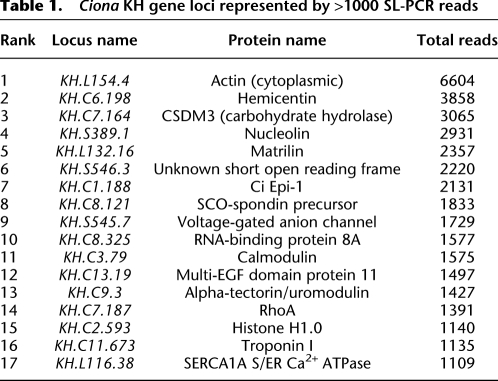
Several lines of evidence, summarized in Supplemental material, section 7, substantiated the expected specificity of SL-PCR amplification for mRNA 5′-segments (including very long mRNAs such as the 15-kb nebulin mRNA; Supplemental Fig. S3), and further established a very high overall sequence quality, with 97% of reads giving high-quality alignments with the version 1 genome (Dehal et al. 2002), and 93% with the KH gene model set (Satou et al. 2008) (Supplemental material, section 4). Moreover, as expected for a natural mRNA population, a diverse range of genes were represented over a wide abundance range, from a single read (1585 KH gene loci) up to >1000 reads (17 KH gene loci listed in Table 1). The most abundantly represented KH gene locus was KH.L154.4 encoding cytoplasmic actin, with 6604 reads, or 2.6% of the total. The total number of KH gene loci matching SL-PCR reads was 8790, which represents ∼58% of the total number of 15,254 KH gene loci in the genome. Given that the proportion of Ciona genes that undergo trans-splicing has been estimated at 50% (Satou et al. 2006), this suggests that our data set effectively samples the trans-spliced gene population.
Table 1.
Ciona KH gene loci represented by >1000 SL-PCR reads
Although all SL-PCR reads can in principle identify trans-spliced genes, we selected for in-depth analysis a restricted subset of reads of particularly high apparent quality and informative structure. This subset, termed Category 1, consisted of reads that started precisely with a perfect SL primer sequence, and AN-start reads that ended precisely with a perfect SL primer reverse-complement sequence (see Methods). Because Category 1 reads include the junction between the spliced leader (SL) sequence and the remainder of the original template mRNA, these reads not only identify trans-spliced mRNAs, but also precisely localize trans-splice acceptor sites. Category 1 reads represented almost one-half of the total number of reads (120,937 or 48.5%) and included 86% (7534/8790) of the total number of KH models present in the global read set. Most of the remaining reads fell into Category 2 (having imperfect SL primer sequences at the start/end) or Category 3 (AN-start reads terminating prior to the SL site at the far end; see Supplemental Fig. S2). The analyses described in this study were limited to Category 1, except where indicated.
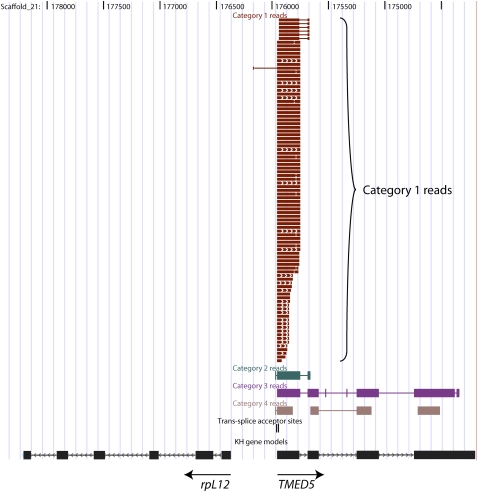
The association of Category 1 reads with mRNA extreme 5′-ends was evident by inspection of alignments in the UCSC Genome Browser setting (http://genome.ucsc.edu/index.html). Figure 2 shows a genomic region containing two divergently transcribed genes, one encoding ribosomal protein L12 (rpL12), the other a protein similar to mammalian TMED5. A total of 231 SL-PCR reads, including 93 Category 1 reads, mapped to this region, all associated with the TMED5 gene. All the Category 1 reads mapped to exon 1, in a few cases extending to exon 2. Moreover, 86/93 Category 1 reads identified precisely the same TMED5 genomic nucleotide at the upstream end of the exon 1 alignment block. Such high precision at the alignment start is expected because this should correspond to a specific genomic trans-splice acceptor site. Of interest, six of the remaining seven Category 1 reads identified a second nearby nucleotide as an alternative minor acceptor site (see also below concerning alternative trans-splicing). Extensive genome browser–based examination of aligned Category 1 reads showed that association of SL-PCR reads with mRNA 5′-ends and precise agreement among multiply sampled alignment starts were general rules.
Figure 2.
Example of SL-PCR reads mapping to a trans-spliced gene. The figure shows a ∼4-kb region of the Ciona genome (assembly version 1), as viewed in the UCSC Genome Browser (http://genome.ucsc.edu), edited for clarity, with SL-PCR reads mapped to the genome by BLAT. In addition to Category 1 reads, i.e., reads containing a perfect SL primer motif at one end or the other, other read categories shown as separate tracks are Category 2 (imperfect SL primer motif at one end or the other), Category 3 (AN-start reads terminating within the SL-PCR product insert and thus falling short of the SL primer at the far end), and Category 4 (atypical reads that do not start with a recognizable primer motif, or that contain additional unexpected primer motifs within the SL-PCR insert). The Category 1 read track is displayed in “squish” mode and the Categories 2–4 tracks in “dense” mode. Also shown is a track indicating with vertical strokes the locations of 8929 trans-splice acceptor sites mapped to the genome on the basis of Category 1 reads, two of which map close together at the 5′-end the TMED5 (transmembrane emp24 protein transport domain containing 5) gene.
