Abstract
We describe a statistical and comparative-genomic approach for quantifying error rates of genome sequence assemblies. The method exploits not substitutions but the pattern of insertions and deletions (indels) in genome-scale alignments for closely related species. Using two- or three-way alignments, the approach estimates the amount of aligned sequence containing clusters of nucleotides that were wrongly inserted or deleted during sequencing or assembly. Thus, the method is well-suited to assessing fine-scale sequence quality within single assemblies, between different assemblies of a single set of reads, and between genome assemblies for different species. When applying this approach to four primate genome assemblies, we found that average gap error rates per base varied considerably, by up to sixfold. As expected, bacterial artificial chromosome (BAC) sequences contained lower, but still substantial, predicted numbers of errors, arguing for caution in regarding BACs as the epitome of genome fidelity. We then mapped short reads, at approximately 10-fold statistical coverage, from a Bornean orangutan onto the Sumatran orangutan genome assembly originally constructed from capillary reads. This resulted in a reduced gap error rate and a separation of error-prone from high-fidelity sequence. Over 5000 predicted indel errors in protein-coding sequence were corrected in a hybrid assembly. Our approach contributes a new fine-scale quality metric for assemblies that should facilitate development of improved genome sequencing and assembly strategies.
Genome sequence assemblies form the bedrock of genome research. Any errors within them directly impair genomic and comparative genomic predictions and inferences based upon them. The prediction of functional elements or the elucidation of the evolutionary provenance of genomic sequence, for example, relies on the fidelity and completeness of these assemblies. Imperfections, such as erroneous nucleotide substitutions, insertions or deletions, or larger-scale translocations, may misinform genome annotations or analyses (Salzberg and Yorke 2005; Choi et al. 2008; Phillippy et al. 2008). Insertion and deletion (indel) errors are particularly hazardous to the prediction of protein-coding genes since many introduce frame-shifts to otherwise open reading frames. Noncoding yet functional sequence can be identified from a deficit of indels (Lunter et al. 2006), but only where this evolutionary signal has not been obscured by indel errors. Several high-quality reference genomes currently exist, and many errors in initial draft genome sequence assemblies have been rectified in later more finished assemblies. However, because of the substantial costs involved, among the mammals only the genomes of human, mouse, and dog have been taken (or are being taken) toward “finished” quality, defined as fewer than one error in 104 bases and no gaps (International Human Genome Sequencing Consortium 2004; Church et al. 2009). It is likely that other draft genome assemblies will remain in their unfinished states until technological improvements substantially reduce the cost of attaining finished genome quality.
Genome assemblies have been constructed from sequence data produced by different sequencing platforms and strategies, and using a diverse array of assembly algorithms (e.g., PCAP [Huang et al. 2003], ARACHNE [Jaffe et al. 2003], Atlas [Havlak et al. 2004], PHUSION [Mullikin and Ning 2003], Jazz [Aparicio et al. 2002], and the Celera Assembler [Myers et al. 2000]). The recent introduction of new sequencing technologies (Mardis 2008) further complicates genome assemblies, as each platform exhibits read lengths and error characteristics very different from those of Sanger capillary sequencing reads. These new technologies have also spawned additional assembly and mapping algorithms, such as Velvet (Zerbino and Birney 2008) and MAQ (Li et al. 2008). Considering the methodological diversity of sequence generation and assembly, and the importance of high-quality primary data to biologists, there is a clear need for an objective and quantitative assessment of the fine-scale fidelity of the different assemblies.
One frequently discussed property of genome assemblies is the N50 value (Salzberg and Yorke 2005). This is defined as the weighted median contig size, so that half of the assembly is covered by contigs of size N50 or larger. While the N50 value thus quantifies the ability of the assembler algorithm to combine reads into large seamless blocks, it fails to capture all aspects of assembly quality. For example, artefactually high N50 values can be obtained by lowering thresholds for amalgamating smaller blocks of—often repetitive—contiguous reads, resulting in misassembled contigs, although approaches to ameliorate such problems are being developed (Bartels et al. 2005; Dew et al. 2005; Schatz et al. 2007). Some validation of the global assembly accuracy, as summarized by N50, can be achieved by comparison with physical or genetic maps or by alignment to related genomes. Contiguity can also be quantified from the alignment of known cDNAs or ESTs. More regional errors can be indicated by fragmentation, incompleteness, or exon noncollinearity of gene models, or by unexpectedly high read depths that often reflect collapse of virtually identical segmental duplications.
In addition to these problems, N50 values fail to reflect fine-scale inaccuracies, such as substitution and indel errors. Quality at the nucleotide level is summarized as a phred score, with scores exceeding 40 indicating finished sequence (Ewing and Green 1998) and corresponding to an error rate of less than one base in 10,000. Once assembled, a base is assigned a consensus quality score (CQS) depending on its read depth and the quality of each base contributing to that position (Huang and Madan 1999). Finally, assessing sequence error has traditionally relied on comparison with bacterial artificial chromosome (BAC) sequence. Discrepancies between assembly and BAC sequences are assumed to reflect errors in the draft sequence, although a minority may remain in the finished BAC sequence.
Here, we introduce a statistical and comparative genomics method that quantifies the fine-scale quality of a genome assembly and that has the merit of being complementary to the aforementioned approaches. Instead of considering rates of nucleotide substitution errors in an assembly, which are already largely indicated by CQSs, the method quantifies genome assembly quality by the rate of insertion and deletion errors in alignments. This approach estimates the abundance of indel errors between aligned genome pairs, by separating these from true evolutionary indels.
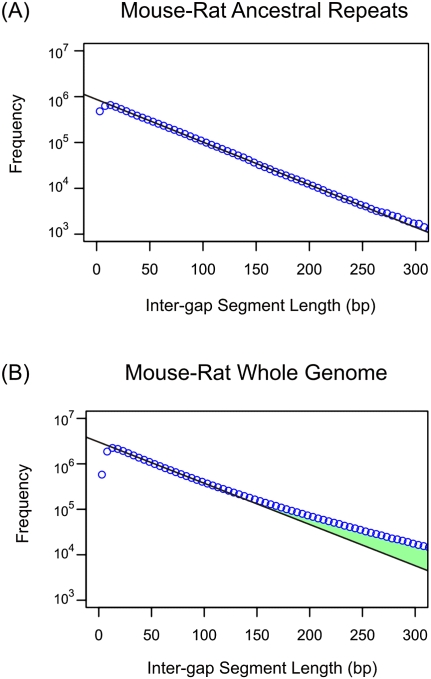
Previously, we demonstrated that in the absence of selection, indel mutations leave a precise and determinable fingerprint on the distribution of ungapped alignment block lengths (Lunter et al. 2006). These block lengths, which represent distances between successive indel mutations (represented as gaps within genome alignments), we refer to as intergap segment (IGS) lengths. Under the neutral indel model, these IGS lengths are expected to follow a geometric frequency distribution whenever sequence has been free of selection. There is substantial evidence that the large majority of mammalian genome sequence has evolved neutrally (Mouse Genome Sequencing Consortium 2002; Lunter et al. 2006). More specifically, virtually all transposable elements (TEs) have, upon insertion, subsequently been free of purifying selection (Lunter et al. 2006; Lowe et al. 2007). This absence of selection manifests itself in IGS in ancestral repeats (those TEs that were inserted before the common ancestor of two species), closely following the geometric frequency distribution expected of neutral sequence (Fig. 1A).
