Abstract
We are entering the era of personalized genomics as breakthroughs in sequencing technology have made it possible to sequence or genotype an individual person in an efficient and accurate manner. Preliminary results from HapMap and other similar projects have revealed the existence of tremendous genetic variations among world populations and among individuals. It is important to delineate the functional implication of such variations, i.e. whether they affect the stability and biochemical properties of proteins. It is also generally believed that the genetic variation is the main cause for different susceptibility to certain diseases or different response to therapeutic treatments. Understanding genetic variation in the context of human diseases thus holds the promise for “personalized medicine.” In this work, we carried out a genome-wide analysis of single nucleotide polymorphisms (SNPs) that could potentially influence protein phosphorylation characteristics in human. Here, we defined a phosphorylation-related SNP (phosSNP) as a non-synonymous SNP (nsSNP) that affects the protein phosphorylation status. Using an in-house developed kinase-specific phosphorylation site predictor (GPS 2.0), we computationally detected that ∼70% of the reported nsSNPs are potential phosSNPs. More interestingly, ∼74.6% of these potential phosSNPs might also induce changes in protein kinase types in adjacent phosphorylation sites rather than creating or removing phosphorylation sites directly. Taken together, we proposed that a large proportion of the nsSNPs might affect protein phosphorylation characteristics and play important roles in rewiring biological pathways. Finally, all phosSNPs were integrated into the PhosSNP 1.0 database, which was implemented in JAVA 1.5 (J2SE 5.0). The PhosSNP 1.0 database is freely available for academic researchers.
As we are entering the age of “personalized genomics,” it is expected that the knowledge of human genetic polymorphisms and variations could provide a foundation for understanding the differences in susceptibility to diseases and designing individualized therapeutic treatments (1, 2). Recent progress of the International HapMap Project and similar projects (3–5) has provided a wealth of information detailing tens of millions of human genetic variations between individuals, including copy number variations (4) and single nucleotide polymorphisms (SNPs) (1,5). It was estimated that ∼90% of human genetic variations are caused by SNPs (2). For example, changes to amino acids in proteins, such as the non-synonymous SNPs (nsSNPs) in the gene coding regions, could account for nearly half of the known genetic variations linked to human inherited diseases (6). In this regard, numerous efforts have been made to elucidate how nsSNPs generate deleterious effects on the stability and function of proteins and their roles in cancers and diseases (7–11). For example, the SNPeffect database was developed as a comprehensive resource of the molecular phenotypic effects of human nsSNPs (7, 8). Later, several databases, including SNP500Cancer (9), PolyDoms (10), and Diseasome (11), were constructed for dissecting potentially cancer- or disease-related nsSNPs. An nsSNP might change the physicochemical property of a wild-type amino acid that affects the protein stability and dynamics, disrupts the interacting interface, and prohibits the protein to form a complex with its partners (12–15). Alternatively, nsSNPs could also influence post-translational modifications (PTMs) of proteins (e.g. phosphorylation) by changing the residue types of the target sites or key flanking amino acids (16–18).
In eukaryotes, phosphorylation is one of the most important PTMs of proteins that plays essential roles in most biological pathways and regulates cellular dynamics and plasticity (19–24). Generally in vivo, different protein kinases (PKs) could recognize distinct short peptide motifs or patterns and attach phosphate moieties to Ser, Thr, or Tyr residues. Conventional experimental identifications and recent advances in high throughput MS techniques have generated a large number of phosphorylated substrates with confirmed phosphorylation sites. From primary scientific literature, Phospho.ELM 8.1 collected >4,600 experimentally verified phosphorylated proteins with 14,518 Ser, 2,914 Thr, and 2,217 Tyr sites (19). With a similar strategy, Li et al. (25) collected 87,068 experimentally verified phosphorylation sites of 24,705 substrates from the scientific literature and MS-derived experiments. More recently, Tan et al. (26) compiled a large data set with 23,979 non-redundant human phosphorylation sites from several phosphorylation databases. Besides experimental methods, a variety of computational approaches were developed to predict protein phosphorylation sites. For example, we previously constructed a highly accurate software (GPS 2.0) to predict kinase-specific phosphorylation sites in hierarchy (22). The latest compendium of computational resources for protein phosphorylation was manually collected and is available upon request.
Recently, more and more experimental observations have suggested that nsSNPs could indirectly or directly disrupt the original phosphorylation sites or create new sites (supplemental Table S1). For example, human OGG1 (RefSeq accession number NM_002542) harbors an nsSNP of S326C (dbSNP accession number rs1052133), which changes the phosphorylation status of OGG1 and disrupts its nucleolar localization during the cell cycle (27). This nsSNP was further reported as a risk allele for a variety of cancers (27). In 2005, Li et al. (28) observed that the P47S nsSNP (rs1800371) of p53 (NM_000546) strongly compromises the phosphorylation level of its adjacent residue Ser-46 by p38 MAPK and reduces the ability of p53 to induce apoptosis up to 5-fold. Moreover, the D149G nsSNP (rs1801724) of p21WAF1/CIP1 (NM_078467) could attenuate Ser-146 phosphorylation by PKCδ to resist tumor necrosis factor α-induced apoptosis and play an important role in cancer development (29). More recently, Gentile et al. (30) predicted 16 nsSNPs that potentially influence the phosphorylation status of human ion channel proteins. For example, the human ether-a-gogo-related gene 1, ERG1/KCNH2/Kv11.1 (NM_000238) channel protein, has a K897T nsSNP (rs1805123), which creates a new AKT phosphorylation site to prolong the QT interval of cardiac myocytes (30). In this regard, comprehensive studies of nsSNPs that alter protein phosphorylation will be helpful to further the understanding of how genetic polymorphisms are involved in regulating biological pathways and processes and how they affect susceptibility to diseases and to determine human population diversity and phenotypic plasticity.