Figure 2 also illustrates the preferential derivation of SL-PCR products from some genes as opposed to others. The association of SL-PCR reads with the TMED5 gene and not the rpL12 gene is not a reflection of mRNA abundance. On the contrary, the rpL12 gene appears to be expressed at ∼10-fold higher levels than the TMED5 gene, based on the occurrence of 29 rpL12 5′-ESTs and only three TMED5 5′-ESTs in the Ciona tailbud embryo conventional cDNA EST library of Satou et al. (2003) (http://hoya.zool.kyoto-u.ac.jp/download.html). This suggests a >1000-fold preferential derivation of SL-PCR products from TMED5 mRNA. This enrichment presumably reflects a different trans-splicing status of these two genes, with TMED5 being a trans-spliced gene and rpL12, like many ribosomal protein genes (see below), being undetectably trans-spliced. Additional evidence for the high specificity of SL-PCR for trans-spliced mRNAs is the virtually complete absence from the global set of 249,239 reads of SL-PCR products corresponding to mitochondrial transcripts or to ribosomal RNAs. These are very abundant RNA species that are not expected to undergo trans-splicing (see Supplemental material, section 7).
Mapping genomic trans-splice acceptor sites
Consistent with the expectation that sequences immediately adjacent to the SL primer in SL-PCR products would correspond to genomic splice acceptor sites, we found that (1) >99% of SL-primer-trimmed Category 1 reads mapped to the version 1 genome, (2) in 92% of cases the top-scoring BLAT alignment started from the first base of the SL-trimmed read, and (3) 98% of such first-base-aligned reads matched genomic sites immediately downstream from AG dinucleotides.
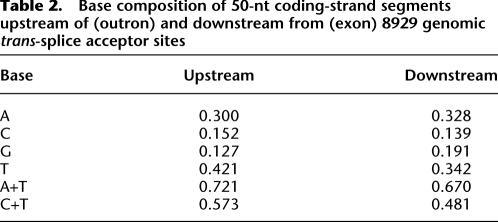
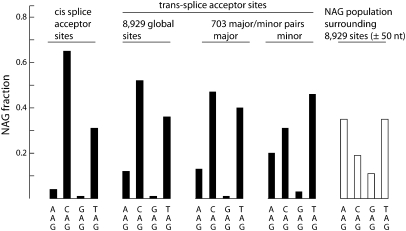
We compiled a list of 8929 stringently defined candidate trans-splice acceptor sites, by recovering top-scoring, first-base-aligned, AG-adjacent alignments of primer-trimmed Category 1 reads (see Methods). For precise mapping purposes the “site” is considered to be the genomic nucleotide corresponding to the first base of the SL-primer-trimmed read, and immediately 3′ of the genomic AG dinucleotide. The genomic DNA sequences upstream of and downstream from these 8929 sites are expected to correspond to outrons and to trans-splicing target exons, respectively. As shown in Table 2, 50-nt upstream segments were more A+T-rich than 50-nt downstream segments, 72.1% A+T versus 67.0% A+T, consistent with reports that in the nematode Caenorhabditis, outrons (and introns) are A+T-rich compared with exons (Csank et al. 1990; Conrad et al. 1991). Upstream segments of 50 nt or 20 nt were also more pyrimidine-rich (∼57% C+T) than corresponding downstream segments (∼48% C+T), consistent with the presence of a pyrimidine-rich sequence immediately upstream of the acceptor site, a known feature of vertebrate cis-splice acceptor sites (Black 2003). The composition of the NAG trinucleotide marking the trans-splice acceptor site showed a marked CAG > TAG > AAG >> GAG profile (Fig. 3), matching that previously established for vertebrate cis-splice acceptor sites (Mount 1982; Aebi et al. 1986; Akerman and Mandel-Gutfreund 2006), and differing from the overall regional NAG composition: TAG, AAG > CAG > GAG. Finally, YUNAY, the consensus motif for vertebrate RNA splicing branchpoints (Gao et al. 2008), was present in most outron segments and was 1.7-fold more abundant in the 50-nt upstream of the acceptor sites than in the 50-nt downstream (16,260 versus 9337 occurrences). These features are fully consistent with those expected of bona fide trans-splice acceptor sites.
Table 2.
Base composition of 50-nt coding-strand segments upstream of (outron) and downstream from (exon) 8929 genomic trans-splice acceptor sites
Figure 3.
NAG composition analysis. The histograms show the NAG composition of eukaryotic cis-splice acceptor sites (Mount 1982), of the 8929 stringently defined Ciona candidate trans-splice acceptor sites, and of the major and minor acceptor sites in 703 closely spaced major:minor pairs from the larger set of 8929 sites. Also shown, for contrast (white bars), is the NAG composition for all AGs occurring in sequences flanking the 8929 sites (±50 nt). Note the similarity of the trans-splice acceptor site NAG composition pattern to that of vertebrate cis-splice acceptor sites. The NAG composition pattern of minor sites reflected that of major sites but in a less pronounced form (see also Supplemental material, section 8).
The 8929 stringently defined candidate trans-splice acceptor sites mapped to 1237 genome sequence scaffolds (including the 297 largest scaffolds) which together represent >90% of the 116.7 Mb version 1 genome assembly. Thus, trans-spliced genes are widely distributed throughout the genome. Due to alternative trans-splicing (see below), the 8929 acceptor sites represent <8929 genes; we estimate a minimum of 6407 and a likely maximum of ∼7900 (Supplemental material, section 8). This represents a large majority of the 8790 genes represented by the global SL-PCR read set.
Closely spaced alternative trans-splice acceptor sites
In examining the genomic distribution of candidate trans-splice acceptor sites, we found that many same-strand nearest-neighbor pairs were unexpectedly close: 14% were ≤50 nt apart. In contrast, very few (<0.1%) opposite-strand nearest-neighbors were ≤50 nt apart. The most frequent same-strand nearest-neighbor interval was 3 nt (Supplemental Fig. S2; and see Fig. 4). Clearly, many sites are too close together to represent distinct genes but apparently are alternative acceptor sites within individual genes, presumably in many cases associated with a single splicing branchpoint. (As indicated in Supplemental material, section 8, we could exclude as unlikely the possibility that closely spaced sites might result not from alternative trans-splicing, but from the presence of minor alleles in which a cryptic acceptor site has been activated by mutational loss of a sole “normal” site.) Most ≤50-nt intervals reflected the presence of a single minor satellite acceptor site close to a major site, the latter being used, on average, 16-fold more frequently (the example of the TMED5 gene in Fig. 2 would thus be typical). Downstream minor sites were 1.8-fold more abundant than upstream minor sites, reflecting a 1.97-fold higher frequency of AG dinucleotides downstream, which in turn reflects the higher purine (i.e., lower C+T) content of the downstream segments (see Table 2).
