Abstract
P450cam from Pseudomonas putida is the best-characterized member of the vast family of cytochrome P450s, and it has long been believed to have a more rigid and closed active site relative to other P450s. Here we report X-ray structures of P450cam crystallized in the absence of substrate and at high and low [K+]. The camphor-free structures are observed in a distinct open conformation characterized by a water-filled channel created by the retraction of the F and G helices, disorder of the B′ helix and loss of the K+ binding site. Crystallization in the presence of K+ alone does not alter the open conformation, while crystallization with camphor alone is sufficient for closure of the channel. Soaking crystals of the open conformation in excess camphor does not promote camphor binding or closure, suggesting resistance to conformational change by the crystal lattice. This open conformation is remarkably similar to that seen upon binding large tethered substrates, showing that it is not the result of a perturbation by the ligand. Re-dissolved crystals of the open conformation are observed as a mixture of P420 and P450 forms, which is converted to the P450 form upon addition of camphor and K+. These data reveal that P450cam can dynamically visit an open conformation that allows access to the deeply buried active site without being induced by substrate or ligand.
Cytochrome P450s are ubiquitous heme monooxygenases that activate O2 for oxygen atom insertion into a wide variety of substrates (1). Over 10000 forms of P450s have been identified in bacteria, archaea, plants, fungi, and all higher eukaryotes (2). Examples include major drug metabolizing enzymes of the liver, and biosynthetic enzymes involved in steroid and prostaglandin pathways. These enzymes all share a common protein fold first observed for the camphor metabolizing P450cam from Pseudomonas putida (3, 4).
The large diversity in the specificity or promiscuity of P450s is believed to be due largely to variations in the structure of the substrate binding channel, which is defined by elements including the F and G helices, FG loop and regions near the B′ helix, which fold over and around the heme and the I helix to enclose the substrate and position it for attack by the ferryl heme center. Sequence variation of these elements result in P450s with structurally distinct substrate binding channels, and large differences in these regions have been observed in the structures of bacterial (5), microsomal (6), and mitochondrial P450s (7), supporting the view that the different conformations of the F, G and B′ helices are responsible for the large diversity in size, shape and specificity of the substrate binding cavity.
Conformational dynamics and plasticity are also recognized to play an important role in P450 substrate access and recognition. Both P450 EryK (8) and PikC (9) have been shown to coexist in open and closed states in the absence of substrate. In addition, conformational changes upon substrate binding have been seen for several P450s. Changes were seen in the F and G helices of P450 BM-3 (10) and CYP158A2 (11), or in the BC loop of CYP119 (12), CYP105P1 (13), and CYP130 (14). More dramatic changes involving both the F and G helices and the BC loop were observed in CYP2B4 (15) upon substrate binding. In other examples, a combination of disorder in the BC loop and retraction of the F and G helices was seen (9, 16). Finally, both the BC loop and C helix were observed to become partially disordered in CYP51 upon ligand association (17).
P450cam is the most well-characterized of all P450s, and in many ways it has served as the archetypal model for all P450s, particularly those with high substrate specificity. However, P450cam has remained somewhat of an outlier with regard to the role of conformational flexibility and dynamics, as there has been a general belief that the active site of P450cam is more static and shielded from solvent than other P450s (5). This is largely due to the report of a camphor-free structure obtained by soaking dithiothreitol (DTT) out the active site of crystals, and which showed a closed conformation that is very similar to the camphor-bound form (3, 4, 18). Small angle x-ray scattering (19) and hydrostatic pressure experiments (20–22) have also been used to support the view that substrate-free P450cam exists in a closed conformation. However, it is clear that the structure must undergo significant conformational changes before substrate can enter the solvent-inaccessible binding site. Thus, either the substrate must induce a conformational change in the protein to initiate its binding, or the protein must dynamically visit open conformations in the absence of substrate to allow its entry into the deeply buried channel.
Previous studies have shown that P450cam can be trapped in a range of open conformations in response to binding large tethered adamantane probes (23–25). These substrate analogs bind with the adamantane at the camphor-binding site and accommodate the flexible linkers by adopting states in which the F and G helices are retracted from their position above the closed substrate-binding cavity. It was suggested that these open structures represent substrate access pathways into the buried active site, but it has not been clear if these open conformations are relevant to native function or if they represent unnatural states held open by the presence of the artificial ligand.
Here, we report the crystal structures of P450cam crystallized in the absence of camphor and at high and low [K+], and in the presence of camphor at low [K+]. Both of the camphor-free structures are observed in an open conformation that is distinctly different from the previously observed closed camphor-free state. The implications of this open state to conformational dynamics associated with substrate recognition and binding are discussed.
EXPERIMENTAL PROCEDURES
Protein preparation
The pETCAM-C334A vector was constructed by inserting P450cam (a gift from Prof. T. Poulos) DNA into the pET15b vector (Novagen) between NcoI and XhoI restriction enzyme sites and introducing the C334A mutant using QuickChange (Stratagene). This construct expresses a full-length protein with a mutation at Cys334 to Ala. C334A mutation has been shown to incease protein stability by blocking intermolecular dimerization, but does not effect protein activity (26). P450cam (C334A) was overexpressed in E. coli BL21(DE3) from vector pETCAM-C334A grown for 24h at 25 °C after induction by 0.4 mM IPTG and addition of 1 mM 5-aminolevulinic acid. Cell pellets were resuspended in 50 mM KPi, pH 6.0, 1 mM camphor, 0.1 mM heme, 0.5 mg/mL lysozyme and 50 μg/ml DNAase before lysis in a French press. Cell lysates, clarified by centrifugation, were purified on a DEAE anion exchange column, which was initially equilibrated with 50 mM KPi, pH 6.0 and 1 mM camphor. The column was washed with the same buffer plus 50–100 mM KCl, and the protein was eluted at 300 mM KCl. The pooled fractions were concentrated by ultrafiltration before loading onto a Sephacryl S-200 size exclusion column equilibrated with 50 mM KPi, pH 6.0 and 300 mM KCl. The protein fractions with A417/A280 above 1.45 were pooled and exchanged into 50 mM KPi, pH 6.0, 1 mM camphor, 30 mM β-mercaptoethanol, and frozen at −80 °C for further experiments.