Figure 1.
Genomic distribution of intergap segment lengths in mouse-rat alignments for ancestral repeats (A) and whole-genome sequences (B). Frequencies of IGS lengths are shown on a natural log scale. The black line represents the prediction of the neutral indel model, a geometric distribution of IGS lengths; observed counts (blue circles) are accumulated in 5 bp bins of IGS lengths. Within mouse-rate ancestral repeat sequence, the observations fit the model accurately for IGS between 10 bp and 300 bp. For whole-genome data, a similarly close fit is observed for IGS between 10 bp and 100 bp. Beyond 100 bp, an excess of longer IGSs (green) above the quantities predicted by the neutral indel model can be observed, representing functional sequence that has been conserved with regards to indel mutations. The depletion of short (<10 bp) IGS reflects a “gap attraction” phenomenon (Lunter et al. 2008).
Within conserved functional sequence, on the other hand, deleterious indels will tend to have been purged, hence IGS lengths frequently will be more extended compared with neutral sequence. This results in a departure of the observed IGS length distribution from the geometric distribution (Fig. 1B), the extent of which allows the amount of functional sequence shared between genome pairs to be estimated accurately (for further details, see Lunter et al. 2006).
In any alignment, a proportion of gaps will represent true evolutionary events, whereas the remainder represent “gap errors” that inadvertently have been introduced during sequencing and assembly. Causes of assembly errors, such as insufficient read coverage or mis-assembly, are often regional and thus may be expected to result in clustering of errors. In contrast, from the results of comparisons between species such as human and mouse, true evolutionary indel events appear to be only weakly clustered, for instance, through a dependence of indel rate on G+C content (Lunter et al. 2006). Indels may cluster because of recurrent and regional positive selection of nucleotide insertions and/or deletions. Nevertheless, these effects are unlikely to be sufficiently widespread to explain the high rates of indel clustering (up to one indel per 4 kb) that we discuss later. Indels may also cluster because of mutational biases that are independent of G+C, although we know of no such short-distance effects (see Discussion). This reasoning provided the rationale for seeking to exploit the neutral indel model to estimate the number of gap errors in alignments of two assemblies. Purifying selection on indels and clustered indel errors contribute to largely distinct parts of the observed IGS histogram: The former increases the representation of long IGS (Fig. 1B), whereas the latter cause short IGS to become more prevalent than expected.
Nevertheless, owing to the considerable divergence between human and mouse, the probability of a true indel greatly exceeds assembly indel error rates (5 × 10−2 versus 10−3 to 10−4 per nucleotide) (see below) (Lunter et al. 2006). In short, the large number of true indel events renders the proportion of gap errors so low as to be inestimable. Even for more closely related species, such as mouse and rat (Fig. 1A), neutral sequence is estimated to contain one true indel per 50 bases, which is also approximately 100-fold higher than the frequency of indel errors we will report later. Consequently, indel errors will be most easily discerned between genome assemblies from yet more closely related species. Few species pairs, whose divergence within neutral sequence is low (<5%), have yet been sequenced. Nevertheless, recent reductions in sequencing costs are likely to result in substantial numbers of closely related genomes being sequenced in the near future.
For this analysis, we took advantage of the newly available genome assembly of the Sumatran orangutan (Pongo pygmaeus abelii), sequenced using a conventional capillary sequencing approach (Orangutan Genome Sequencing Consortium, in prep.; D Locke, pers. comm.), and its alignment to other closely related great ape genome assemblies, namely, those of human (Homo sapiens) and chimpanzee (Pan troglodytes). The latter two genomes have been sequenced to finished quality and sixfold coverage, respectively (see Methods) (International Human Genome Sequencing Consortium 2004; The Chimpanzee Sequencing and Analysis Consortium 2005), whereas the effective coverage of the Sumatran orangutan is lower at approximately fourfold (Orangutan Genome Sequencing Consortium, in prep.).
We were able to take advantage of a data set of short reads at approximately 10-fold statistical coverage from a single Bornean orangutan (Pongo pygmaeus pygmaeus) that was shotgun-sequenced using the Illumina short read platform as part of the orangutan sequencing project (Orangutan Genome Sequencing Consortium, in prep.). This substantial read depth afforded us an opportunity to quantify the improvement to traditional capillary-read assemblies from the mapping of short sequence reads. Using a sequence mapper (Stampy) that was specifically designed for high sensitivity and accuracy in the presence of indels as well as substitution mutations (see Methods) (GA Lunter and M Goodson, in prep.), we placed these reads onto the Sumatran orangutan genome assembly. Using this assembly as a template, we called indels and substitutions and, from these, derived a templated assembly of the Bornean individual. This assembly is expected to contain polymorphisms specific to the Bornean individual and also to correct many fine-scale substitution and indel errors present in the Sumatran capillary-read assembly. The assembly will be syntenic with the Sumatran assembly, rather than following the Bornean genome where structural variants exist. Moreover, in regions where the Sumatran genome is divergent or contains many errors, reads will not be mapped; such regions will be excluded from the templated assembly. Using our indel error statistics, we show that this templated assembly improves on the original assembly in terms of accuracy by effectively separating low-fidelity from high-fidelity sequence.
Results
Indel errors in primate genome assemblies
The neutral indel model accurately predicts the relative frequencies of ungapped alignment blocks (or IGS) of different sizes in neutrally evolved sequence. This neutral contribution to the IGS histogram can be modeled accurately and, moreover, separates the contributions arising from clusters of gaps, on one hand, and purifying selection on the other. Clusters of wrongly inserted or deleted nucleotides would cause an excess of short IGS over the number predicted by the neutral indel model. By quantifying this excess, the proportion ɛ, average density D, and number Ng, of clustered erroneous gaps in genome alignments can be estimated (see Methods). In practice, an estimate of ɛ will exceed the true value by an amount that reflects biological indel clusters. However, for draft but not always for finished-quality assemblies, contributions to these statistics predominantly are from indel errors, rather than true indels (see below). In a pairwise genome alignment ɛ, D, and Ng refer to the total number of clustered gaps present in the two assemblies. By polarizing the gaps in three-way genome alignments, it becomes possible to compute these statistics for each of the three assemblies separately and thus quantify the fine-scale quality of single genome assemblies.