Figure 4.
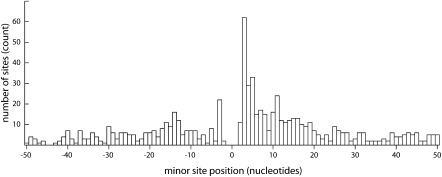
Spatial distribution of satellite minor sites relative to major sites. Among the 703 major/minor acceptor site pairs the distance of the minor site from the major was recovered and results in the region ±50 nt are shown. Negative values are upstream of and positive values are downstream from the major site.
In slightly more than half the major/minor site pairs (403/703 = 57%) the minor site represented the closest AG dinucleotide to the major site. In the remaining 43% of cases, there were closer AGs that were not utilized as acceptor sites, and in 27% of cases (190 cases) the splicing machinery “skipped over” one or more AGs located between the major and minor sites. Thus proximity of an AG dinucleotide to a major site, though important, is not the sole determinant of minor site use. An additional factor was minor site NAG trinucleotide composition, which differed from the regional overall NAG composition, and resembled that of major sites, or of the 8929 sites overall, but with a reduced stringency of preferences (Fig. 3).
The distribution of minor site distances upstream and downstream of the major site showed strong maxima at 3 nt (Fig. 4) similar to the distribution of alternative cis-splicing sites observed among human genes (Akerman and Mandel-Gutfreund 2006; Dou et al. 2006; Hiller and Platzer 2008), but unlike the latter, did not show peaks at ±3n-nt (n > 2) intervals. Mechanistic implications of these similarities and differences are considered in the Discussion.
Frequently and infrequently trans-spliced genes
Finding a gene represented by one or more SL-PCR products does not necessarily imply that the gene is expressed uniquely through the trans-splicing pathway. It is possible that individual genes could generate both trans-spliced and non-trans-spliced mRNAs, e.g., by alternative promoter use. To assess trans-splicing frequencies/efficiencies, we compared the representation of individual genes in the trans-spliced mRNA subpopulation (represented by Category 1 SL-PCR reads) with their representation in the global mRNA population as reflected in the 30,278-member 5′-EST data set of Satou et al. (2003), which is based on oligo(dT)-primed conventional cDNA cloning of tailbud embryo RNA (tailbudESTs). Although these tailbudESTs are 5′-incomplete in terms of mRNA structure, and therefore do not reveal whether the original template mRNA molecule was trans-spliced or not, it is expected that all mRNAs regardless of trans-splicing status would be represented by EST counts in proportion to their abundances.
The number of KH gene loci represented in the tailbudEST library and/or in the Category 1 SL-PCR read set (normalized to the same population sampling depth) was 6277 and, as described in Methods, we calculated for each of these gene loci an estimate, termed the corrected relative abundance ratio (cRAR), of the trans-spliced mRNA fraction. cRAR is structured such that cRAR = 0 for undetectably trans-spliced mRNAs, and, on average, cRAR = 1 for efficiently trans-spliced mRNAs (i.e., those that generate almost exclusively trans-spliced mRNAs). Unexpectedly, there were a significant number of small but nonzero values—e.g., 1207 loci had 0 < cRAR < 0.1, and 60 loci had 0 < cRAR < 0.01, the latter suggesting the possibility of genes that generate on the order of 99% non-trans-spliced mRNA molecules and 1% trans-spliced mRNA molecules.
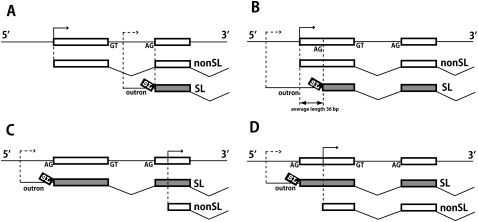
We critically assessed the interpretation that genes with low cRAR values generate predominantly non-trans-spliced mRNAs by referring to our previous independent oligocapping 5′-RACE EST data (oligocapESTs) (Satou et al. 2006). In that 2078 EST data set, 102 genes were represented by ≥3 oligocap ESTs, all non-trans-spliced. Thus, these are abundantly expressed genes that clearly generate mostly non-trans-spliced mRNAs. As expected, we found that many of these genes (68/102) gave cRAR values of 0 in our current analysis, i.e., they were not represented among our Category 1 SL-PCR reads. However, a significant minority (34/102) were represented in our Category 1 SL-PCR reads, and most of these genes gave low cRAR values; 30/34 gave cRAR values between 0 and 0.1. Moreover, the comparatively small numbers of SL-PCR products we detected for these predominantly non-trans-spliced genes represented bona fide trans-splicing events. Close examination of the 30 genes with cRAR between 0 and 0.1 showed that in 21 cases the Category 1 SL-PCR products corresponded to trans-splicing to the acceptor site at the beginning of exon 2 (Fig. 5A), and in the remaining nine cases trans-splicing was to a site upstream of the 5′-end of the non-trans-spliced molecules (Fig. 5C,D). Thus these SL-PCR products did not arise by mispriming on the majority non-trans-spliced mRNAs produced from these loci, but from a minor fraction of authentic trans-spliced mRNA molecules. We use the term “infrequently trans-spliced” to refer to such loci that generate mostly non-trans-spliced mRNAs but also a smaller number of trans-spliced mRNAs.
Figure 5.