Crystallization, data collection and crystal structure determination
Camphor was removed from protein stock solutions by buffer exchange into 50 mM Tris, pH 7.4 by gel filtration through two sequential PD-10 columns (GE healthcare). For camphor-bound P450cam, 2 mM camphor was mixed with 1 mM P450cam before crystallization. Crystals of P450cam were grown by sitting-drop vapor diffusion at 6 °C. Substrate-free crystals grew from 50 mM Tris, pH 7.4, 12–22% polyethylene glycol 8000 +/−200 mM K+. Camphor-bound crystals grew from 50 mM Tris, pH 7.4, 12–22% polyethylene glycol 8000 and 2–4 mM camphor. The crystals were transferred to cryoprotectant buffer consisting of 50 mM Tris, pH 7.4, 18% polyethylene glycol 8000 and 25% polyethylene glycol 600 +/− 200 mM K+, mounted on nylon loops and flash frozen at 77K.
X-ray diffraction data were collected at 100 K using beamline 7-1 or 11-1 at the Stanford Synchrotron Radiation Laboratory. Data were processed using Scala (27). Initial models for the closed and open structures were derived from those of camphor-bound P450cam (PDB entry 5CP4) and the most open conformation for a series of tethered substrate analogs (unpublished results), respectively. Molecular replacement was done using Molrep (28), and model fitting and refinement were done with Coot (29) and Refmac5 (30). The final models were validated using the programs Procheck (31), Sfcheck (32) and Molprobity (33). Statistics for data collection and refinement are shown in Table 1. The atomic coordinate and structures factors were deposited in the Protein Data Bank under the codes 3L61, 3L62 and 3L63. All structural graphics were generated using the Pymol Molecular Graphics System (34).
Table 1.
Data collection, refinement statistics and validation
| Data set | camphor-free, low [K+] | camphor-free, 200 mM [K+] | camphor-bound, low [K+] |
|---|---|---|---|
| Data collection | |||
| wavelength (Å) | 0.979 | 0.979 | 0.0979 |
| unit cell (Å) | 65.6, 73.5, 91.9 | 65.6, 73.8, 92.4 | 35.4, 98.2, 54.2 |
| β = 103.9 | |||
| space group | P212121 | P212121 | P21 |
| resolution range (Å) | 49.0–1.70 (1.79–1.70)a | 46.2–1.50 (1.58–1.50)a | 27.8–1.50 (1.58–1.50)a |
| No. of total reflections | 175296 | 506840 | 211818 |
| No. of unique reflections | 49496 | 72274 | 57384 |
| Completeness (%) | 99.8 (99.9) | 99.7 (100.0) | 100.0 (100.0) |
| Rmerge (%) | 6.2 (41.7) | 6.0 (54.1) | 6.6 (52.9) |
| <I/σ(I)> | 13.6 (2.9) | 19.4 (3.9) | 12.8 (2.3) |
| Wilson B-value (Å2) | 17.8 | 17.6 | 14.2 |
| Refinement statistics | |||
| resolution range (Å) | 10–1.7 | 10–1.5 | 10–1.5 |
| No. of reflections used | 46706 | 68435 | 54283 |
| Free R reflections (%) | 5.0 | 5.0 | 5.0 |
| R/Rfree | 0.190/0.224 | 0.197/0.216 | 0.164/0.196 |
| rmsd bond length (Å) | 0.0121 | 0.0093 | 0.0096 |
| rmsd bond angle (deg) | 1.377 | 1.284 | 1.407 |
| Ramachandran analysis (%) | |||
| No. of residues in | |||
| favored regions | 98.5 | 99.0 | 98.5 |
| allowed regions | 1.5 | 1.0 | 1.5 |
| outlier regions | 0.0 | 0.0 | 0.0 |
| PDB entry | 3L62 | 3L61 | 3L63 |
Data for the outermost shell are given in the parentheses
UV-visible spectroscopy
Open-form crystals, which were grown in the absence of camphor, were initially dissolved in 50 mM Tris, pH 7.4. The re-dissolved protein samples were divided into two aliquots. One aliquot was diluted into 50 mM Tris, pH 7.4, while the other into 50 mM Tris, pH 7.4, 0.5 mM camphor and 200 mM KCl. UV-visible spectra of ferric and ferrous CO-bound forms were recorded on a HP 8453 UV-visible spectrophotometer. For spectra of ferrous-CO bound form, the protein sample was bubbled with CO gas before addition of aliquots of a fresh solution of sodium dithionite (5–10 mM final). For single crystal spectroscopy, absorbance spectra were recorded of crystals mounted on nylon loops in an Oxford cryo-stream operating at 100 K using an in-house system based on an Ocean Optics USB4000 spectrometer.