Previously, Savas and Ozcelik (16) carried out a small scale prediction to identify 15 nsSNPs that might create or remove potential phosphorylation sites in 14 DNA repair- and cell cycle-related proteins. Later, Yang, et al. mapped 109,262 nsSNPs to experimentally verified phosphorylation sites taken from the NCBI dbSNP database (31) and observed that 64 known phosphorylation sites might be removed by nsSNPs (17). Recently, Ryu et al. (18) took 33,651 human sequence variations (∼26% as nsSNPs) from the Swiss-Prot/UniProt database and carried out a large scale survey of potential phosphovariants, which were defined as amino acid variations that might influence protein phosphorylation status.
In this work, we performed a genome-wide analysis of genetic polymorphisms that influence protein phosphorylation in humans. We collected 91,797 nsSNPs from NCBI dbSNP Build 130 (31). The human mRNA/protein sequences were taken from RefSeq Build 31 (32). We used GPS 2.0 software (22) to predict potential kinase-specific phosphorylation sites for human proteins and nsSNP sites. Here, we defined a phosphorylation-related SNP (phosSNP) as an nsSNP that might influence protein phosphorylation status. We classified all phosSNPs into five groups. The first three types (I, II, and III) were similarly defined as described previously (18), including change of an amino acid with Ser/Thr/Tyr residue or vice versa to create a potential new (Type I (+)) or remove an original phosphorylation site (Type I (−)), variations to create (Type II (+)) or remove adjacent phosphorylation sites (Type II (−)), and mutations to induce changes of PK types in adjacent phosphorylation sites (Type III) (18). Also, we observed that an amino acid substitution among Ser, Thr, or Tyr might also induce a change of PK types for the phosphorylation site; i.e. the target site might still be phosphorylated but by a different type of kinase (Type IV). Moreover, we defined the Type V phosSNP as a variation that results in a stop codon, which might remove its following phosphorylation sites in the protein C terminus. Unexpectedly, we computationally detected 69.76% of nsSNPs as potential phosSNPs (64,035) in 17,614 proteins. In this regard, we proposed that most nsSNPs might affect protein phosphorylation and play important roles in rewiring the biological pathways. Interestingly, we observed 74.58% of phosSNPs as Type III phosSNPs (47,760), suggesting that nsSNPs induce changes of PK types of adjacent phosphorylation sites in an indirect manner rather than creating or removing a phosphorylation site directly. Taken together, our results represent a useful resource for future disease diagnostics and provide a basis for better and individualized treatment. Finally, all phosSNP data were integrated into he PhosSNP 1.0 database, which was implemented in JAVA 1.5 (J2SE 5.0). The PhosSNP 1.0 supports Windows, Unix/Linux, and Mac and is freely available for academic researchers.
EXPERIMENTAL PROCEDURES
Preparation of nsSNP Data
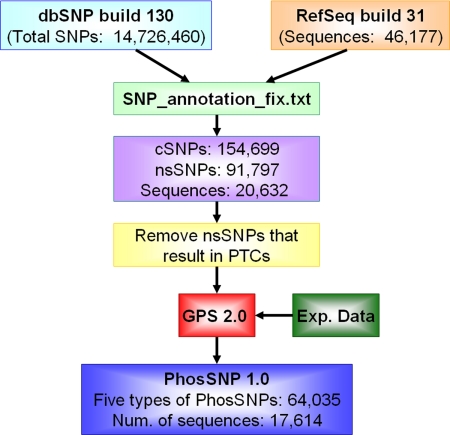
We downloaded 14,726,460 human SNP data (dbSNP Build 130, released on June 18, 2008) (31) from the NCBI FTP server. We then extracted 368,140 SNPs in the coding regions (cSNPs) after removing other entries in which the SNP characters were presented in lowercase letters (SNPs in non-coding sequences). We then downloaded the human RefSeq Build 31 from NCBI (released on October 28, 2008) (32) containing 46,177 mRNAs and their corresponding proteins. We downloaded the “SNP_annotation_fix.txt” (released on September 30, 2008, RefSeq), which contains precalculated mapping results between dbSNP and human RefSeq. The nsSNP was defined as a cSNP changing a single nucleotide that could cause an amino acid substitution and subsequent functional alteration in its protein product (1). Based on the mapping file, we used the bl2seq program from the NCBI BLAST package (33) to compare RefSeq proteins with their dbSNP data in a pairwise manner. Then the positions and allele changes of nsSNPs could be easily found. In total, we detected 154,699 cSNPs, including 91,797 nsSNPs in 20,632 proteins.
Removing nsSNPs That Result in Premature Termination Codons (PTCs)
In the above data set, there were 3,448 nonsense nsSNPs that changed amino acids into stop codons. A large proportion of nonsense nsSNPs might result in PTCs, which trigger the nonsense-mediated mRNA decay (NMD) pathway and inhibit the production of proteins (34–36). Because the PTC-containing mRNA sequences were not translated at all, such nonsense nsSNPs will have no effect on protein phosphorylation. In this work, we adopted a previously used NMD rule to detect potential PTCs (36). The nsSNPs located >50 nt upstream of the 3′-most exon-exon junction were regarded as potential PTCs and removed before further analysis. The exon and intron information was taken from the annotation file (human.rna.gbff.gz) of the human RefSeq Build 31. In total, there were only 592 non-PTC nsSNPs identified.