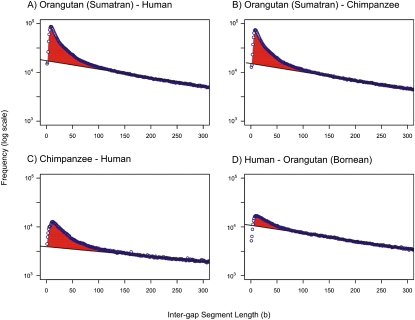
The neutral indel model was first applied to pairwise BLASTZ alignments of genome assemblies from three primates: human, Sumatran orangutan, and chimpanzee (Fig. 2). Genome sequence was filtered to exclude TEs and unplaced sequence: The former show systematic variations in indel rate that the method is sufficiently sensitive to identify (see Methods), while alignments of the latter are less reliable. For each pairwise alignment, the IGS frequency distribution is exceptionally well approximated by the neutral indel model, particularly for extended segments between 150 bp and 300 bp in length, the region over which the model is calibrated. Strikingly, for shorter IGSs, there is a considerable excess of segments over what is expected from an accurate assembly of predominantly neutrally evolving sequence. It is this excess that is used subsequently to estimate rates of clustered gap errors.
Figure 2.
Quantifying gap errors in pairwise alignments of primate genome assemblies. Frequency histograms (natural log scale) of IGS lengths between whole-genome alignments of Sumatran orangutan and human assemblies (A), Sumatran orangutan and chimpanzee assemblies (B), chimpanzee and human assemblies (C), and the human assembly and the Bornean orangutan template assembly (D) created from short reads at 10-fold coverage (see Methods). Repetitive sequence and sequence not placed on chromosomes were excluded (see Methods). Black lines represent the neutral indel model predictions calculated from observed frequencies of IGS lengths (blue circles) between 150 and 300 bases. In all four examples, the expected number of short IGSs is in excess (red) of the number predicted by the neutral indel model. These excesses of short IGSs are due, at least in part, to clusters of gaps representing missing or erroneously inserted sequence, and represent artefacts of the sequencing and assembly process. In alignments of the Sumatran orangutan with human and chimpanzee assemblies, Ng is estimated at 1.3 × 106 and 1.7 × 106, respectively. For alignments of chimpanzee and human, far fewer errors are seen (Ng = 0.3 × 106), suggesting that the anomalies observed in A and B largely reflect inaccuracies in the Sumatran orangutan genome assembly. This is further substantiated by the results for the Bornean orangutan template assembly, which is expected to be more accurate than the Sumatran assembly (Fig. 4).
The largest excess is observed between the alignment of Sumatran orangutan and chimpanzee genome sequences (Fig. 3), from which we estimate an indel error rate of D = 1.2 errors per kilobase (Ng = 1.4 × 106; ɛ = 25%). In other words, one in four gaps in alignments between these two genome assemblies is inferred to have arisen from sequencing or assembly artefacts rather than representing true indel events. Gap errors are only marginally less frequent when we examine the alignment of Sumatran orangutan and human assemblies (D = 0.99 errors per kilobase; Ng = 1.4 × 106; ɛ = 24%) (Fig. 3) but are considerably less frequent in the alignment of chimpanzee and human assemblies (D = 0.28 errors per kilobase; Ng = 3.0 × 105; ɛ = 12%) (Fig. 3). The smaller estimated gap error rate in alignments not involving the Sumatran orangutan assembly suggests that this sequence harbors a larger number of errors compared with either of the chimpanzee or human assemblies. The small difference in estimated gap error rates between chimpanzee and human assemblies when aligned to the Sumatran orangutan assembly indicates that, as expected based on level of curation and hand editing of the clone by clone data, the human genome assembly is of higher fidelity than the chimpanzee assembly (primarily a whole-genome shotgun assembly, with only a relatively small number of finished BACs).
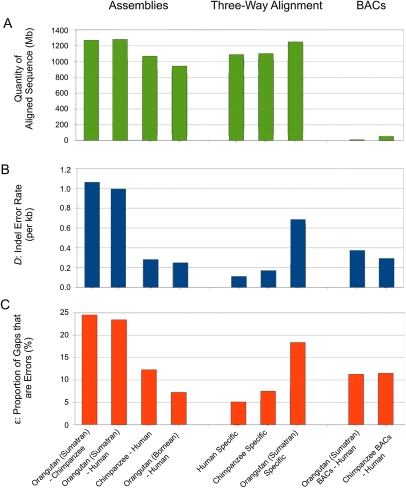
Figure 3.
Inferred gap errors are abundant within low coverage regions of the orangutan assembly and are more scarce in both BAC sequence and a hybrid build of capillary sequence (Sumatran) and Illumina (Bornean) orangutan sequence reads. Histograms showing quantities of aligned sequence (A), frequencies of gap errors (B), and proportions of gaps inferred as errors (ɛ) (C), for diverse aligned assemblies. With whole-genome assembly alignments of primates, high error rates are observed for both alignments that contain the Sumatran orangutan assembly. In contrast, the chimpanzee–human alignment contains relatively few errors. When analyzing only the BAC sequences contributed to the Sumatran orangutan assembly, and aligned to human, the indel error rate D is reduced by over twofold. In contrast, alignments between chimpanzee BAC or whole-genome sequence show similar indel error rates. The increased prevalence of gap errors in the Sumatran orangutan assembly is further demonstrated in lineage-specific analysis of a three-way alignment of primate genome assemblies. Analysis of the Bornean build of the orangutan genome using Illumina shotgun reads (fourth column from left) shows a much reduced indel error rate compared with the original Sumatran assembly.
Lineage-specific analysis
The previous analysis provided information on error rates for assembly pairs, from which some conclusions about individual assemblies could be derived. Nevertheless, it would be preferable to have direct rate estimates for individual assemblies. By using orangutan as an outgroup to human and chimpanzee, it is possible, using parsimony, to infer in which of these two lineages each gap arose, regardless of whether it reflects a true mutational event or else an assembly artifact. The same approach also identifies indels that arose either in the Sumatran orangutan lineage or in the human/chimpanzee ancestral lineage leading up to their split (Fig. 4). The use of parsimony is justified by the modest numbers of gaps within alignments between the three species.