RESULTS
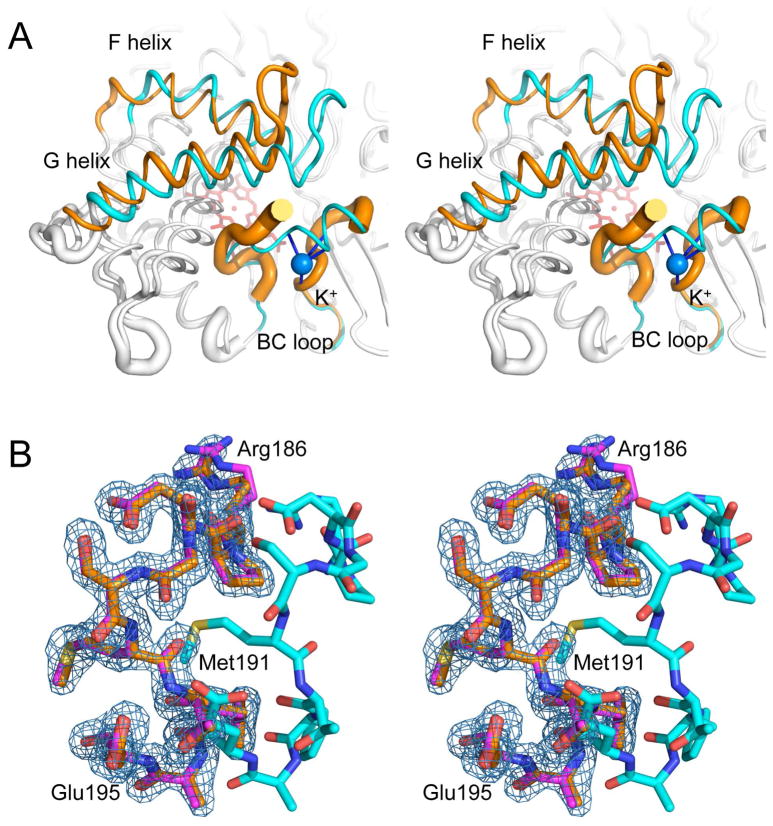
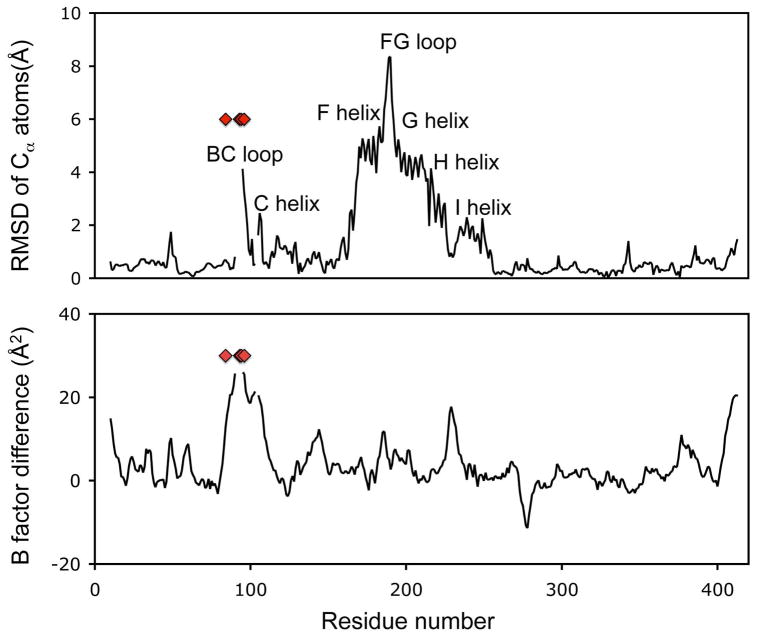
P450cam crystallized in the absence of camphor or other distal heme ligands and at low concentration of monovalent cations was observed in an open conformation characterized by retraction of F and G helices and FG loop, and loss of order in the B′ helix and bound K+ (Figure 1A). The RMSD between the closed and open conformation was 1.8 Å with the greatest displacement at the FG loop (Figure 2), which showed a 9 Å retraction relative to the closed camphor-bound state, while the F and G helices were displaced by about 5–6 Å. The FG loop showed well defined electron density (Figure 1B) and the large movements of this segment were not associated with significantly increased B values (Figure 2). In contrast, the B′ helix, comprising residues 91 to 94, and the K+ ion, were completely disordered in the open camphor-free state.
Figure 1.
Superimposed structures of the closed (PDB entry 3L63) and the open (PDB entry 3L62) forms of P450cam. A) Stereo view of the two conformational forms showing B-values as backbone tube thickness. Only the F and G helices and BC loop are colored. The closed and open forms are shown in cyan and orange, respectively. A blue sphere shows K+. B) 2Fo-Fc electron density map contoured at 1σ of the FG loop for the open structure (PDB entry 3L62). The corresponding model is shown in orange, overlaid on the models for the other open conformation (PDB entry 3L61) in magenta and the closed conformation (PDB entry 3L63) in cyan.
Figure 2.
Per residue RMSD and B factor difference between the open and closed conformations of P450cam. K+ coordination residues are marked by red diamonds.
Additional structures were solved to determine the individual roles played by camphor and K+ ion in the conformational state of the enzyme. The 1.5 Å structure obtained from a crystal grown in the absence of camphor but in the presence of 200 mM K+ was essentially identical to open state seen at low [K+], and did not result in increased ordering of the B′ helix, occupation of the K+ site, or closure of the active site channel (Figure 1B). A third structure at 1.5 Å of P450cam crystallized in the presence of 2 mM camphor and at low [K+] was observed in a completely closed conformation (Figure 1,3), with well defined density for camphor at the active site. In addition, electron density is observed at the K+ binding site of the B′ helix in spite of attempts to exclude monovalent cations during crystallization. When the structural model was refined with a full occupancy of K+ at the coordination site, the Fo-Fc map showed negative electron density around the K+. Thus, the site might be occupied by Na+ or K+ at lower occupancy. Nonetheless, this structure was essentially identical to the previously reported structure of camphor bound enzyme (3, 4). We also considered the possibility that soaking crystals of the open conformation in high concentrations (8 mM) of camphor might reveal camphor bound to the open conformation. However, no significant difference density was seen in the soaked crystals (data not shown), indicating that that camphor has little intrinsic affinity for the open conformation.