Prediction of Kinase-specific Phosphorylation Sites Using GPS 2.0
Previously, we developed a well tested rule to classify PKs into a hierarchical structure with four levels, including group, family, subfamily, and single PK (37). Then we developed a software package, GPS 2.0, that contains 144 serine/threonine and 69 tyrosine PK groups and could predict kinase-specific phosphorylation sites for 408 human PKs in hierarchy (22). In this work, a PK type was defined as a unique PK group in GPS 2.0. To reduce the false positive rate, the high threshold was chosen in this work (false positive rates of 2% for serine/threonine kinases and 4% for tyrosine kinases) (22). In GPS 2.0, we defined a phosphorylation site peptide, PSP(m, n), as a Ser, Thr, or Tyr amino acid flanked by m residues upstream and n residues downstream (22). The PSP(7, 7) was adopted in GPS 2.0 (22).
Computational Detection of Potential phosSNPs Using GPS 2.0
Previous experimental and computational studies proposed that various PKs recognize distinct linear motifs around target sites for precise modifications (38–42). In this regard, an nsSNP located at the phosphorylated position or in near flanking regions might influence the protein phosphorylation status. In this work, we defined a potential phosSNP as an nsSNP located in a PSP(7, 7).
First, we took the 20,632 RefSeq proteins as the benchmark sequence data. Then we made changes to a protein sequence, one of its nsSNPs at a time, to prepare a variant sequence. The variant proteins were integrated together as the variant sequence data. We directly used GPS 2.0 with the high threshold to scan benchmark proteins and variant proteins, respectively. By comparing results of the two data sets, the phosSNPs with their corresponding types could be easily detected based on definitions.
Detection of Potential phosSNPs with Experimentally Verified Phosphorylation Sites
Currently, there are a number of phosphorylation prediction databases constructed. For example, Phospho.ELM 8.1 collected ∼4,600 experimentally verified phosphorylated substrates with 14,518 Ser, 2,914 Thr, and 2,217 Tyr sites from the scientific literature (19), whereas SysPTM contains 87,068 known phosphorylation sites in 24,705 proteins (25). However, most phosphorylation sites in these databases are not human-specific. Recently, Tan et al. (26) compiled a large data set with 23,979 experimentally verified human phosphorylation sites, including one His, 11,731 Ser, 2,964 Thr, and 9,283 Tyr sites. In this work, the one His site was discarded, whereas the remaining 23,978 human phosphorylation sites were adopted for detection of potential phosSNPs by exact string matching (26). As previously described (26), we used the PSP(7, 7) of these phosphorylation sites to identify identical hits in the benchmark sequence data and the variant sequence data, respectively. By comparison, the potential phosSNPs were identified and classified into different types based on GPS 2.0 predictions.
Database Construction
Our aim is to develop an integrated platform for computational analysis of PTMs. We chose the JAVA (J2SE) language for its excellent portability under different operating systems. For example, the self-developed tools used in this work, including GPS 2.0 (22) and DOG 1.0 (43), were implemented in JAVA. In this work, we also developed the PhosSNP 1.0 database with JAVA 1.5 (J2SE 5.0). The local packages of the PhosSNP 1.0 database support three major operating systems, including Windows, Unix/Linux, and Mac. The usage of the PhosSNP 1.0 database is available in the user manual. The database will be continuously updated twice per year when new phosphorylation sites, dbSNP, and other SNP data become available.
RESULTS
Genome-wide Identification of Potential phosSNPs in Human
The procedure of predicting potential phosSNPs in human is shown in Fig. 1. First, we collected 14,726,460 SNPs from dbSNP (31) Build 130 and 46,177 human mRNA sequences with their corresponding proteins from RefSeq (32) Build 31. With the precalculated SNP annotation file, we detected 154,699 cSNPs, including 91,797 nsSNPs in 20,632 proteins (Fig. 1). In our results, we observed that there were 3,448 nonsense nsSNPs that changed amino acids into stop codons. Previously, it was proposed that nonsense mutations or nsSNPs might result in PTCs, which could trigger the NMD pathway to prohibit the expression of proteins (34–36). In this regard, we detected PTCs with a previously adopted NMD rule: nsSNPs should locate >50 nt upstream of the 3′-most exon-exon junction (36). All PTCs were removed from the data set for further analysis (Fig. 1). Previously, we developed a kinase-specific predictor called GPS 2.0, which included 144 serine/threonine and 69 tyrosine PK groups (22). In this work, GPS 2.0 with high confidence level (false positive rates of 2% for serine/threonine kinases and 4% for tyrosine kinases) (22) was directly used to predict potential kinase-specific phosphorylation sites for human RefSeq proteins and nsSNP data, respectively. The two results were compared to identify potential phosSNPs. All phosSNPs were classified into five types as defined based on the definitions listed below (Fig. 2): (i) Type I, an nsSNP at a phosphorylatable position that directly creates (Type I (+)) or removes (Type I (−)) the phosphorylation site; (ii) Type II, an nsSNP that creates (Type II (+)) or removes (Type II (−)) one or multiple adjacent phosphorylation sites; (iii) Type III, an nsSNP that induces changes of PK types for one or multiple adjacent phosphorylation sites; (iv) Type IV, an nsSNP at a phosphorylation site that induces a change of PK types for the phosphorylation site; and (v) Type V, a stop codon nsSNP that removes downstream phosphorylation sites in the protein C terminus.
Fig. 1.
Computational procedure of phosSNPs detection. In addition to ab initio prediction of kinase-specific phosphorylation sites using GPS 2.0 (22), we also detected potential phosSNPs by exact string matching with 23,978 experimentally identified human phosphorylation sites (Exp. Data) from a recent analysis (26). In total, there were 64,035 potential phosSNPs identified in 17,614 sequences.