Production of both trans-spliced and non-trans-spliced mRNAs from individual genes. Each panel depicts one of four gene expression patterns observed. The top line in each panel illustrates the genomic exon organization of the 5′ part of the gene, with transcription starts shown as arrows. Below each genome depiction are lines showing trans-spliced (SL) and non-trans-spliced (nonSL) pre-mRNAs, and indicating their cis- and trans-splicing patterns. Transcription start sites for non-trans-spliced mRNAs (solid arrow) are precisely mapped from 5′-RACE data (Satou et al. 2006). Transcription start sites for trans-spliced mRNAs (dashed arrow) are not precisely mapped, but must be located upstream of the trans-splice acceptor sites (identified in this study). Note that in C and D the trans-spliced and non-trans-spliced mRNAs must be generated from different promoters. In A and B, a one-promoter hypothesis is not ruled out, but the two-promoter hypotheses illustrated are plausible, based on previous studies of Caenorhabditis genes (Choi and Newman 2006) and on the very short outron lengths a single-promoter hypothesis would require to explain the pattern in B in many cases. The data were derived from a set of 107 genes that were represented among the non-trans-spliced oligocap 5′-RACE ESTs reported in Satou et al. (2006), and also among Category 1 reads in this study. Some of these genes generated more than two mRNA types, and thus displayed more than one of four patterns; the number of occurrences of each pattern in the whole set of 107 genes is: (A) 49; (B) 38; (C) 15; (D) 20.
Because the great majority of the 34 validated infrequently trans-spliced genes discussed above gave 0 < cRAR < 0.1, it was convenient to use cRAR = 0.1 as an empirical dividing line between infrequently trans-spliced and frequently trans-spliced genes. To independently assess the trans-splicing status of cRAR > 0.1 genes, we referred again to our previous oligocapEST data (Satou et al. 2006). The genes in the cRAR > 0.1 category were represented by 336 oligocapESTs, a large majority of which (286 = 85%) were trans-spliced. This result confirmed that, collectively, genes in the cRAR > 0.1 category indeed generate a majority of trans-spliced mRNAs. For further analysis (see below), we can also recognize, within the class of frequently trans-spliced genes, an empirically defined subset of “efficiently” trans-spliced genes that have higher cRAR values, i.e., cRAR > 0.9 (653 genes), with the expectation that genes that generate almost entirely trans-spliced mRNAs would be concentrated in this subset.
Our analysis indicated that the newly recognized class of infrequently trans-spliced genes is a significant component of the genome. Among the 6277 genes for which we could calculate cRAR, 1207 (19%) were infrequently trans-spliced (0 < cRAR < 0.1), 3038 (48%) were frequently trans-spliced (cRAR > 0.1), and 2032 (33%) were undetectably trans-spliced (cRAR = 0). Thus, among cRAR-calculable genes, infrequently trans-spliced genes account for approximately one-fifth of the total number, and an even larger fraction (1207/4245 = 28%) of those represented by SL-PCR products. Assuming the same proportion applies to our global 249,239 read data set, the 8790 total genes represented therein would include ∼2500 infrequently trans-spliced genes and ∼6300 frequently trans-spliced genes.
Trans-splicing and gene function
Our deep sampling of the population of trans-spliced genes permitted investigation of possible correlates between trans-splicing status and gene function as assessed by Gene Ontology (GO) annotation (The Gene Ontology Consortium 2000; http://www.geneontology.org/).
We found a small number (two to four) of GO terms were significantly overrepresented (false discovery rate [FDR] < 0.05) in the infrequently trans-spliced (0 < cRAR < 0.1) and undetectably trans-spliced (cRAR = 0) gene subpopulations (Supplemental Table S6). Three of these overrepresented GO terms were related to ribosomes/translation/nucleic acid binding, consistent with our previous findings (Satou et al. 2006) that ribosomal protein genes were strongly overrepresented among non-trans-spliced mRNAs. Indeed, we found, by direct inspection, that of 77 ribosomal protein genes in the cRAR-calculable set of 6277 genes, 43 had cRAR = 0 and 33 had 0 < cRAR < 0.1.
A much larger number of GO terms, 119, were overrepresented (FDR < 0.05) among the set of 653 efficiently trans-spliced genes (cRAR > 0.9) (Supplemental Tables S7–9). Although numerous and drawn from all three GO ontologies (Cellular Component, Molecular Function, and Biological Process) the overrepresented GO terms clearly fell into a limited number of closely interrelated themes. A total of 84 terms were primarily membrane-related, including 24 Cellular Component terms concerning the plasma membrane/endomembrane system, 19 Molecular Function terms concerning ATP-dependent transmembrane Ca2+ transport, and 41 Biological Process terms concerning Ca2+ transport/homeostasis (19 terms), cell–cell signaling/secretion/transmembrane signal transduction (11 terms), membrane organization/transport/endocytosis (seven terms), and cell adhesion (four terms). An additional theme, actin cytoskeleton, was represented by a set of five Biological Process terms, and by the related term “cytoskeletal protein binding” among a set of Molecular Function terms relating to protein binding/transport. Finally, and perhaps related to the foregoing specific membrane and cytoskeletal themes, a total of 14 Biological Process terms concerned protein transport/assembly and general transport/localization themes. The themes were robust in their support by multiple overrepresented GO terms, including 13 terms with FDR < 0.001, and 50 terms with FDR < 0.01, and also by their stability upon repeat analysis with cRAR thresholds 0.5 or 0.1 (see Supplemental material, section 10). It is interesting to note that these overrepresented GO term themes form a biologically coherent set concerning functionally integrated cellular features (Supplemental Figure S5). Plasma and endomembrane systems, Ca2+ homeostasis/regulation, cell–cell signaling, and cortical cytoskeleton are strongly interlinked elements in the behavior of eukaryotic cells.