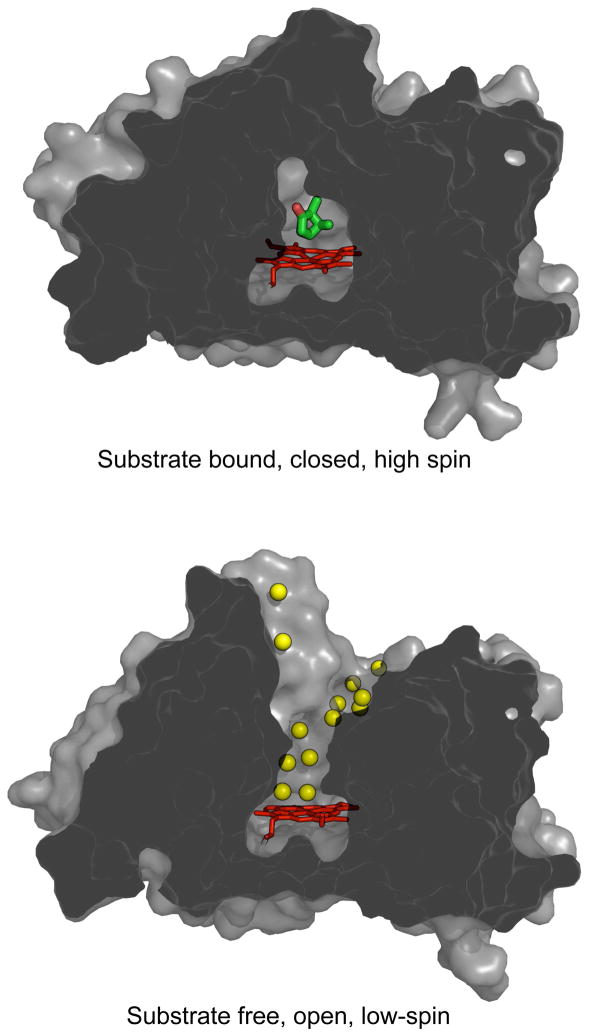
Figure 3.
Cross-section view of the solvent accessible surface calculated for the closed (PDB entry 3L63) and the open (PDB entry 3L62) forms of P450cam. Heme and camphor at the active site are shown in red and green, respectively. Ordered waters seen in the channel are shown as yellow spheres.
The open camphor-free conformation results in a deep water filled channel, exposing the substrate-binding site to solvent (Figure 3). At least 12 ordered water molecules are observed within the open channel which are excluded in the closed camphor-bound state. Included in this set of ordered waters are the distal heme-bound water responsible for substrate induced spin-state conversion, and a water near the position of the “catalytic” water observed in the Fe+2-O2 complex (35, 36) (Figure 4). Accommodation of this “catalytic” water is possible due to the larger space in the distal cavity and a small rotation of Thr-252 side-chain. The locations of these channel waters are distinct from those previously observed to occupy the camphor site in the closed camphor-free form (18). However, an unidentified small peak at 2 σ was seen at the camphor site that was too large for a water and to small for camphor. Possible candidates for this density are Tris and β-mercaptoethanol from the original protein stock and the crystallization buffer. The smaller β-mercaptoethanol fits better to the electoron density, but the identity of this peak remains uncertain. Additional electron density was also observed at ~ 1 σ throughout the open channel that may represent disordered solvent.
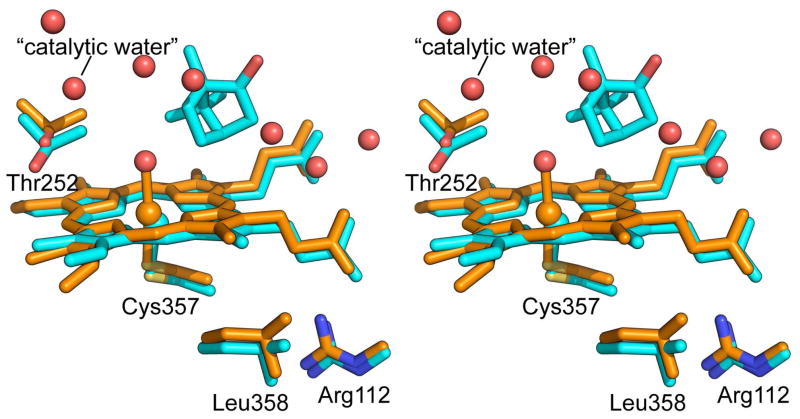
Figure 4.
Movements of the heme and nearby residues in the open conformation (PDB entry 3L62) relative to the closed form (PDB entry 3L63). The closed and open forms are colored in cyan and orange, respectively. Camphor is only present in the closed form. Waters shown in red are only present in the open form. In the closed camphor-bound state the heme is 5-coordinate with only Cys357 as proximal ligand, while in the open conformations the heme is 6-coordinate, with a distally coordinated water shown bound to the iron.
As shown in Figure 4, subtle structural differences between the open and closed conformations extend to proximal heme face. Both the Arg112 and Leu358 side chains move slightly toward the heme in the open conformation, making contact with the heme D ring. Small changes were also seen in the geometry of the heme which shows increased ruffling. The iron in the open conformation moves 0.7 Å toward the heme plane and the axial Cys ligand follows this movement. While the Fe-S bond length was refined to be ~0.05 Å shorter in the open relative to the closed conformations (Table 2), this difference may be within the uncertainty of refinement precision. Overall, these proximal changes are very similar in nature to those seen in the L358P mutant which was proposed to mimic changes induced by Pdx binding (37).
Table 2.
Geometry around the heme of P450cam
| Structure | camphor-bound, low [K+] | camphor-free, low [K+] | camphor-free, 200 mM [K+] |
|---|---|---|---|
| Fe–OH2 (Å) | NA | 2.25 | 2.17 |
| Fe–S (Å) | 2.38 (2.37)a | 2.31 | 2.33 |
| Fe–NA (Å) | 2.07 (2.05) | 2.02 | 2.04 |
| Fe–NB (Å) | 2.04 (2.05) | 2.04 | 2.01 |
| Fe–NC (Å) | 2.08 (2.05) | 2.04 | 2.07 |
| Fe–ND (Å) | 2.11 (2.06) | 2.05 | 2.05 |
| ∠ NA–Fe–S (°) | 100.4 (102.3) | 96.9 | 96.8 |
| ∠ NB–Fe–S (°) | 90.1 (92.3) | 88.5 | 88.8 |
| ∠ NC–Fe–S (°) | 94.8 (93.9) | 88.1 | 87.4 |
| ∠ ND–Fe–S (°) | 105.6 (103.5) | 96.1 | 95.6 |
Data from structure of PDB entry 2CPP (4) is shown in parentheses.