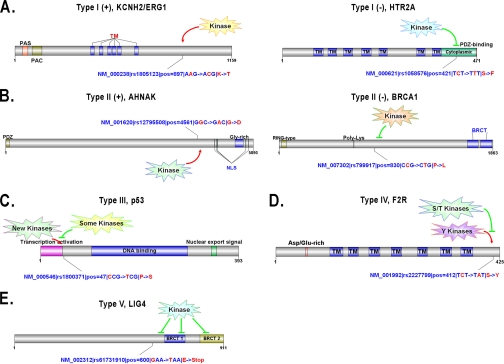
Fig. 2.
Five types of phosSNPs with typical examples. A, Type I PhosSNP. The K897T nsSNP of KCNH2/ERG1 creates a new AKT-specific phosphorylation site (Type I (+)), whereas the S421F nsSNP of HTR2A removes the phosphorylation site at Ser-421 (Type I (−)). B, Type II PhosSNP. The G4561D nsSNP of AHNAK might render its nearby Thr-4564 residue as a potential phosphorylation site (Type II (+)), whereas the P830L nsSNP of BRCA1 might prohibit Ser-832 phosphorylation (Type II (−)). C, Type III PhosSNP. The P47S nsSNP of p53 might induce changes of PK types for multiple adjacent phosphorylation sites. D, Type IV PhosSNP. The S412Y nsSNP of F2R might induce a change of its upstream serine/threonine PKs into tyrosine PKs. E, Type V PhosSNP. The E600Stop nonsense nsSNP might remove its following phosphorylation sites. TM, transmembrane.
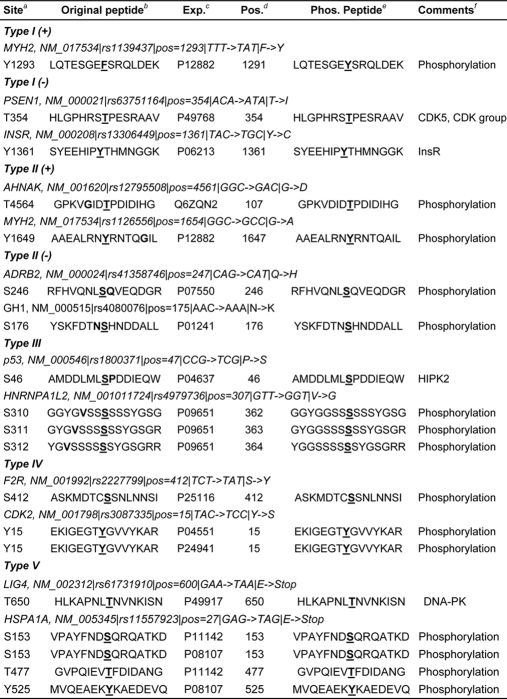
In total, we analyzed 64,035 potential phosSNPs in 17,614 proteins (Fig. 1 and Table I). Besides ab initio prediction of kinase-specific phosphorylation sites, the experimentally verified human phosphorylation sites were also used. We took 23,978 experimentally identified human phosphorylation sites from a recent analysis (26). By exact string matching with the PSP(7,7) (26), we detected 2,004 potential phosSNPs in 1,528 proteins (Table I). Several examples uncovered from experimental data are shown in Table II. For example, an F1293Y nsSNP (rs1139437) of human MYH2 (Myosin-2, NM_017534) changes the peptide “LQTESGEFSRQLDEK” into “LQTESGEYSRQLDEK”, which is identical to the PSP(7,7) of the experimentally verified Tyr-1291 site in human MYH1 (Myosin-1; UniProt accession number P12882) (Table II). In this regard, this F1293Y nsSNP is a Type I (+) phosSNP. Furthermore, the Presenilin-1 (PSEN1; NM_000021) was experimentally identified to be phosphorylated by CDK5 or CDK group PKs at Thr-354, whereas a T354I nsSNP (rs63751164) might remove the site (Table II). Thus, this nsSNP was classified as a Type I (−) phosSNP (Table II). The detailed data statistics for detection of potential phosSNPs is shown in Table I. More detailed information on data processes are presented under “Experimental Procedures.”
Table I. The data statistics for phosSNPs detection. We used GPS 2.0 to predict kinase-specific phosphorylation sites after taking into account of the nsSNPs. A large data set including 23,978 experimentally identified human phosphorylation sites was also used to scan potential phosSNPs (Exp. results).
| PhosSNP 1.0 | GPS 2.0 results |
Exp. results |
|||
|---|---|---|---|---|---|
| Proteins | PhosSNPs | Proteins | PhosSNPs | ||
| Total | 17,614 | 64,035 | 1,528 | 2,004 | |
| Type I | All | 10,449 | 16,954 | 225 | 172 |
| (+) | 7,048 | 8,866 | 2 | 1 | |
| (−) | 6,422 | 8,315 | 223 | 171 | |
| Type II | All | 12,207 | 24,721 | 193 | 174 |
| (+) | 8,301 | 12,659 | 3 | 2 | |
| (−) | 9,028 | 14,367 | 190 | 172 | |
| Type III | 16,054 | 47,760 | 1,333 | 1,699 | |
| Type IV | 1,023 | 873 | 22 | 19 | |
| Type V | 448 | 442 | 50 | 48 | |
Table II. Examples of our analysis results using experimentally determined phosphorylation data. a. Site, the position of potential or experimentally verified phosphorylation site in benchmark or variant proteins; b. Original peptide, the original PSP(7, 7) in benchmark/native proteins; c. Exp., experimentally identified phosphorylated substrate; d. Pos. the of the experimentally identified phosphorylation site; e. Phos. Peptide, the exactly matched PSP(7,7) in experimental substrate; f. Comments on matched substrates (with or without PK information).