Discussion
Our study provides an overview of SL trans-splicing whose breadth and depth are unprecedented. Comprehensive validation analyses confirmed that the vast majority of random-primed SL-PCR products did, as expected, derive from 5′-segments of trans-spliced mRNA molecules, including extremely long molecules such as nebulin mRNA (∼15 kb). Thus random-primed SL-PCR is an efficient and highly specific method for characterizing trans-spliced genes. It should be applicable, with appropriate primer design, to any organism in which SL trans-splicing occurs. It is readily adapted to ultra-high throughput DNA sequencing technologies.
Frequently and infrequently trans-spliced genes
Our data indicate that at least 8790 KH gene loci (58% of the total number of 15,254 KH gene loci) are capable of generating trans-spliced mRNAs in tailbud embryos. However, not all of these gene loci are frequently trans-spliced genes. An unexpected discovery of our work was that a significant number of genes that generate predominantly non-trans-spliced mRNA molecules also generate a detectable number of trans-spliced mRNA molecules. We estimate that such infrequently spliced genes account for ∼2500 of the 8790 genes represented in our SL-PCR data. However, because many of these were represented by a single read, it is very likely that a larger SL-PCR sample would have found more. Indeed it seems probable that a sufficiently large SL-PCR sample would be found to include molecules representing almost every gene in the genome, including those that generate trans-spliced mRNAs only very rarely. In the limit, such extensive sampling could eliminate the undetectably trans-spliced category (apart perhaps from a small number of genes with TOP, or similar specialized, regulation; see below).
With this new insight, our previous qualitative categorization of Ciona genes simply as trans-spliced versus non-trans-spliced, with a very small fraction of “dual” genes generating comparable numbers of trans-spliced and non-trans-spliced mRNAs (Satou et al. 2006), is seen to be an incomplete description. The great depth of our current data has revealed that there are many genes that give rise to both trans-spliced and non-trans-spliced mRNAs over a very wide range of fractional trans-splicing rates. Thus, a more accurate classification would be according to a quantitative measure of trans-spliced mRNA fraction for each locus. As a step in this direction, we introduce the terminology of frequently trans-spliced versus infrequently trans-spliced genes.
The wide range of fractional trans-splicing rates among Ciona genes could be explained by a simple model based on multiple promoters. (Genes with multiple promoters have been shown to be very common in, e.g., the human genome; Kimura et al. 2006.) Depending on their locations within the gene in relation to the distribution of splice donor and acceptor sites, different transcription start sites could generate transcripts that differ in their abilities to undergo trans-splicing. Experimental studies in nematodes have indicated that whether an acceptor site participates in cis- or trans-splicing is largely determined by the presence or absence of an upstream donor site (Conrad et al. 1991, 1993, 1995). Thus, genes could contain one promoter that generates a “donor-site-first” transcript that is efficiently cis-spliced, and another promoter that generates an “acceptor-site-first” transcript that is efficiently trans-spliced. Depending on the relative strengths of the two types of promoter, different genes could generate a mixture of trans-spliced and non-trans-spliced mRNAs in any proportion. We now have data for 107 genes that give rise to both trans-spliced and non-trans-spliced mRNAs and a substantial fraction of these genes (33%) show patterns C and D illustrated in Figure 5C,D, which explicitly require distinct promoters. Patterns A and B (Fig. 5A,B) do not require distinct promoters, but the dual promoter models shown in Figure 5 are plausible and consistent with previous studies of Caenorhabditis genes (Choi and Newman 2006).
The production of alternative trans-spliced and non-trans-spliced mRNAs by individual Ciona genes could be of biological or regulatory significance. Distinct predicted proteins are encoded by the trans-spliced and non-trans-spliced mRNAs of 38 of the 107 genes discussed above, including genes showing each of the patterns A–D in Figure 5.
Concerning the frequently trans-spliced gene class, we estimate that the 8790 genes represented in our global SL-PCR data include ∼6300 frequently trans-spliced genes. Because our prior oligo-capping EST study (Satou et al. 2006) leads us to expect a genome-wide total of ∼7600 such genes (i.e., one-half of the 15,254 total number of KH gene loci) we believe our data have sampled a large majority (∼80%) of the genomic population of frequently trans-spliced genes.
Closely spaced acceptor sites in trans- and cis-splicing
In addition to identifying trans-spliced genes, our SL-PCR sequencing approach precisely maps genomic trans-splice acceptor sites. Detailed analysis of 8929 trans-splice acceptor sites revealed a novel phenomenon: a significant level of alternative trans-splicing consisting of minor acceptor sites as satellites of nearby major sites. Our current data reveal alternative sites within ∼1000 trans-spliced genes, but because many satellite minor sites were represented by a single read this is a minimum estimate of the number of genes having alternative trans-splicing.
Short-interval alternative acceptor sites are also a prominent feature of cis-splicing (Zavolan et al. 2003; Dou et al. 2006). Because most alternative cis-splicing events join protein-coding exons, the outcomes may be sculpted by processes related to translation, such as nonsense-mediated mRNA decay or evolutionary selection operating at the level of protein structure/function. Indeed, some cases of short-interval alternative cis-splice acceptor use are functionally significant and evolutionarily conserved (Akerman and Mandel-Gutfreund 2006; Hiller and Platzer 2008). However, it has been suggested that most alternate use of short-interval acceptor sites in cis-splicing reflects a stochastic aspect of the splicing mechanism, with some intrinsic flexibility in the choice of acceptor site AG (Chern et al. 2006; Dou et al. 2006; Hiller and Platzer 2008). The short-interval alternative trans-splicing revealed by our data concerns sites within the 5′-untranslated region of mRNAs, and would have no expected impact on mRNA degradation or on the structure of the encoded proteins. Thus our finding of extensive use of nearby alternative trans-splice acceptor sites strongly supports the hypothesis that much of the observed short-interval alternative cis-splicing is stochastic (and neutral) microheterogeneity.