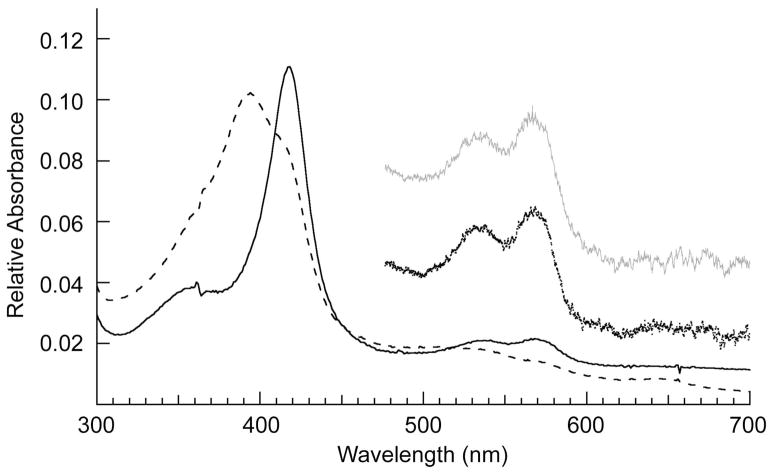
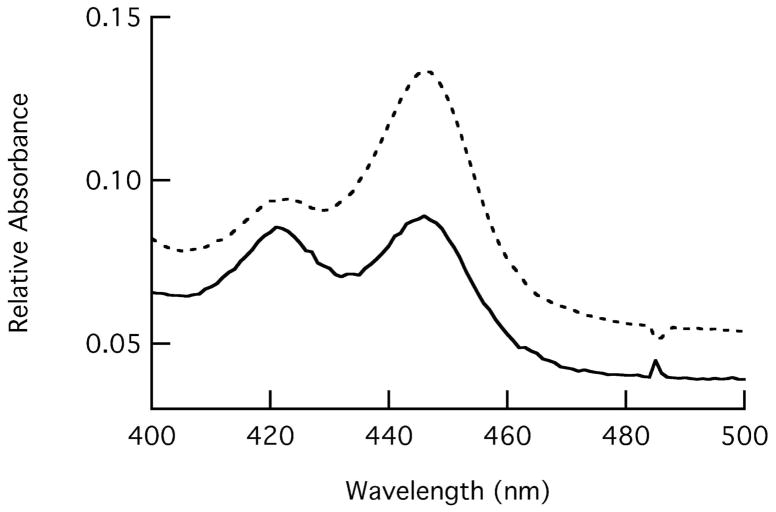
Spectra of frozen single crystals at 100K of P450cam crystallized in camphor-free, low [K+] and in camphor-free, 200 mM [K+] conditions both exhibit α/β bands at 568 nm and 535 nm which are characteristic of the low-spin state expected from the observed water coordination to heme (Figure 5). When these open-state crystals are re-dissolved in 50 mM Tris, pH 7.4, they remain in a low-spin state, while addition of camphor (0.5 mM final) and K+ (200 mM final) to this solution causes conversion to high-spin, as seen by the significant attenuation of the α/β bands, appearance of the charge transfer band at 645 nm and a shift of the Soret from 418 nm to 393 nm (Figure 5). This shows that the open form of P450cam seen in the crystal is capable of binding camphor to reversibly generate the closed camphor-bound state. Finally, the ferrous CO complex produced from open state crystals re-dissolved in 50 mM Tris, pH 7.4 without camphor or K+ is observed to exist as a mixture of P450 and P420 forms, while addition of 0.5 mM camphor and 200 mM K+ to this sample of re-dissolved crystals shows that it has converted to predominantly the P450 form (Figure 6).
Figure 5.
UV-Visible spectra of P450cam derived from open form crystals. Single crystal UV-visible spectra at 100K of P450cam grown in camphor-free buffer 50 mM Tris, pH 7.4 and 200 mM KCl (black dots) and grown in camphor-free buffer 50 mM Tris, pH 7.4 (gray dots). Spectra are also shown for re-dissolved open-form crystals in 50 mM Tris, pH 7.4 (solid line) and in 50 mM Tris, pH 7.4, 200 mM KCl and 0.5 mM camphor (dashed line).
Figure 6.
UV-Visible spectra of the ferrous CO complex for P450cam derived from open-form crystals. The open-form crystals were dissolved in 50 mM Tris, pH 7.4 (solid line) or in 50 mM Tris, pH 7.4, 200 mM KCl and 0.5 mM camphor (dashed line) before conversion to the ferrous CO complex.
DISCUSSION
A number of significant conclusions can be made from the simple observation that P450cam crystallized in the absence of camphor exists in an open conformation.
The open conformation described here would appear to be at variance with the previous report of the substrate-free structure which was observed in a closed conformation (18). However, it is noted that this closed camphor-free structure was obtained by first crystallizing the protein with DTT bound to the heme in place of camphor and subsequently soaking DTT out of the crystals. As such, it is possible that the closed DTT bound conformation was trapped by the crystal lattice, preventing its conversion to the open conformation. Recently, a closed P450cam structure partially bound to camphor was reported, but this crystal was also grown with excess DTT in addition to 1 mM camphor and 200 mM K+ (38). Consistent with this, we find in this study that crystals of the open camphor-free protein, when soaked in excess camphor, fail to convert to the closed conformation, which is observed when the same protein is crystallized in the presence of camphor. Again, this suggests that crystal packing resists the conversion between open and closed conformations.