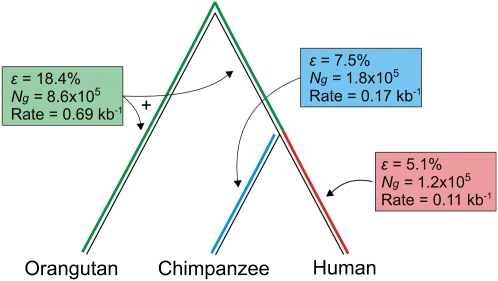
Figure 4.
Estimates of lineage-specific indel errors in great ape genome assemblies. Using a three-way alignment of the Sumatran orangutan, chimpanzee, and human genome assemblies, it is possible to estimate the quantity of lineage-specific indel errors for the chimpanzee and human assemblies. We infer that the remaining indel errors are present in the Sumatran orangutan genome assembly.
We first estimated the total number of indels for each lineage and then used the neutral indel model to separate numbers of indels reflecting true evolutionary events from others representing error (see Methods). This analysis confirmed the previous ranking of the three assemblies by quality. The human genome sequence exhibits the highest fidelity (D = 0.11 errors per kilobase; Ng = 1.2 × 105; ɛ = 5%) (Fig. 3), which is expected considering the greater attention it has received during its transition toward a finished state (International Human Genome Sequencing Consortium 2004). Importantly, this highest-quality assembly still harbors a small number of clustered indels, which may represent errors, clusters of true indels, or a combination of these. The estimate of D thus represents an upper bound to the indel error rate in this assembly (see Discussion).
Compared with human, the chimpanzee genome assembly contains about 50% more predicted errors (D = 0.17 errors per kilobase; Ng = 1.8 × 105; ɛ = 7%), while the Sumatran orangutan genome assembly contains over sixfold more (D = 0.69 errors per kilobase; Ng = 8.6 × 105; ɛ = 18%). The species-specific error rates for human and chimpanzee add up to the estimated pairwise rate (D = 0.28 kb−1), but the pairwise rates for alignments that include the Sumatran orangutan genome (D = 0.99 kb−1 and D = 1.2 kb−1) exceed the respective sums of estimated species-specific rates (D = 0.80 kb−1 and D = 0.86 kb−1). The reduced error rates inferred from three-way alignments likely reflect the preferential exclusion of loci at which any of the three assemblies contains unreliably assembled or badly placed sequence. In addition, the parsimony approach used in generating the three-way alignment may have collapsed true gaps, particularly in regions with high gap densities, thereby further reducing the estimated error rate.
Analysis of BAC sequence
BAC sequence is often considered to be of high fidelity. For the Sumatran orangutan–human alignment, segments enriched with inferred indel errors are substantially less frequent in sequence assembled from finished orangutan BAC clones. For the whole-genome alignment, the neutral indel model leads us to expect 2.90 × 105 short IGS between 5 and 25 bases in length, whereas substantially more (1.04 × 106; 14.0 Mb) short IGS segments between 5 and 25 bases in length are actually observed. Consequently, ∼72% of these short IGS are predicted to be flanked by gap errors rather than by true indels. BAC clones comprise ∼0.5% of the autosomal portion of the genome (15.0 Mb), yet only 0.25% (35 kb) of these short IGS intersect with finished BAC sequence: short IGS are thus only half as abundant in these BAC sequences as elsewhere in the genome. These findings that BAC sequences are indeed of high quality thus present an opportunity to assess the utility of the proposed method for estimating genome assembly quality. We estimated the gap error rate within Sumatran orangutan finished BAC clones that align to human sequence and lie outside of TEs (7.5 Mb) to be D = 0.37 errors kb−1 (ɛ = 11%), over twofold lower than the error rate observed in whole-genome alignments (Fig. 3B). The 95% confidence interval of this value of D is broad (0.24–0.48), owing to the limited quantity of Sumatran orangutan BAC sequences available, relative to the narrow interval observed from the whole-genome analysis (D 95% confidence interval = 0.980–1.002). Nevertheless, despite their low values of D and wide confidence intervals, the present data suggest that errors persist within these high-quality BAC sequences.
Next, we applied our method to 49.4 Mb of nonrepetitive human-alignable chimpanzee BAC clones. This resulted in a similar error rate of D = 0.25 per kilobase (ɛ = 12%) for these clones, which represents only a marginal reduction compared with the chimpanzee–human whole-genome error rate of D = 0.28 errors per kilobase (ɛ = 13%). Again, the 95% confidence interval for D from chimpanzee BAC clones was broader (0.23–0.28) than for the whole-genome value (0.22–0.24) due to lower quantities of aligned sequence. This is consistent with previous declarations that over 98% of the initial (panTro1) chimpanzee genome assembly is of comparable quality to that of finished sequence, having less than one error per 104 bases (The Chimpanzee Sequencing and Analysis Consortium 2005). The quality of Sumatran orangutan and chimpanzee BACs does not reach the level observed for human genome sequence, which consists entirely of sequence from BAC clones. This is likely due to the genome being covered multiply with BAC clones, which reduces the likelihood of the few errors occurring within them being retained in the final assembly, and the extensive curation of the human genome sequence that has been undertaken to meet the Bermuda standards (http://www.genome.gov/10001812) of less than one error in 104 bases and no gaps.
Comparison of assembly methods
In order to demonstrate the use of the neutral indel model as a tool for assessing genome assembly methods, we took advantage of the availability of two different assemblies of the rhesus macaque (Macaca mulatta) genome sequence (Gibbs et al. 2007), both of which had been aligned to the human genome sequence assembly using BLASTZ (Schwartz et al. 2003). The first was an intermediate assembly produced using the PCAP algorithm (Huang et al. 2003). The second was the published rhesus macaque assembly (Gibbs et al. 2007) that was created by merging three intermediate assemblies produced by the PCAP (Huang et al. 2003) assembler, together with the Celera (Myers et al. 2000) and Atlas (Havlak et al. 2004) algorithms. Both rhesus macaque assemblies were produced using the same capillary read data with approximately sixfold statistical coverage. The PCAP assembly, of which 1060 Mb was nonrepetitive and alignable with human, had an estimated indel error rate of D = 0.42 per kilobase (Ng = 4.5 × 105; ɛ = 6.9%). The published merged assembly, of which 1275 Mb was nonrepetitive and human alignable, showed a lower error rate of D = 0.35 per kilobase (Ng = 4.4 × 105; ɛ = 5.9%). This merged assembly had been subjected to additional quality control measures and had a frequency of erroneous indels comparable to that observed between the chimpanzee and human assemblies (D = 0.28 per kilobase).