Type I phosSNPs: Directly Adding or Removing Phosphorylation Sites at Phosphorylated Positions
As an nsSNP, a non-phosphorylated amino acid could be changed into a Ser, Thr, or Tyr, which might create a new phosphorylation site and be modified by some PKs (Type I (+)). Also, a phosphorylated Ser/Thr/Tyr residue could be changed into another amino acid type to disrupt an original phosphorylation site (Type I (−)). In this regard, Type I phosSNPs play direct roles to add or remove phosphorylation sites. In our results, there were 16,954 potential Type I phosSNPs in 10,449 sequences (26.48%) identified (Table I).
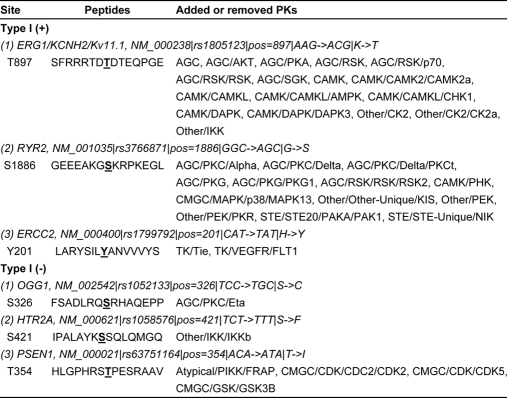
From our results, we randomly selected several instances of Type I (+) and Type I (−) phosSNPs for detailed analysis (Table III). Three typical instances for Type I (+) phosSNPs are shown in Table III. For example, the human ERG1/KCNH2/Kv11.1 (NM_000238), an ether-a-go-go-related gene, is a potassium channel and is crucial for rhythmic excitability of the cardiac muscle and the pituitary (30) (Fig. 2A). The wild-type ERG1 could be activated by thyroid hormone through a protein phosphatase, PP5-mediated dephosphorylation, whereas a K897T nsSNP allele creates a new AKT-specific phosphorylation site. The AKT-mediated phosphorylation of K897T ERG1 will inhibit its activity to prolong the QT interval of cardiac myocytes (30). Here, we successfully predicted that the K897T nsSNP creates a potential phosphorylation site, which might be phosphorylated by a variety of PKs, including AGC/AKT (Table III). Also, it was proposed that a G1886S nsSNP of RYR2 (NM_001035) might generate a protein kinase C phosphorylation site (44). And our prediction result was consistent with the previous study (Table III) (44). Previously, Savas and Ozcelik (16) performed a small scale analysis to identify 15 nsSNPs that might create or remove potential phosphorylation sites in 14 DNA repair- and cell cycle-related proteins. One of the DNA repair-related genes (16), ERCC2 (NM_000400), has a H201Y nsSNP to create a potential tyrosine phosphorylation site recognized by TK/Tie and TK/VEGFR/FLT1 (Table III).
Table III. Several examples for type I (+) and type I (−) phosSNPs, respectively. The potentially phosphorylated or phosSNP positions were marked in red.
We also present three typical examples of Type I (−) phosSNPs (Table III). As described in the Introduction, the S326C nsSNP of human OGG1 (NM_002542) removes the Ser-326 phosphorylation site, disrupts it nuclear localization during the cell cycle, and affects susceptibility to a variety of cancers, although it is still not known which PKs could phosphorylate the Ser-326 site (27). Here, we predicted that the Ser-326 might be phosphorylated by AGC/PKC/Eta (Table III). Such a prediction will be helpful for further experimental verification. Also, it was reported that the 5-HT2A serotonin receptor (HTR2A; NM_000621) has an nsSNP at the Ser-421 locus (S421F) that removes the Ser-421 phosphorylation site and significantly attenuate agonist-mediated desensitization of HTR2A (45) (Fig. 2A). Again, the PK types for Ser-421 phosphorylation were also unclear (45). We predicted that the Ser-421 might be phosphorylated by other/IKK/IKKb (Table III). In addition, human PSEN1 (NM_000021) has a T354I nsSNP (Table III). Interestingly, we found that this site was previously verified as a real phosphorylation site from experimental phosphorylation data (Table II).
Type II phosSNPs: Creating or Disrupting Adjacent Phosphorylation Sites
In GPS 2.0, a PSP(7, 7) was defined as a Ser/Thr/Tyr residue flanked by 7 residues upstream and 7 residues downstream (22). In this work, we defined the Type II phosSNP as an nsSNP located in a PSP(7, 7) to render the middle phosphorylation site accessible (Type II (+)) or inaccessible (Type II (−)) by PKs. In total, we detected 24,721 potential Type II phosSNPs (38.61%) from 12,207 sequences (Table I).