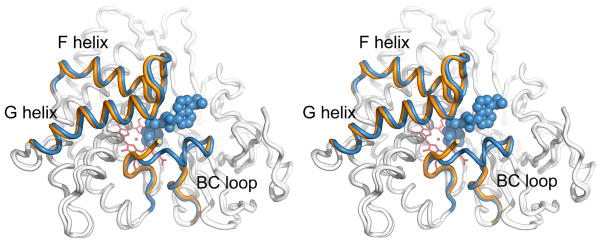
The open camphor-free conformation is very similar to that observed when bound to large tethered substrate analogs containing linkers that prevent simultaneous binding and closure of the channel (23–25). The RMSD between the camphor-free structure reported here and the most open of these tethered substrate bound forms is 0.5 Å (Figure 7). The similarity of these structures shows that the protein samples the same open conformation in the absence of any substrate as when it is bound to the long tethered substrates, showing that the open conformation is not the result of artificially forcing the channel open. Recent observations suggest that CYP119 exists in multiple conformational states that depend on heme coordinating inhibitors, and these multiple conformations persist even at saturating ligand concentrations. This suggests that both ligand association and dissociation are governed by protein conformational dynamics (39, 40)
Figure 7.
Stereo view of the superimposed structures of P450cam in the substrate-free open conformation (orange) (PDB entry 3L62) and the previously reported open state bound to a tethered substrate (blue) (PDB entry 1RF9). Only the F and G helices and BC loop are colored. The tethered substrate, adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide, is shown as a cpk model in blue. The most significant difference in the structures is the increased disorder in the B′ helix for the substrate-free form.
P450cam is the only P450 known to have a specific K+ binding site in the B′ helix. K+ has been shown to enhance camphor binding by 10 fold (41), and to increase the stability of the enzyme (22, 42). However, crystallization in the presence of high [K+] alone does not induce any change in the open conformation. This suggests that K+ binding does not in itself provide sufficient stability to reorder the B′ helix, and its presence alone is not sufficient to drive closure of the active site channel in the absence of camphor. Thus the ordering of the B′ helix and K+ coordination appear driven by substrate binding. As the K+ binding affinity has been estimated to be very weak even in the presence of camphor (~ 10 mM) (41), we initially assumed that two successive gel filtration steps would result in its removal from the camphor bound state. However, the structure of P450cam crystallized in the presence of camphor and at low [K+] is observed in a closed conformation with the K+ site partially occupied, showing that a monovalent cation is still sequestered by the protein or scavenged from the media. This suggests that camphor-driven closure of the substrate channel accompanies cation binding and reordering of the B′ helix, and it thus provides a structural explanation for the known synergistic effect of camphor and K+ binding (41).
In spite of the hydrophobic nature of the exposed substrate binding channel in the open conformation, at least 12 ordered waters and evidence for significantly more disordered solvent molecules are seen in the open conformation of P450cam. This is consistent with the previous report that the volume change induced by osmotic pressure involves reorganization of approximately 19 waters during spin-state conversion (20).
Comparison of the open and closed conformations suggest that multiple small changes propagate to the proximal region near the proposed Pdx binding site that may have functional significance. These changes are very similar to those seen for the L358P mutant, which mimics conformational dynamics seen upon Pdx binding (37, 43). The L358P has 30 times higher binding affinity for Pdx than the WT P450cam (44), and hydroxylates camphor using sodium dithionite as the sole reductant (45). Pdx binding has been shown to induce changes in the spin-state (46) and electronic structure (47) of P450cam, and triggers trapping of water molecules in the P450cam-Pdx complex (48, 49). It has also been shown that Pdx binding causes perturbations at the B′ helix (50) and cis-trans isomerization of the Ile88-Pro89 peptide bond (51). Although we did not see such a transition of the Ile88-Pro89 peptide bond in the open-state crystal structures, given the similarity of changes near proximal site between the L358P mutant and WT compared with the changes between the open and closed form, it is possible that Pdx may modulate structural changes that are coupled to the open-closed transition.
When crystals of the open conformation of P450cam are dissolved in the absence of camphor and at low [K+] and converted to the ferrous CO complex, a mixture of P420 and P450 species is seen, while addition of camphor and K+ results in predominantly the P450 form. It is reported that high pressure reversibly converts P450 BM3 and CYP119 to a P420 form that is associated with a change in molecular volume and spin-state conversion (52, 53). For P450cam, intermediate pressure induces a spin-state change that is reversible but higher pressure induces an irreversible conversion to P420 (54, 55). However, a structural basis for these changes has not been understood. Our results suggest that the open conformation obtained by crystallization of camphor-free enzyme may be associated with a P420 form that reverts to the closed P450 form upon binding substrate. It has been proposed that changes in iron-cysteine coordination may be responsible for P420 forms (56–58), but it is unclear at present if the slightly shorter Fe-S distance seen in the open conformations is significant in this regard (Table 2).
The observation of an open conformation of P450cam unifies a developing consensus that conformational dynamics plays an important role in substrate recognition for most if not all P450s. Recent structural studies have also shown that substrate-free P450 EryK can exist in both open or closed conformations in different salt conditions (8), and two monomers in the asymmetric unit of substrate-free P450 PikC displayed alternate open and closed conformations (9). In both cases, the open state resulted from a retraction of the F and G helices similar to the changes seen for P450cam.
The structures reported here of P450cam in the presence and absence of camphor and K+ ion provide a new picture of substrate access and conformational mobility in this archetypal enzyme. The active site of the closed camphor bound conformation is solvent inaccessible, requiring a protein conformational change to allow substrate binding and product release. However, the previously reported structure of a camphor-free form was seen in a closed conformation (18), and the only open forms observed for this enzyme have resulted from binding of large tethered substrate analogs (23–25). This has given credence to the idea that the substrate channel opens rarely on its own, or requires a secondary camphor binding site on the protein surface as the initial step in a substrate induced mechanism of channel opening (59).