Construction and analysis of a hybrid genome assembly
The original Sumatran orangutan genome sequence was assembled from capillary reads with an effective statistical coverage of fourfold. We thought to apply this approach to additional sequence that was available from the genome of a second individual, a Bornean orangutan. The two orangutan taxa from Borneo and from Sumatra are considered by some to be species (P. pygmaeus and P. abelii, respectively) owing to their substantial divergence resulting from reproductive isolation over several million years, despite their ability to interbreed successfully (Xu and Arnason 1996). An alternative Bornean orangutan genome assembly was created from short 35-bp and 50-bp Illumina reads mapped to the original Sumatran genome assembly (see Methods). As before, we filtered to retain human aligning, nonrepetitive, placed sequence. This provided 944 Mb of Bornean orangutan sequence for analysis, compared with 1279 Mb from the original Sumatran orangutan assembly.
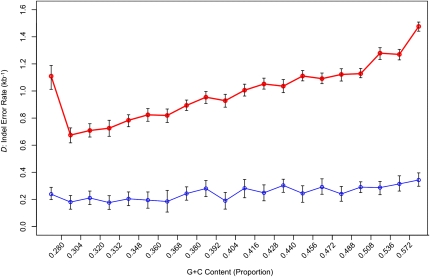
Next, we applied the neutral indel model to alignments containing this Bornean orangutan hybrid assembly. We aligned to the human sequence because owing to its high quality, most of the errors identified will reside within the orangutan sequence. The estimated indel error rate for the hybrid assembly was markedly reduced, relative to that for the original Sumatran orangutan assembly: One anomalous gap was predicted to be present for every 4 kb of sequence (D = 0.25 errors per kilobase; Ng = 2.4 × 105; ɛ = 7.2%). This frequency of indel errors is comparable to that of the chimpanzee assembly (Fig. 3). Comparing the fidelity of the Bornean hybrid assembly with that for the entire original Sumatran assembly, we see a fourfold improvement in D (Fig. 5), whereas the per-base predicted probabilities for true indel events remain largely constant (Supplemental Table 1). For the Sumatran orangutan assembly aligned with human, we noted an association between the G+C content of the sequence and the error rate D (Fig. 5). Errors tended to be highest in high G+C sequence, with the exception of the lowest G+C bin, whose high level of error may accrue from inaccuracies when sequencing homopolymer runs. Capillary read sequencing errors that have previously been associated with G+C-rich motifs (Keith et al. 2004) would explain this bias. In the Bornean hybrid genome assembly, the distribution of errors with respect to G+C is far more uniform (Fig. 5), likely reflecting the incorporation of data from sequence-by-synthesis platforms, as opposed to traditional capillary instruments and dye terminator chemistry.
Figure 5.
Density of gaps that are errors partitioned by genomic G+C content. The y-axis represents the density of indel errors (D) in alignments of nonrepetitive sequence between the human and Sumatran orangutan genome assemblies (red) and the human and Bornean orangutan assembly (blue). Error bars, 95% confidence intervals for the estimation of D, as determined against the neutral indel model calibrated on the frequency of IGS lengths ∼150–300 bp. In the original Sumatran orangutan–human alignments, D appears to be dependent on the G+C content of the sequence, with more anomalous gaps located in G+C-rich regions. D has been significantly reduced in the Bornean genome build, while the estimated per-site probabilities of true indel mutations are comparable to those calculated using the Sumatran genome. As a result, the correlation between G+C and D is less prominent in our build of the Bornean orangutan genome assembly, with a relatively even distribution of gap errors across the 20 G+C bins.
Bornean short reads, as expected, were found to map preferentially to high-quality Sumatran sequence. In 1279 Mb of human aligning, nonrepetitive sequence from the Sumatran orangutan genome assembly, 1238 Mb (96.2%) is of high quality (CQS > 40). Bornean reads were mapped with at least twofold coverage to 1070 Mb of this high-quality sequence (1070/1238 = 86.4%), whereas only 10 Mb (10/41 = 24.4%) of Bornean sequence was mapped to lower-quality sequence (CQS < 0). Indel errors are particularly abundant in the 199 Mb of Sumatran orangutan genome assembly sequence against which insufficient numbers of Bornean orangutan short reads could be mapped. When aligned to the human assembly, the indel error rate for this sequence was D = 4.3 per kilobase (Ng = 8.4 × 105; ɛ = 45.6%), approximately 15-fold higher than for the 1080 Mb of Sumatran sequence mapped with Bornean reads (D = 0.29 errors per kilobase; Ng = 3.1 × 105; ɛ = 8.7%). This result reflects a bias for Bornean orangutan sequence reads to be preferentially mapped to Sumatran orangutan sequence when the latter contains few or no indel errors. Mapping of the Bornean orangutan sequence thus provides both a modest improvement in indel error rates (from 0.29–0.25 errors kb−1) while delineating 1080 Mb of Sumatran sequence of high fidelity from 199 Mb of sequence of low fidelity.
Association between indel errors and CQS
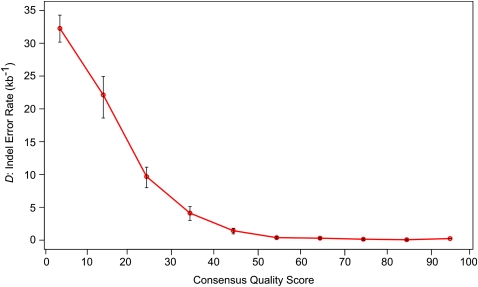
Finally, we compared our metrics for sequence quality against the CQS values of the Sumatran orangutan assembly (Fig. 6). The Sumatran orangutan genome sequence was partitioned into 10 bins, by CQS values, and estimates of the indel error rate D calculated for human alignable nonrepetitive sequence in each bin. As expected, indel errors occur less frequently in assembled sequence with higher CQS (Pearson correlation test: P = 0.004; r = −0.82). For high-quality human-alignable nonrepetitive sequence (CQS > 40; 1238 Mb), the indel error density was reduced threefold (D = 0.34 errors per kilobase; Ng = 4.15 × 105; ɛ = 9.7%) compared with the original unfiltered assembly (D = 0.99 errors per kilobase). For the highest-quality sequence (CQS > 0; 1142 Mb), the error density was further reduced (D = 0.27 errors per kilobase; Ng = 3.08 × 105; ɛ = 8.2%). We conclude that indel error rates in high-quality (CQS > 40) sequence and in BAC sequence are comparable. Finally, because our method quantitatively supports the qualitative CQS values, we may predict that for the 2.72 Gb (88%) of the Sumatran orangutan genome assembly with CQS > 80, error rates will be minimal (D ∼ 0.3 errors per kb).
Figure 6.