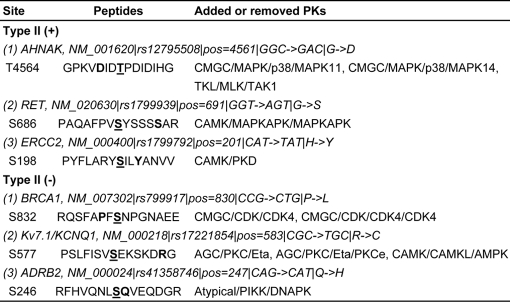
Here, we present several examples for Type II (+) and Type II (−) phosSNPs, respectively (Table IV). A neuroblast differentiation-associated protein, AHNAK (NM_001620) harbors a G4561D nsSNP, which makes its nearby Thr-4564 residue a potential phosphorylation site (Fig. 2B). Interestingly, the PSP(7, 7) of AHNAK Thr-4564 is identical to an experimentally verified phosphopeptide (Thr-107) in an unknown protein (Q6ZQN2) (Table II). In this regard, the AHNAK Thr-4564 might also be a bona fide phosphorylation site. However, the PKs responsible for its modification are not known. In this regard, our predictions will be helpful for further experimental design (Table IV). Previously, Yang et al. (46) proposed that the G691S nsSNP of RET, a proto-oncogene tyrosine-protein kinase, could generate a new phosphorylation site at position 691 and also influence the phosphorylation status of Tyr-687 and Ser-696. All these experimental observations were detected in this work (see PhosSNP database). Moreover, we observed that the G691S nsSNP might create a new phosphorylation site at Ser-686 (Table IV). Again, although the H201Y nsSNP of ERCC2 directly creates a potential phosphorylation site at position 201, it generates an additional potential phosphorylation site at Ser-198 (Table IV).
Table IV. Several examples for type II (+) and type II (−) phosSNPs, respectively. The potentially phosphorylated positions were marked in blue, while the phosSNP positions were marked in red.
As an important DNA repair gene (16), the P830L nsSNP of BRCA1 (NM_007302) might prohibit Ser-832 phosphorylation (Fig. 2B and Table IV). Also, potassium channel Kv7.1/KCNQ1 (NM_000218) has an R583C nsSNP, which might remove a potential phosphorylation site at Ser-577 (Table IV). In addition, the Ser-246 of β2-adrenergic receptor ADRB2 (NM_000024) was experimentally verified as a real phosphorylation site (Table II), whereas its Q247H nsSNP might prevent Ser-246 phosphorylation by atypical/PIKK/DNAPK (Table IV). Further experimental identification needs to be carried out to dissect whether the Ser-246 site is really not phosphorylated in the Q247H allele.
Type III phosSNPs: Inducing Changes of PK Types for Adjacent Phosphorylation Sites
Although there were 518 putative PKs reported in human, the kinase activities and exact substrates of a large proportion of them still remained to be experimentally identified (37). Based on a widely adopted hypothesis that similar PKs recognize similar patterns, GPS 2.0 was developed to classify all human PKs into a hierarchical structure with four levels, including group, family, subfamily, and single PK (22). GPS 2.0 contains 144 serine/threonine and 69 tyrosine PK clusters. Different PK clusters used different training data sets and exhibited different substrate preferences. In this work, we defined a PK type as a unique PK group in GPS 2.0. The Type III phosSNPs were defined as nsSNPs that induce changes of PK types for flanking phosphorylation sites rather than adding or removing phosphorylation sites altogether. In total, we detected 47,760 potential Type III phosSNPs (74.58%) in 16,054 sequences (Table I). In this regard, the Type III phosSNPs might play predominant roles in influencing protein phosphorylation states to rewire signaling pathways.
Here, we present five examples of Type III phosSNPs (Table V). Previously, Li et al. (28) reported that the P47S nsSNP of p53 (NM_000546) strongly diminishes the phosphorylation level of its adjacent Ser-46 by p38 MAPK and reduces the ability of p53 to induce apoptosis up to 5-fold (Fig. 2C). This nsSNP and potential impact on kinase-substrate relationship was also detected in our prediction results (Tables II and V). Also, in addition to removing a potential phosphorylation site at Ser-577 (Table IV), the R583C nsSNP of Kv7.1/KCNQ1 might also change the PK types for Ser-580 and Ser-585, respectively (Table V). As described in the Introduction, the D149G nsSNP of p21WAF1/CIP1 (NM_078467) could attenuate Ser-146 phosphorylation by PKCδ to resist tumor necrosis factor α-induced apoptosis and thus has important implications in cancer development (29). Besides Ser-146, we also found that D149G nsSNP might alter the PK types for Thr-145, Thr-148, and Tyr-151 (Table V). Taken together, our predictions were not only consistent with previous experimental studies but also provided a useful resource for further experimental considerations.
Table V. Several examples for type III phosSNPs. The potentially phosphorylated positions were marked in blue, while the phosSNP positions were marked in red. Only added or removed PK types were shown and included in PhosSNP 1.0 database.
Type IV phosSNPs: Occurring at Phosphorylation Sites to Induce Changes of PK Types
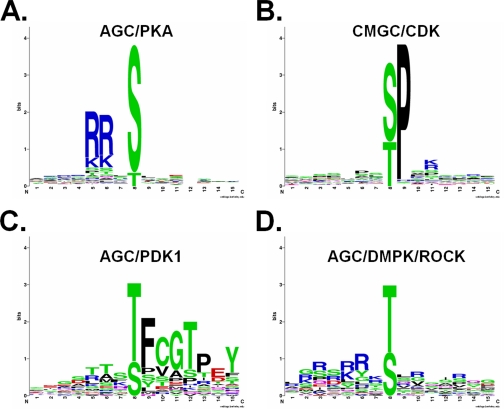
We observed that nsSNPs that occurred at phosphorylation sites could also induce changes of PK types rather than directly adding or removing phosphorylation sites. The human kinome could be classified into serine/threonine PKs and tyrosine PKs (37). The serine/threonine PKs usually recognize specific Ser/Thr residues for modification, whereas tyrosine PKs commonly modify special Tyr residues. In this regard, an nsSNP from Ser/Thr to Tyr or vice versa might change the PK types at the phosphorylated position. Moreover, we collected 377, 347, 36, and 38 experimentally verified phosphorylation sites for AGC/PKA, CMGC/CDK, AGC/PDK1, and AGC/DMPK/ROCK from Phospho. ELM 8.2 (19), respectively (the data set is available upon request). WebLogo software was used to generate sequence logos for the four PK types (Fig. 3). For AGC/PKA and CMGC/CDK, the Ser residue was more preferred to be recognized, whereas the Thr residue was more preferred for AGC/PDK1 and AGC/DMPK/ROCK (Fig. 3). Thus, an nsSNP between Ser and Thr might also change the PK types. In this work, we observed 873 potential Type IV phosSNPs (1.36%) in 1,023 sequences (Table I).