In summary, it now appears that P450cam, along with many other members of the P450 family, displays a dynamic personality, and has revealed that the conformational space sampled by the protein includes an open conformation allowing substrate access to the buried active site.
Acknowledgments
The authors thank Prof. C. D. Stout and Andrew Annalora for helpful discussions.
Abbreviations
- DTT
dithiothreitol
- Pdx
putidaredoxin
- RMSD
root mean square deviation
Footnotes
This work was supported by grant GM41049 from the NIH.
The atomic coordinate and structures factors were deposited in the Protein Data Bank under the codes 3L61 (substrate-free, 200 mM [K+]), 3L62 (substrate-free, low [K+]) and 3L63 (camphor-bound, low [K+]).
References
- 1.Ortiz de Montellano PR. Cytochrome P-450: Structure, Mechanism, and Biochemistry. 3. Kluwer Academics/Plenum; New York: 2004. [Google Scholar]
- 2.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 4.Poulos TL, Finzel BC, Howard AJ. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 5.Pylypenko O, Schlichting I. Structural aspects of ligand binding to and electron transfer in bacterial and fungal P450s. Annu Rev Biochem. 2004;73:991–1018. doi: 10.1146/annurev.biochem.73.011303.073711. [DOI] [PubMed] [Google Scholar]
- 6.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 7.Annalora AJ, Goodin DB, Hong W, Zhang Q, Johnson EF, Stout CD. The crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J Mol Biol. 2009;396:441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savino C, Montemiglio LC, Sciara G, Miele AE, Kendrew SG, Jemth P, Gianni S, Vallone B. Investigating the structural plasticity of a cytochrome P450: Three dimensional structures of P450 EryK and binding to its physiological substrate. J Biol Chem. 2009;284:29170–29179. doi: 10.1074/jbc.M109.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J Biol Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Poulos TL. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 11.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, Yamane K, Adachi S, Shiro Y, Weiss KE, Maves SA, Sligar SG. Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties. J Inorg Biochem. 2002;91:491–501. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 13.Xu LH, Fushinobu S, Ikeda H, Wakagi T, Shoun H. Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state. J Bacteriol. 2009;191:1211–1219. doi: 10.1128/JB.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouellet H, Podust LM, de Montellano PRO. Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J Biol Chem. 2008;283:5069–5080. doi: 10.1074/jbc.M708734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerbe K, Pylypenko O, Vitali F, Zhang W, Rouset S, Heck M, Vrijbloed JW, Bischoff D, Bister B, Süssmuth RD, Pelzer S, Wohlleben W, Robinson JA, Schlichting I. Crystal structure of OxyB, a cytochrome P450 implicated in an oxidative phenol coupling reaction during vancomycin biosynthesis. J Biol Chem. 2002;277:47476–47485. doi: 10.1074/jbc.M206342200. [DOI] [PubMed] [Google Scholar]
- 17.Podust LM, Yermalitskaya LV, Lepesheva GI, Podust VN, Dalmasso EA, Waterman MR. Estriol bound and ligand-free structures of sterol 14alpha-demethylase. Structure. 2004;12:1937–1945. doi: 10.1016/j.str.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Poulos TL, Finzel BC, Howard AJ. Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry. 1986;25:5314–5322. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BA, Sligar SG. Structural studies of cytochrome P-450 using small angle x-ray scattering. J Biol Chem. 1983;258:3599–3601. [PubMed] [Google Scholar]
- 20.Di Primo C, Deprez E, Hoa GH, Douzou P. Antagonistic effects of hydrostatic pressure and osmotic pressure on cytochrome P-450cam spin transition. Biophys J. 1995;68:2056–2061. doi: 10.1016/S0006-3495(95)80384-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher MT, Scarlata SF, Sligar SG. High-pressure investigations of cytochrome P-450 spin and substrate binding equilibria. Arch Biochem Biophys. 1985;240:456–463. doi: 10.1016/0003-9861(85)90050-5. [DOI] [PubMed] [Google Scholar]
- 22.Hui Bin Hoa G, Marden MC. The pressure-dependence of the spin equilibrium in camphor-bound ferric cytochrome-P-450. Eur J Biochem. 1982;124:311–315. doi: 10.1111/j.1432-1033.1982.tb06593.x. [DOI] [PubMed] [Google Scholar]
- 23.Dmochowski IJ, Crane BR, Wilker JJ, Winkler JR, Gray HB. Optical detection of cytochrome P450 by sensitizer-linked substrates. Proc Natl Acad Sci USA. 1999;96:12987–12990. doi: 10.1073/pnas.96.23.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci USA. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hays AMA, Dunn AR, Chiu R, Gray HB, Stout CD, Goodin DB. Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires. J Mol Biol. 2004;344:455–469. doi: 10.1016/j.jmb.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450cam: practical consequences and engineering of a monomeric enzyme. Protein Eng. 1997;10:1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 27.Evans PR. Data reduction. CCP4 Study Weekend Proceedings DL/SCI/R34. 1993:114–122. [Google Scholar]
- 28.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 33.Lovell SC, Davis IW, Adrendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMOL Molecular Graphics System. 2002 www.pymol.org.