Density of gaps that are errors partitioned by consensus quality score. The Sumatran orangutan genome was divided into 10 bins based upon consensus quality score. For each bin, the indel error rate D was calculated for human-alignable nonrepetitive sequence. Error bars, 95% confidence intervals for the estimation of D. This demonstrates that indel errors occur less commonly in sequence with a high-quality score (Pearson correlation test, P = 0.0038; r = −0.82).
Discussion
Applicability of the neutral indel model
We have introduced a model-based, objective, and quantitative method for assessing the fine-scale quality of genome assemblies. The method is independent of the assembly process, unlike other quality metrics such as CQS, statistical read coverage, and N50 contig size. In addition, the model represents an improvement on the CQS metric since it provides error rates without the need for calibration against a gold-standard sequence. The density of gap errors D within genome assemblies provides a new quality metric that assists objective assessment of different sequencing and assembly approaches.
Application of this model is contingent on the availability of genome assemblies from closely related species whose coverage and fidelity must be sufficient to allow their accurate alignment using tools such as BLASTZ (Schwartz et al. 2003). Our approach is less able to quantify gap errors in genomes as more distantly related species are compared, owing to true indel events becoming proportionately greater than gap errors. Rodent genomes, for example, are too distantly related as shown by the lack of excess IGS in alignments to primate genomes (e.g., Lunter et al. 2006). Consequently, this approach is most suited to genome assemblies for species from finely sampled clades, such as catarrhine primates (dS < 0.05) (data not shown) including baboon, vervet monkey, gibbon, and cynomolgus monkey, which together with the marmoset, are among many currently approved sequencing targets. Concentrating genome sequencing efforts on groups of recently diverged species would thus aid not only comparative analyses for biological species (Boffelli et al. 2003) but also the construction of higher-fidelity genome assemblies. The approach is not suitable for analyzing genomes that contain low proportions of neutrally evolving sequence, such as prokaryote genomes, since these do not provide an opportunity for calibrating the neutral indel model. It is expected that as new animal genomes are constructed entirely from short read sequences at high coverage, accurate assessment of their quality, using approaches such as ours, will become increasingly important.
Quantification of clustered indel errors
The method assesses the quality of genome assemblies by estimating the amounts of sequence containing clusters of indels that are in excess of the amounts predicted by the neutral indel model. Based on our observations that (1) the human genome assembly contains the smallest amount of clustered indels of any genome considered here; (2) BAC sequence generally contains fewer indel clusters than the whole-genome assembly of which they form a part; (3) various filtering steps that are expected to reduce the error rate (i.e., consideration only of short read mapped or high CQS or nonrepetitive sequence) reduce the fraction of indel clusters; and (4) indel cluster rates rank genomes as expected by previously perceived quality, we conclude that the rate of indel clusters D provides a valid proxy for assembly quality at the sequence level.
We have provided an initial estimate of D = 0.11 errors per kilobase in the human genome using an orangutan-based three-way alignment. However, this remains an upper-bound value since contributions from true biological clustered indels cannot be discounted. Although indel clustering may arise from recurrent local positive selection, there is scant evidence that this occurs on a genomic scale at a sufficient rate to explain much of the residual indel clustering. Using a human-based three-way alignment for the human lineage–specific analysis, which allows for a more accurate filtering of TEs, we achieve a lower estimate of excess indels of D = 0.057 errors per kilobase. This estimate is approximately 10-fold higher than a previous measure of the discrepancy rate for indels between overlapping clones from the same haplotype (International Human Genome Sequencing Consortium 2004). However, it should be noted that this study only considered overlapping clones if there were no single base mismatches between them. If substitution errors and indel errors during sequencing are correlated, which appears highly likely, this approach would tend to underestimate the true error rate. This therefore suggests that the true error rate will lie somewhere between these two estimates.
As indels are known not to be distributed randomly within TEs (Batzer and Deininger 2002) they have been excluded from our analysis. Moreover, when simple repeat sequences (well-known to be hypermutable) (Dallas 1992) were discarded from our analysis, this had no substantial effect on our estimations of indel error rate (D = 0.91 per kilobase for the orangutan–human comparison; ɛ = 22.4%), although estimates for the human assembly did, however, decrease slightly (from 0.11 to 0.08 errors per kilobase, using orangutan-based alignments). Few other biological processes are known to contribute to short-range correlated indel events, and those that do, such as the regular spacing of indel events next to transcription start sites (Sasaki et al. 2009), appear insufficiently frequent to impact on our results.
If we were to accept the extreme scenario that all indel clusters in the human assembly are of biological origin, we are still able to conclude that BAC sequence from the Sumatran orangutan draft genome assembly contains, in error, a further indel every 10 kb. This is because the inferred rates of indel errors in this BAC sequence is approximately twofold higher than the inferred rate in human sequence, which is also composed of BAC sequence, but incorporated at higher coverage than in the other two draft assemblies.
Our method quantifies only indel errors that are clustered, and will overlook other, more sporadic, errors that lie elsewhere. If, as may be expected, the rate of sporadic indel errors is proportional to that of clustered errors, then we may safely conclude that the human genome assembly harbors the fewest indel errors, followed by the chimpanzee genome assembly and then the Sumatran orangutan assembly.
Short read data and quality scores
We have demonstrated that short-read “next generation” sequencing data from a Bornean orangutan individual can be effectively mapped to much of the Sumatran orangutan assembly, originally constructed from capillary reads, in order to create a Bornean hybrid sequence. In so doing, it was possible to correct a number of presumed errors within the Sumatran genome, while the remaining differences represent true polymorphic indels. In total, there were ∼700,000 differences between the original Sumatran orangutan assembly and the Bornean orangutan hybrid assembly, 87% of which are only of a single nucleotide. Of these “fixed” indels, 5660 were located in human protein coding sequence mapped to orangutan sequence. The short-read Bornean sequences thus will allow many orangutan protein coding gene models to be newly predicted or amended.
The majority of the Sumatran orangutan assembly that is mapped with Bornean orangutan reads is expected to be of high quality, as indeed is indicated by its enrichment in high-CQS sequence. In contrast, the small minority (19.7%) of the assembly that has not been mapped at sufficient coverage with Bornean orangutan reads will be of lower quality. Consequently, mapping of short read data provides an efficient approach to separating high-quality from low-quality regions of an assembly. In our study, we examined the use of Illumina short read data, although 454 Life Sciences (Roche) and Applied Biosystems SOLiD System data could be used also for this purpose.
High fidelity assembly of genomes
Our results show that the Sumatran orangutan draft genome assembly is exceeded in accuracy by that for chimpanzee and the essentially finished assembly for human. Nevertheless, in the 1279 Mb of this orangutan assembly, ∼70% of inferred errors are concentrated in the 3.2% of the assembly that is of low quality (CQS < 40). This demonstrates that much of this assembly, and the conclusions that have been derived from it (Orangutan Genome Sequencing Consortium, in prep.), are of high fidelity.