Fig. 3.
Different PKs exhibit different substrate preferences on Ser or Thr residue. From the Phospho.ELM 8.2 database (19), we collected 377, 347, 36, and 38 experimentally verified phosphorylation sites for four well studied PK groups, including AGC/PKA, CMGC/CDK, AGC/PDK1, and AGC/DMPK/ROCK, respectively. For AGC/PKA (A) and CMGC/CDK (B), the Ser is the preferred residue, whereas the Thr residue is more preferred for AGC/PDK1 (C) and AGC/DMPK/ROCK (D).
Again, we selected five examples for Type IV phosSNPs (Table VI). For example, the human proteinase-activated receptor F2R (NM_001992) was experimentally verified to be phosphorylated at Ser-412 (Table II), whereas the S412Y nsSNP might induce a change of its upstream serine/threonine PKs into tyrosine PKs (Fig. 2D and Table VI). Also, CDK2 has a verified phosphorylation site of Tyr-15, and this site is conserved in human (P04551) and fission yeast (P04551) (Table II). The Y15S nsSNP of human CDK2 might change the PK types at position 15 (Table VI). In addition, the T345S nsSNP of HLA class I histocompatibility antigen HLA-A (NM_002116) might add several additional PK types for the site (Table VI). Although both Type III and Type IV phosSNPs change PK types, we did not mix them together because they occur at distinct positions of phosphorylation site peptides.
Table VI. Several examples for type IV phosSNPs. The potentially phosphorylated or phosSNP positions were marked in red. Only added or removed PK types were shown and included in PhosSNP 1.0 database.
Type V phosSNPs: Removing Following Phosphorylation Sites by Nonsense SNPs
Although a large proportion of nonsense nsSNPs might result in PTCs to trigger the NMD pathway and prohibit the expression of proteins (34–36), there are some nsSNPs located ≤50 nt upstream of the 3′-most exon-exon junction that might result in a truncated protein without the following phosphorylation sites in the protein C terminus (Type V phosSNPs). In total, there were 442 Type V nsSNPs from 448 sequences detected (Table I).
Five instances for Type V phosSNPs are shown in Table VII. For example, the human DNA ligase LIG4 (NM_002312) was experimentally verified to be phosphorylated at Thr-650 (Table II), whereas its E600Stop nonsense nsSNP might remove its downstream phosphorylation sites, including the Thr-650 site (Fig. 2E and Table VII). Also, human HSPA1A (NM_005345) has two verified phosphorylation sites, including Ser-153 and Tyr-525 (in P08107), whereas the Ser-153 is also conserved in its paralog HSPA8 (P11142) (Table II). HSPA8 has an additionally known phosphorylation site at Thr-477, which is also conserved in HSPA1A (Table II), and the E27stop nsSNP of HSPA1A will remove the Ser-153, Thr-477, and Tyr-525 sites (Table VII).
Table VII. Several instances for type V phosSNPs. The potentially removed phosphorylation sites were shown.
| PhosSNPs | Removed Phosphorylation sites | Num. |
|---|---|---|
| LIG4, NM_002312 rs61731910 pos = 600 GAA->TAA E->Stop | Y616, T650, S668, T670, S672, Y688, Y698, S734, Y761, Y765, S779, T788, S794, Y801, Y803, S811, T817, Y819, S822, Y823, S861, T881, S892, T895, S897, Y909 | 26 |
| TWISTNB, NM_001002926 rs61734275 pos = 320 GAA->TAA E->Stop | S328, S335 | 2 |
| HSPA1A, NM_005345 rs11557923 pos = 27 GAG->TAG E->Stop | T38, Y41, T66, S106, T111, Y115, S120, T140, Y149, S153, T158, T177, T226, S254, T265, T273, S275, S276, S277, T278, S281, S286, Y294, T295, S296, T298, S362, T411, S418, T419, T425, T430, S432, T450, S462, T477, T495, T502, T504, S511, Y525, S537, Y545, S551, S563, Y611, S633, T636 | 48 |
| MC4R, NM_005912 rs13447340 pos = 320 TAT->TAG Y->Stop | S329, S330, Y332 | 3 |
| MYBBP1A, NM_014520 rs62620242 pos = 1256 CAG->TAG Q->Stop | S1267, T1269, S1290, S1293, S1303, S1308, S1310, S1314 | 8 |
DISCUSSION
Although only a very small proportion of human SNPs are nsSNPs (<1%), these nsSNPs could change amino acids; affect protein stability, function, and modification; and play important roles in regulating susceptibility to a variety of diseases and cancers (6–18). In 2006, Erxleben et al. (the Armstrong group) (47) first introduced the term “phosphorylopathy” to describe human genetic variation that results in aberrant regulation of protein phosphorylation. Later, they carried out a small scale prediction to identify 16 nsSNPs that potentially influence the phosphorylation status of human ion channel proteins; a K897T nsSNP (rs1805123) of human ERG1/KCNH2/Kv11.1 (NM_000238) channel protein was experimentally verified to create a new AKT phosphorylation site to prolong the QT interval of cardiac myocytes (30). In this regard, genome-wide prediction of phosphorylopathies in human might provide a highly valuable resource for further experimental identifications.