A marked similarity between alternative cis-splicing and the alternative trans-splicing we document here is a clear predominance of alternative splicing at the +3 and −3 positions vis-à-vis the major site, corresponding to the motif NAGNAG, so-called tandem alternative acceptor sites (Akerman and Mandel-Gutfreund 2006; Dou et al. 2006; Hiller and Platzer 2008). For cis-splicing, it has been suggested that the ±3-nt preponderance may result in part from post-splicing events such as nonsense-mediated mRNA decay (or evolutionary selection operating at the protein level) because only 3n-nt spacing would preserve the translational reading frame in the alternatively spliced RNAs (Dou et al. 2006; Hiller and Platzer 2008). However, the fact that we also see a predominance of 3-nt spacing in trans-splicing in non-protein-coding 5′-untranslated regions suggests this pattern does not arise from such post-splicing events, but directly reflects spliceosomal mechanisms. Given a requirement for a dinucleotide splice signal (i.e., AG), the closest two acceptor sites can in theory be ±2 nt, i.e., AGAG. However in this sequence the second site NAG identity would be GAG, which is strongly disfavored (Mount 1982; Smith et al. 1993). Thus, the shortest practical spacing is ±3 nt, as in NAGNAG, and it appears that the probability of alternative acceptor site use diminishes with increasing distance beyond this functional minimum for both cis- and trans-splicing.
At slightly longer intervals than ±3 nt, alternative cis-splicing shows clear peaks in the frequency distribution at 3n-nt (n > 2), i.e., at 9, 12, 15, and 18 nt (Dou et al. 2006). In our alternative trans-splicing data (Fig. 4) we see no such peaks, despite a very good depth of data. Thus it is likely that, as suggested by Dou et al. (2006) the cis-splicing 3n (n > 2) pattern results from loss of frame-shifting splice events by nonsense-mediated decay or evolutionary selection at the protein level.
Trans-splicing and gene function
An important issue for understanding gene expression in trans-splicing organisms is to elucidate aspects of gene structure or function that may influence the functional impact of trans-splicing versus conventional (non-trans-spliced) gene expression. In the case of operons, the functional relevance of trans-splicing is known: It plays an essential role in generating translatable mRNAs from downstream cistrons (Blumenthal and Gleason 2003). However, the majority of frequently trans-spliced genes do not reside in operons, and the functional relevance of trans-splicing versus conventional expression in the case of monocistronic genes is not well understood. The extensive sampling of gene populations in our present data permitted us to explore this issue by asking whether trans-splicing status is correlated with gene function as assessed through GO annotation.
In a previous study (Satou et al. 2006), we discovered that the GO term “structural constituent of ribosome” was overrepresented in the non-trans-spliced gene class, largely reflecting the preferential occurrence in this class of ribosomal protein genes. Our current results, with a ∼10-fold larger gene population, confirm the overrepresentation of this and several apparently related GO terms in the population of undetectably and infrequently trans-spliced genes. However, despite the much larger sample size, our current study did not reveal additional overrepresented GO term themes in these gene classes that might suggest the overrepresentation of additional functional gene sets.
The functional significance of the preferential occurrence of ribosomal protein genes in the infrequently and/or undetectably trans-spliced gene classes has not been interpreted, but it likely reflects the prevalence of terminal oligo-pyrimidine (TOP) translational regulation in mRNAs encoding ribosomal proteins and translation factors (Meyuhas 2000). The TOP mechanism requires the presence of a pyrimidine-rich sequence immediately adjacent to the mRNA cap structure (Meyuhas 2000). Because trans-splicing removes the pre-mRNA's original 5′-end (as the outron) it would result in the loss of the TOP sequence and the loss of the translational regulatory control. Selection to maintain the TOP sequence and translational control mechanism could account for the low or undetectable levels of trans-splicing of ribosomal protein genes in Ciona.
Our previous study (Satou et al. 2006) found no GO terms significantly overrepresented in a small sample (∼330) of trans-spliced genes, but in a related sample of similar size Sierro et al. (2009) reported that the terms “mitochondrion,” “protein transport,” and “cell cycle” were overrepresented in the trans-spliced gene class. A significant new finding of our current study was the identification of a large number of GO terms (>100) overrepresented among efficiently trans-spliced and frequently trans-spliced gene classes. These terms fell into several functionally interrelated themes featuring the plasma membrane/endomembrane system, transmembrane Ca2+ transport, cell–cell signaling/secretion/transmembrane signal transduction, membrane organization/transport/endocytosis, cell adhesion, and cytoskeleton. In agreement with the results of Sierro et al. (2009), we found the term “protein transport” overrepresented. However, we did not observe overrepresentation of terms related to mitochondria or to cell cycle, nor did Sierro and colleagues observe overrepresentation of membrane- or Ca2+-transport-related terms. These differences may reflect the ∼10-fold difference in the size of the global gene sets in the two studies.
The overrepresented GO terms are numerous but are tightly focused on a small number of functionally integrated cellular systems. Plasma and endomembrane systems, Ca2+ homeostasis and regulation, and cytoskeleton are functionally integrated, for example, in synaptic transmission and in receptor-mediated endocytosis, two processes identified among overrepresented terms in the Biological Process ontology. Moreover these systems correspond to a coherent larger theme, namely, the cytoplasmic features most characteristic of the eukaryotic cell. Though this is a broad theme, it is represented with high specificity among the overrepresented GO terms. For example, GO terms relating to another eukaryotic cell feature, the nucleus, were completely absent from the list of overrepresented terms, and there was also a virtually complete absence of GO terms relating to eukaryotic cytoplasmic features shared with prokaryotic cells, e.g., energy and intermediary metabolism.
The biological significance of this gene function/trans-splicing correlation is not yet clear. It is not a reflection of operon organization, because we found no GO terms were overrepresented among the 820 KH operon-downstream-cistron genes represented among the 6277 cRAR-calculable genes (data not shown). Rather, the overrepresentation of GO term themes among frequently trans-spliced genes is driven by monocistronic genes. We do not have a mechanistic or evolutionary hypothesis to explain why these particular GO term themes would be overrepresented among efficiently trans-spliced monocistronic genes. However, it presumably reflects the existence of one or more aspects of gene structure/regulation that are correlated with gene function and that have an impact on, or are differentially impacted by, trans-splicing versus conventional expression. It will be an important subject for future research to establish whether a similar pattern applies in other trans-splicing organisms, and ultimately to elucidate the underlying genetic mechanisms.