We have described how the neutral indel model can assess sequence fidelity within whole genomes or selected portions, such as BACs; within an assembly; and within a single assembly aligned to one or two other closely related genome assemblies. Differences in fidelity can now be quantified within, and between, genome assemblies. This should now allow objective comparisons to be made between assembly algorithms, between sequencing technologies, and between different assembly regions.
Methods
Sequences and annotation
Five sets of BLASTZ whole-genome alignments were acquired from UCSC Genome Informatics (http://genome.ucsc.edu): mouse/rat (mm8vsRn4), orangutan/human (ponAbe2vsHg18), orangutan/chimpanzee (ponAbe2vsPanTro2), chimpanzee/human (panTro2vsHg18), and human/rhesus macaque (hg19vsRheMac2). An analysis of an early draft chimpanzee assembly has been published (The Chimpanzee Sequencing and Analysis Consortium 2005). The panTro2 assembly we investigate here is of higher sixfold coverage. An additional whole-genome alignment of human and the intermediate PCAP assembly of rhesus macaque (Gibbs et al. 2007) was produced using the UCSC BLASTZ whole-genome alignment process (Schwartz et al. 2003), the tools for which are available from UCSC Genome Informatics. Sequence not placed on chromosomes was excluded, because alignment of these regions is often inaccurate. The repetitive portion of the genome was identified using annotations from RepeatMasker (http://www.repeatmasker.org) and discarded. CQSs for the Sumatran orangutan genome were acquired from UCSC Genome Informatics. The locations of BAC clones incorporated into the orangutan and chimpanzee genome assemblies were provided by the orangutan and chimpanzee genome sequence consortia, respectively. Annotations of human coding sequence (NCBI36.54) were acquired from Ensembl (http://www.ensembl.org). These were mapped to the Sumatran orangutan genome using the Lift-Over function on Galaxy (http://main.g2.bx.psu.edu/), where a minimum of 10% of annotated bases were required to map to be included.
IGS length histograms
IGSs are defined as gap-delimited ungapped segments of aligned sequence from genome assemblies of two species. In the case that a genomic region has been excluded from the analysis (e.g., because of a TE annotation), any gaps within the excluded region were ignored: The IGS consists of the ungapped segments on either side of the excluded region, each delimited by a single gap and considered as one contiguous segment. The neutral indel model provided a fit to the observed histogram of IGS counts against length, by weighted linear regression on the log frequencies, with weights derived from the expected sampling error per length bin (modeled as a binomial distribution) in log-space. For histograms of primate alignments, the length intervals over which this regression was performed were determined by maximizing the coefficient of determination over a range of IGS length intervals. This procedure was performed independently for each of 20 data sets consisting of sequence binned by G+C content (see below). Limits were placed on the length intervals we considered so that the start of the regression would begin over IGSs 150–160 bp in length, and end in IGSs 300–310 bp in length. These limits prevented the regression from fitting to frequencies of shorter and longer IGSs, which exhibit substantial contributions from indel errors and functional sequence, respectively. The resulting regression line represents the expected counts under the neutral indel model. To estimate Ng, we accumulated the difference between the observed and expected IGS counts for small IGS lengths, starting from the smallest IGS lengths that exceeded the expectation in order to account for low IGS counts due to gap attraction (Lunter et al. 2008). The proportion ɛ of indels that are errors was calculated by dividing Ng by the total number of IGSs in the whole of the alignment being analyzed. The indel error rate D was calculated by dividing Ng by the total number of aligning bases (which is equal to the total number of nucleotides covered by IGSs).
For the lineage-specific analyses based on the three way alignment (see Creation of a Three-Way Alignment section), parsimony was used to infer in which of the three lineages each indel event had occurred. The neutral indel model was then applied, in turn, to each individual genome sequence assembly within the alignment, with IGSs now defined as stretches of sequence between adjacent indels occurring in each particular lineage. For each sequence in the alignment, Ng was estimated as the difference between observed and expected counts of short IGSs. The proportion ɛ was calculated by dividing the species-specific estimate of Ng through the total number of IGSs for that species. The indel error rate D was calculated by dividing Ng by the total number of alignable non-indel bases in each lineage.
Accounting for variation in indel rates
The neutral indel model hinges on two key assumptions: that true indel events occur independently of one another and that indel events occur uniformly across neutral sequence. While the first of these assumptions is likely to be true, there are three factors affecting the uniformity of indel occurrence across the genome, all of which were accounted for before we began our analyses. First, the rate of neutral indel occurrences correlates with local G+C content (Lunter et al. 2006), with higher frequencies of indels located within sequences showing extremes of G+C content. We account for this variation in indel distribution by partitioning genomic sequence into 20 bins based on the proportion of G+C content in windows 250 bases in length. Thresholds of these bins are adjusted so that the genome is apportioned equally among these bins, and indel probabilities are calculated accordingly for each bin. Due to its small size, BAC sequence data were partitioned among five rather than 20 G+C bins. Second, indels are not randomly distributed within primate TEs. These sequences contain multiple homopolymer runs that are prone to mutations and that appear at specific intervals from one another (Batzer and Deininger 2002), leading to systematic preferences in IGS lengths within this subset. To avoid this complication, sequence annotated as originating from mobile elements was excluded from our analysis. Finally, we restrict our analysis to autosomal sequence, so as to prevent complications arising from variation in indel rates as a result of differences in germline history.
Regions to be excluded from the analyses were determined from annotations of one “primary” sequence in each alignment. In the majority of our analyses, this was chosen to be the Sumatran orangutan sequence, because alignments were being partitioned using known properties of the orangutan genome assembly (e.g., BAC clone locations and CQS values). Estimates of indel error vary slightly when different choices of primary sequence are made, largely as a result of uneven quality and coverage of TE annotations among the various genome assemblies. When the human genome assembly is used as the primary sequence in an alignment, a more complete TE annotation is available. This results in larger quantities of aligned sequence being excluded from the analysis.
Creation of a three-way alignment
The three-way alignment among great ape genome assemblies was created from the two pairwise alignments by a local realignment pipeline implemented in Python. First, two parallel streams of human-based pairwise alignment columns were synchronized and naïvely spliced together into a stream of triple alignment columns. This stream was fed first into a repositioner, which was run twice in series, then through a postprocessor, and finally to a formatter. The repositioner considers at most three gaps in a window of 200 bp, and merges abutting identical-length insertions and deletions that may result from the naïve splicing. In addition, it considers a range of re-positioned gaps to enable further merging, using a score function that includes penalties for lineage-specific gap opening and sequence mismatches. It was found that after two rounds of repositioning, no further improvements could be obtained. The postprocessor flags gaps extending beyond 200 bp in any of the three species, which likely represent either synteny breaks, TE insertions, or missing sequence. Finally, the formatter breaks the stream of alignment columns up into MAF-formatted blocks, removing long gaps from the output. The implementation is available upon request.