- 35.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 36.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam. Implications for the dioxygen activation mechanism. J Biol Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 37.Nagano S, Tosha T, Ishimori K, Morishima I, Poulos TL. Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding. J Biol Chem. 2004;279:42844–42849. doi: 10.1074/jbc.M404217200. [DOI] [PubMed] [Google Scholar]
- 38.Sakurai K, Shimada H, Hayashi T, Tsukihara T. Substrate binding induces structural changes in cytochrome P450cam. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:80–83. doi: 10.1107/S1744309108044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lampe JN, Brandman R, Sivaramakrishnan S, Ortiz de Montellano PR. 2D NMR and All-atom molecular dynamics of cytochrome P450 CYP119 reveal hidden conformational substates. J Biol Chem. 2010 doi: 10.1074/jbc.M109.087593. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampe JN, Floor SN, Gross JD, Nishida CR, Jiang Y, Trnka MJ, Ortiz de Montellano PR. Ligand-induced conformational heterogeneity of cytochrome P450 CYP119 identified by 2D NMR spectroscopy with the unnatural amino acid (13)C-p-methoxyphenylalanine. J Am Chem Soc. 2008;130:16168–16169. doi: 10.1021/ja8071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Primo C, Hui Bon Hoa G, Douzou P, Sligar S. Mutagenesis of a single hydrogen bond in cytochrome P-450 alters cation binding and heme solvation. J Biol Chem. 1990;265:5361–5363. [PubMed] [Google Scholar]
- 42.Manna SK, Mazumdar S. Reversible inactivation of cytochrome P450 by alkaline earth metal ions: auxiliary metal ion induced conformation change and formation of inactive P420 species in CYP101. J Inorg Biochem. 2008;102:1312–1321. doi: 10.1016/j.jinorgbio.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 43.OuYang B, Pochapsky SS, Pagani GM, Pochapsky TC. Specific effects of potassium ion binding on wild-type and L358P cytochrome P450cam. Biochemistry. 2006;45:14379–14388. doi: 10.1021/bi0617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tosha T, Yoshioka S, Takahashi S, Ishimori K, Shimada H, Morishima I. NMR study on the structural changes of cytochrome P450cam upon the complex formation with putidaredoxin. Functional significance of the putidaredoxin-induced structural changes. J Biol Chem. 2003;278:39809–39821. doi: 10.1074/jbc.M304265200. [DOI] [PubMed] [Google Scholar]
- 45.Tosha T, Yoshioka S, Ishimori K, Morishima I. L358P mutation on cytochrome P450cam simulates structural changes upon putidaredoxin binding: the structural changes trigger electron transfer to oxy-P450cam from electron donors. J Biol Chem. 2004;279:42836–42843. doi: 10.1074/jbc.M404216200. [DOI] [PubMed] [Google Scholar]
- 46.Lipscomb JD. Electron paramagnetic resonance detectable states of cytochrome P-450cam. Biochemistry. 1980;19:3590–3599. doi: 10.1021/bi00556a027. [DOI] [PubMed] [Google Scholar]
- 47.Unno M, Christian JF, Benson DE, Gerber NC, Sligar SG, Champion PM. Resonance Raman investigations of cytochrome P450(cam) complexed with putidaredoxin. J Am Chem Soc. 1997;119:6614–6620. [Google Scholar]
- 48.Aoki M, Ishimori K, Fukada H, Takahashi K, Morishima I. Isothermal titration calorimetric studies on the associations of putidaredoxin to NADH-putidaredoxin reductase and P450cam. Biochim Biophys Acta. 1998;1384:180–188. doi: 10.1016/s0167-4838(98)00017-x. [DOI] [PubMed] [Google Scholar]
- 49.Furukawa Y, Morishima I. The role of water molecules in the association of cytochrome P450cam with putidaredoxin. An osmotic pressure study. J Biol Chem. 2001;276:12983–12990. doi: 10.1074/jbc.M010217200. [DOI] [PubMed] [Google Scholar]
- 50.Pochapsky SS, Pochapsky TC, Wei JW. A model for effector activity in a highly specific biological electron transfer complex: the cytochrome P450(cam)-putidaredoxin couple. Biochemistry. 2003;42:5649–5656. doi: 10.1021/bi034263s. [DOI] [PubMed] [Google Scholar]
- 51.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450cam. Structure. 2008;16:916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davydov DR, Hui Bon Hoa G, Peterson JA. Dynamics of protein-bound water in the heme domain of P450BM3 studied by high-pressure spectroscopy: comparison with P450cam and P450 2B4. Biochemistry. 1999;38:751–761. doi: 10.1021/bi981397a. [DOI] [PubMed] [Google Scholar]
- 53.Tschirret-Guth RA, Koo LS, Hoa GH, Ortiz de Montellano PR. Reversible pressure deformation of a thermophilic cytochrome P450 enzyme (CYP119) and its active-site mutants. J Am Chem Soc. 2001;123:3412–3417. doi: 10.1021/ja003947+. [DOI] [PubMed] [Google Scholar]
- 54.Hui Bon Hoa G, Di Primo C, Geze M, Douzou P, Kornblatt JA, Sligar SG. The formation of cytochrome P-450 from cytochrome P-420 is promoted by spermine. Biochemistry. 1990;29:6810–6815. doi: 10.1021/bi00481a008. [DOI] [PubMed] [Google Scholar]
- 55.Martinis SA, Blanke SR, Hager LP, Sligar SG, Hoa GH, Rux JJ, Dawson JH. Probing the heme iron coordination structure of pressure-induced cytochrome P420cam. Biochemistry. 1996;35:14530–14536. doi: 10.1021/bi961511u. [DOI] [PubMed] [Google Scholar]
- 56.Hanson LK, Eaton WA, Sligar SG, Gunsalus IC, Gouterman M, Connell CR. Origin of anomalous soret spectra of carboxycytochrome P-450. J Am Chem Soc. 1976;98:2672–2674. doi: 10.1021/ja00425a050. [DOI] [PubMed] [Google Scholar]
- 57.Jung C. Quantum chemical explanation of the hyper spectrum of the carbon-monoxide complex of cytochrome-P-450. Chem Phys Lett. 1985;113:589–596. [Google Scholar]
- 58.Perera R, Sono M, Sigman JA, Pfister TD, Lu Y, Dawson JH. Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc Natl Acad Sci USA. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao H, McCullough CR, Costache AD, Pullela PK, Sem DS. Structural evidence for a functionally relevant second camphor binding site in P450cam: model for substrate entry into a P450 active site. Proteins. 2007;69:125–138. doi: 10.1002/prot.21508. [DOI] [PubMed] [Google Scholar]