Contribution to genetic knowledge of Ciona as a model organism
Our study provides several types of data that will be useful for future genomic analyses of Ciona, including the sequence data of 5′-segments of trans-spliced mRNAs, the list of trans-spliced genes including those specifically identified as frequently trans-spliced or infrequently trans-spliced, and the genomic locations of the 8929 stringently defined trans-splice acceptor sites. Access to these resources is outlined in Supplemental material, section 11.
Methods
SL-PCR and DNA sequencing
Isolation of RNA from tailbud embryos of Ciona intestinalis, SL-PCR conditions, and estimation of SL-PCR product size distribution by plasmid cloning are detailed in Supplemental material, section 1. SL-PCR products were ligated to 454 Adaptors A and B and subjected to bead-coupled emulsion-amplification (Margulies et al. 2005) and were sequenced in two runs using the 454 Life Sciences (Roche) GS20 sequencer and protocol with 100 four-nucleotide flow cycles. A total of 249,239 reads passing the instrument quality criteria were obtained. Preprocessing of reads to remove terminal nucleotides designated “N,” present on ∼2% of reads, and estimation of total numbers of reads beginning with the SL-PCR primers SL (124,175 reads) and AN (121,680 reads) are described in Supplemental material, sections 2 and 3. Sorting and filtering the global read set into Categories 1–4 are detailed in Supplemental material, section 5, and trimming of SL and AN primer motifs from read starts/ends is described in Supplemental material, section 6.
Mapping the global set of 249,239 reads to the genome and to gene models
Alignments were by BLAT (Kent 2002) using reads as query and with parameters tileSize = 11, stepSize = 5, minMatch = 2, and minScore = 20. The top-scoring hit(s) was(were) reported for each read query and the output list was filtered so that only hits with a stretch of ≥20 perfectly aligned residues were retained. The distribution of BLAT topscores in alignments with genome version 1 (median 169, mean 150, and mode 186) was parallel to, and only slightly lower than, the distribution of primer-trimmed read lengths (median 188, mean 167, and mode 199). Because a perfect full-length match would give a BLAT score equal to the insert length, this indicates that in general the observed alignment scores were ∼90% of the maximum possible score for the corresponding query lengths. For 95% of reads that hit the genome the second-ranked-hit score was <95% of the topscore, indicating mapping to a unique genomic site.
Genome
To count the number of reads hitting the genome, only the topscore hit was returned for each query (in the case of tied topscore hits, only the first-listed hit was retained). We used the version 1 (December 2002) assembly (Dehal et al. 2002) for detailed analysis because with the version 2 (March 2005) assembly a ∼5% smaller number of reads gave first-base alignments with sites immediately adjacent to AG dinucleotides.
Gene models
The topscore hit was reported for each read query, or in the case of ties, all equal topscore hits were reported. To count the number of reads that hit one or more models, the read name was recovered for each hit and the list of names was made nonredundant and was counted. To count the number of model hits, the model name was recovered for each hit and the list of names was made nonredundant and was counted. In the case of KH models, which are organized both as individual transcript models and “gene loci” which generate one or multiple alternative transcripts, we also collapsed the transcript model data down into gene loci based on the nomenclature convention that all alternative transcripts from a given gene locus share the first three KH model name-fields (Satou et al. 2008). We counted the number of reads hitting each KH gene locus in such a way that a read giving tied topscore hits to several loci would be counted for each locus, but that reads hitting multiple transcript models within a gene locus would be counted only once for that locus. Supplemental Table S2 summarizes gene model set analyses.
Trans-splice acceptor sites in the genome defined by Category 1 reads
We used primer-trimmed Category 1 reads as BLAT queries against the version 1 genome assembly. To define and count the number of trans-splice acceptor sites, we recovered the topscore hit, or all tied topscore hits, for each read. Sites represented by a single read were accepted as candidates only if the alignment score was ≥95% of the theoretical maximum score for that read query-length. The list of sites was filtered to retain only cases where the alignment block started with the first base of the trimmed read, and where the genomic DNA sequence immediately upstream of that base was AG. This yielded 8929 sites.
Genome Browser
The sequences of Categories 1–4 reads were loaded as separate custom tracks in the Ciona intestinalis version 1 (December 2002) assembly UCSC Genome Browser (http://genome.ucsc.edu/; Kent et al. 2002). These custom tracks are available as outlined in Supplemental material, section 11, which also describes access to SL-PCR reads mapped to the KH assembly (Satou et al. 2008).
Estimating fractional trans-splicing efficiency (cRAR)
Efficiencies of trans-splicing were assessed by comparing gene representations in Category 1 SL-PCR products (120,937 reads) and in the Kyoto tailbud embryo 5′-EST library (30,278 ESTs; http://hoya.zool.kyoto-u.ac.jp/download.html; Satou et al. 2003). To count representation, Category 1 reads or tailbudESTs were used as query in BLAST search of the KH transcript model set (Satou et al. 2008) and for each query read/EST the name of the model giving the topscore hit was recovered (in the case of a tie the first model reported was recovered). Transcript models were collapsed down to independent gene loci based on their sharing the first three KH model name-fields and the number of Category 1 reads, and Kyoto tailbudESTs mapping to each gene locus were counted and expressed in terms of relative abundance, i.e., number of counts for that gene divided by the total number of reads/ESTs in the corresponding library. For each gene we computed a corrected Relative Abundance Ratio (cRAR) as follows: cRAR = [(relative abundance in Category 1)/(relative abundance in tailbudEST)]/3.7. The correction factor 3.7 was introduced because only ∼27% of the global population of mRNA molecules are trans-spliced (Satou et al. 2006), so that the relative abundance of an efficiently trans-spliced mRNA species should be 3.7-fold higher among SL-PCR products than in the tailbudEST library. Thus, the cRAR formula is designed so that an efficiently trans-spliced gene should (on average) give cRAR = 1.0 and an undetectably trans-spliced gene would give cRAR = 0. Additional details, including normalization of library sampling depths, are given in Supplementary material, section 9.