Creation of the Bornean orangutan genome sequence
A Bornean orangutan genome template assembly was created by mapping Illumina single and paired-end reads of 35 and 50 bp of a Bornean orangutan individual, with 10-fold total coverage, onto the existing Sumatran genome assembly. The first stage involved mapping reads using Stampy (GA Lunter and M Goodson, in prep.). Full details on the algorithm will be published elsewhere; in brief, Stampy hashes the genome using 15-bp words, against which lookups are performed for every 15-bp subsequence of a read, and each of their 45 neighbors at edit distance 1. A list of potential candidates is created by filtering for a similar genomic environment, duplicate removal, prealignment by a banded linear-gap-penalty aligner that considers 1- and 2-bp gaps, and finally scoring using affine gap penalties. The algorithm guarantees that the candidate list includes the correct location whenever a read has either three or fewer substitutions, or a 1- or 2-bp indel and one substitution, in the first 34 bp and degrades gracefully beyond this. For paired-end reads, the algorithm next builds paired-end candidates using the results from the single-end stage. Pairs are scored and ordered by likelihood using a model that includes priors for substitutions, indels, library separation, large (<10 kb) indel events, and structural variation. Finally, reads are scored and realigned using a probabilistic aligner, identifying nonuniquely placed indels. On simulated human data with polymorphisms and empirical read errors, Stampy correctly maps 97% of paired-end reads and, conditional on accessibility, has a sensitivity for identifying indels in both single- and paired-end maps of up to 96%, depending on the indel size.
A Bornean orangutan template assembly was created from the mapped reads. First, a filtering step retained uniquely mapable and not excessively divergent reads using the following criteria: Map accuracy Phred scores between 10 and 90 for single reads, and between 10 and 140 for paired-end reads; the read or reads show at most three single-nucleotide variants and one indel; the distance between paired reads is at most 700; and the total coverage is between three and 20. Single nucleotide variants relative to the reference sequence were identified and applied to the Bornean assembly by a simple majority vote. Indels in reads were considered only when placed at least 10 bp from either end, owing to the lower sensitivity for inferring indels toward the ends of reads, and called when supported by at least two reads, taking account of nonunique placement. Because of these criteria, indels could be called effectively only in regions with better than twofold coverage of reads, after removing their 10-bp flanks. Whenever this “effective” coverage, so defined, was insufficient for calling indels, the reference sequence was copied instead and marked as lower case. The resulting hybrid Bornean template assembly is expected, conditional on sufficient effective coverage, to contain a fraction of the species-specific and the Bornean individual's polymorphic single-nucleotide variants and indels, but to be largely free from indel errors that were inadvertently included in the Sumatran assembly, as these would appear as homozygous variants with respect to that assembly. Of 2.79 Gb of the original Sumatran orangutan assembly, 2.24 Gb had sufficient effective coverage. After filtering to retain human aligning, nonrepetitive, placed sequence, 944 Mb of Bornean orangutan sequence remained for analysis.
Acknowledgments
We thank the Medical Research Council (S.M., C.P.P., and G.L.) and the Wellcome Trust (G.L.) for funding. We thank The Genome Center at Washington University for use of the orangutan genome assembly (http://genome.wustl.edu/genomes/view/pongo_abelii/), and the consortium, Elaine R. Mardis, and Richard K. Wilson for helpful discussions. We thank Oliver A. Ryder and Leona G. Chemnick at San Diego Zoo Conservation for providing samples for the Bornean orangutan individual.
Footnotes
[Supplemental material is available online at http://www.genome.org. Software is available at http://genserv.anat.ox.ac.uk/downloads/software/indelerror/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.096966.109.
References
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 [DOI] [PubMed] [Google Scholar]
- Bartels D, Kespohl S, Albaum S, Druke T, Goesmann A, Herold J, Kaiser O, Puhler A, Pfeiffer F, Raddatz G, et al. 2005. BACCardI—a tool for the validation of genomic assemblies, assisting genome finishing and intergenome comparison. Bioinformatics 21: 853–859 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394 [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Choi JH, Kim S, Tang H, Andrews J, Gilbert DG, Colbourne JK 2008. A machine-learning approach to combined evidence validation of genome assemblies. Bioinformatics 24: 744–750 [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas JF 1992. Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mamm Genome 3: 452–456 [DOI] [PubMed] [Google Scholar]
- Dew IM, Walenz B, Sutton G 2005. A tool for analyzing mate pairs in assemblies (TAMPA). J Comput Biol 12: 497–513 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Havlak P, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Weinstock GM, Gibbs RA 2004. The Atlas genome assembly system. Genome Res 14: 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A 1999. CAP3: A DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang J, Aluru S, Yang SP, Hillier L 2003. PCAP: A whole-genome assembly program. Genome Res 13: 2164–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, Zody MC, Lander ES 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res 13: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JM, Cochran DA, Lala GH, Adams P, Bryant D, Mitchelson KR 2004. Unlocking hidden genomic sequence. Nucleic Acids Res 32: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D 2007. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci 104: 8005–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Ponting CP, Hein J 2006. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput Biol 2: e5 doi: 10.1371/journal.pcbi.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Rocco A, Mimouni N, Heger A, Caldeira A, Hein J 2008. Uncertainty in homology inferences: Assessing and improving genomic sequence alignment. Genome Res 18: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2008. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9: 387–402 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z 2003. The Phusion assembler. Genome Res 13: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204 [DOI] [PubMed] [Google Scholar]
- Phillippy AM, Schatz MC, Pop M 2008. Genome assembly forensics: Finding the elusive mis-assembly. Genome Biol 9: R55 doi: 10.1186/gb-2008-9-3-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Yorke JA 2005. Beware of mis-assembled genomes. Bioinformatics 21: 4320–4321 [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, Ogawa M, Matsushima K, Gu SG, Kasahara M, Ahsan B, et al. 2009. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323: 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Phillippy AM, Shneiderman B, Salzberg SL 2007. Hawkeye: An interactive visual analytics tool for genome assemblies. Genome Biol 8: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W 2003. Human-mouse alignments with BLASTZ. Genome Res 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Arnason U 1996. The mitochondrial DNA molecule of Sumatran orangutan and a molecular proposal for two (Bornean and Sumatran) species of orangutan. J Mol Evol 43: 431–437 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]