GO term analysis
GO term analysis was based on the use of Blast2GO (Conesa et al. 2005; http://blast2go.bioinfo.cipf.es/home) to recover GO annotations for the set of 6277 cRAR-calculable KH gene loci. Blast2GO was able to recover GO terms for 4023 genes in the 6277-gene master set. GOSSIP (Bluthgen et al. 2005; http://gossip.gene-groups.net/) was used to analyze the distribution of GO terms between subsets of this master gene set. The operator defines a test gene subset and GOSSIP assesses whether any terms are overrepresented in the test gene set by comparison with a reference gene set (master set minus test set) using Fisher's exact test to estimate the FDR, which is a measure of the probability that the observed enrichment for each individual GO term is a false-positive finding. We defined test gene sets on the basis of cRAR values (see above), a measure of trans-spliced mRNA fraction. Blast2GO was able to recover GO terms for 432 of the subset of 653 efficiently trans-spliced genes (cRAR > 0.9).
Acknowledgments
This work was supported by Canadian Institutes of Health Research grant MOP-77708 (K.E.M.H., K.D.). We thank Wayne Sossin for pointing out how TOP regulation can explain why ribosomal protein genes would not be trans-spliced. We also thank Stephen Kenton for preparing the primary data submission to the NCBI Short Read Archive.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRX006190.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.100271.109.
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C 1986. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47: 555–565 [DOI] [PubMed] [Google Scholar]
- Agabian N 1990. Trans splicing of nuclear pre-mRNAs. Cell 61: 1157–1160 [DOI] [PubMed] [Google Scholar]
- Akerman M, Mandel-Gutfreund Y 2006. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res 34: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Gleason KS 2003. Caenorhabditis elegans operons: Form and function. Nat Rev Genet 4: 110–118 [DOI] [PubMed] [Google Scholar]
- Bluthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D 2005. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform 16: 106–115 [PubMed] [Google Scholar]
- Chern TM, van Nimwegen E, Kai C, Kawai J, Carninci P, Hayashizaki Y, Zavolan M 2006. A simple physical model predicts small exon length variations. PLoS Genet 2: e45. doi: 10.1371/journal.pgen.0020045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Newman AP 2006. A two-promoter system of gene expression in C. elegans. Dev Biol 296: 537–544 [DOI] [PubMed] [Google Scholar]
- Clayton CE 2002. Life without transcriptional control? From fly to man and back again. EMBO J 21: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M 2005. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T 1991. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol 11: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T 1993. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J 12: 1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Lea K, Blumenthal T 1995. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA 1: 164–170 [PMC free article] [PubMed] [Google Scholar]
- Csank C, Taylor FM, Martindale DW 1990. Nuclear pre-mRNA introns: Analysis and comparison of intron sequences from Tetrahymena thermophila and other eukaryotes. Nucleic Acids Res 18: 5133–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE 1996. Spliced leader RNA trans-splicing in metazoa. Parasitol Today 12: 33–40 [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157–2167 [DOI] [PubMed] [Google Scholar]
- Dou Y, Fox-Walsh KL, Baldi PF, Hertel KJ 2006. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA 12: 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Kallesoe T, Reinhardt R, Chourrout D, Thompson EM 2004. Spliced-leader RNA trans splicing in a chordate, Oikopleura dioica, with a compact genome. Mol Cell Biol 24: 7795–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36: 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium 2000. Gene Ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KEM 2005. SL trans-splicing: Easy come or easy go? Trends Genet 21: 240–247 [DOI] [PubMed] [Google Scholar]
- Hiller M, Platzer M 2008. Widespread and subtle: Alternative splicing at short-distance tandem sites. Trends Genet 24: 246–255 [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y 2006. Regulatory blueprint for a chordate embryo. Science 312: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. 2006. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16: 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 267: 6321–6330 [DOI] [PubMed] [Google Scholar]
- Mount SM 1982. A catalogue of splice junction sequences. Nucleic Acids Res 10: 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Robin F, Lemaire P 2006. Cellular morphogenesis in ascidians: How to shape a simple tadpole. Curr Opin Genet Dev 16: 399–405 [DOI] [PubMed] [Google Scholar]
- Nilsen TW 1993. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol 47: 413–440 [DOI] [PubMed] [Google Scholar]
- Nilsen TW 2001. Evolutionary origin of SL-addition trans-splicing: Still an enigma. Trends Genet 17: 678–680 [DOI] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Kohara Y, Satoh N 2003. Large scale EST analyses in Ciona intestinalis: Its application as Northern blot analyses. Dev Genes Evol 213: 314–318 [DOI] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N 2005. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zoolog Sci 22: 837–843 [DOI] [PubMed] [Google Scholar]
- Satou Y, Hamaguchi M, Takeuchi K, Hastings KEM, Satoh N 2006. Genomic overview of mRNA 5′-leader trans-splicing in the ascidian Ciona intestinalis. Nucleic Acids Res 34: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, et al. 2008. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol 9: R152. doi: 10.1186/gb-2008-9-10-r152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Li S, Suzuki Y, Yamashita R, Nakai K 2009. Spatial and temporal preferences for trans-splicing in Ciona intestinalis revealed by EST-based gene expression analysis. Gene 430: 44–49 [DOI] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13: 4939–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Steele RE 2001. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci 98: 5693–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe AE, Meedel TH, Hastings KEM 2001. mRNA 5′-leader trans-splicing in the chordates. Genes & Dev 15: 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa HJ, Kawamura K, Yamamoto H, Takagi T 2002. The structural organization of ascidian Halocynthia roretzi troponin I genes. J Biochem 132: 135–141 [DOI] [PubMed] [Google Scholar]
- Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y, Gaasterland T 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res 13: 1290–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]