In this work, we conducted a systematic analysis to detect nsSNPs that potentially influence protein phosphorylation status. The term phosphorylopathy was refined as phosSNP. Interestingly, we observed that ∼69.8% of nsSNPs are potential phosSNPs (64,035) in 17,614 proteins (Table I). In particular, ∼74.5% of phosSNPs are Type III phosSNPs (47,760), which induce changes of PK types for adjacent phosphorylation sites rather than creating or removing phosphorylation sites (Table I). In this regard, most nsSNPs might regulate protein phosphorylation dynamics and play ubiquitous roles in rewiring the biological pathways. Our results could be a useful resource for future experimental identification and disease diagnostics and provide helpful information for better and individualized treatment.
From the H-Invitational Database (H-InvDB), we found that there were ∼43,000 gene clusters identified (48). However, the human RefSeq Build 31 contains 46,177 mRNAs. Thus, there might be multiple mRNAs for a unique gene cluster in the RefSeq data set. For example, there were three distinct mRNAs for ERG1/KCNH2/Kv11.1 gene (NM_000238, NM_172056, and NM_172057) (supplemental Table S2). The three mRNAs could be translated into highly similar but slightly different proteins. More importantly, the precalculated SNP mapping information for the three mRNAs was not identical, and not a single entry has the full SNP annotations (supplemental Table S2). To overcome this problem, we decided to keep all human RefSeq mRNAs for the analysis without removing any redundancy.
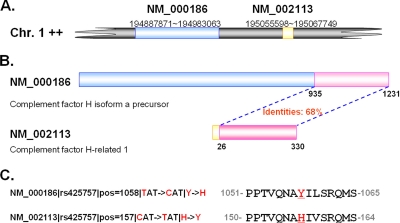
Also, because of close sequence similarity between paralogous genes, we observed that some SNP annotations could be mapped onto multiple genes. For example, one nsSNP (rs425757) could be mapped on either complement factor H (CFH; NM_000186) or complement factor H-related 1 (CFHR1; NM_002113) (Fig. 4). Using the BLAT from University of California Santa Cruz (49), we found that both of these genes are localized on Chromosome 1 (Fig. 4A) and share 68% sequence identity in their C terminus (Fig. 4B). The nsSNP in complement factor H and complement factor H-related 1 is Y1058H and H157Y, respectively (Fig. 4C). Because the SNPs in both genes are potential phosSNPs, we include both entries in this work.
Fig. 4.
One nsSNP could be mapped on different genes. A, the human CFH and CFHR1 were located on Chromosome (Chr.) 1. B, by sequence comparison, two proteins share 68% identities in their C terminus. C, one nsSNP (rs425757) in complement factor H and complement factor H-related 1 is Y1058H and H157Y, respectively.
Here, we defined a PK type as a unique PK group in GPS 2.0 (22). In GPS 2.0, we classified human PKs together with their verified phosphorylation sites into a hierarchical structure with four levels, including group, family, subfamily, and single PK (22). Thus, some lower PK clusters could be included in their upper level groups. However, because different PK clusters used different training data sets, each PK group exhibits a distinct substrate preference. And in our result (Tables V and VI), the added or removed PK types were clearly predicted. And for Type III and Type IV phosSNPs, only added or removed PK types were included in the PhosSNP 1.0 database.
We also investigated experimentally verified human phosphorylation sites in this work. Previously, the SNPeffect database was developed as a comprehensive resource of molecular phenotypic effects of human nsSNPs (7, 8). Several well studied PTMs, including phosphorylation, glycosylation, myristoylation, farnesylation, glycosylphosphatidylinositol anchor attachment, and geranylgeranylation, were extensively considered (7, 8). In the SNPeffect database, the experimental phosphorylation data were taken from PhosphoBase (40), which contains 1,052 phosphorylation sites. Here, we used a far larger data set, including 23,978 known human phosphorylation sites from a previous study (26). By exact string matching (26), we detected 2,004 potential phosSNPs in 1,528 proteins (Tables I and II). However, most of these results still need further experimental validation. For example, although human PSEN1 (NM_000021) was experimentally verified to be phosphorylated at Thr-354 by CDK5 or CDK group PKs (Table II), it remains to be confirmed whether the T354I nsSNP has made Presenilin-1 unable to be phosphorylated by CDK5 or CDK group PKs. Also, the functional consequence of such an nsSNP should be experimentally elucidated. Moreover, although it was proposed that the P47S nsSNP of p53 (NM_000546) strongly reduces the phosphorylation level of its adjacent Ser-46 by p38 MAPK (Table II), whether the P47S nsSNP also diminishes the modification by CMGC/CDK/CDK4, CMGC/CDK/CDK4/CDK4, CMGC/CDK/CDK7, CMGC/MAPK/p38/MAPK13, and STE/STE7/MAP2K7 and renders the Ser-46 accessible by STE/STE7/MAP2K6 (Table V) has not been examined. Taken together, the phosSNPs detected from experimental phosphorylation data provide a useful reference for further experimental design.
Finally, the PhosSNP 1.0 database was implemented in JAVA 1.5 (J2SE 5.0). The local packages of the PhosSNP 1.0 database are freely available for academic researchers and support major operating systems, including Windows, Unix/Linux, and Mac.
Supplementary Material
Acknowledgments
We thank Dr. Francesca Diella (European Molecular Biology Laboratory) for always providing the new data set of Phospho.ELM database during the past 5 years. We are also grateful for the two anonymous reviewers, whose suggestions have greatly improved the presentation of this manuscript.
* This work was supported in part by the National Basic Research Program (973 project) (Grants 2006CB933300, 2007CB947401, 2007CB914503, and 2010CB912103), Natural Science Foundation of China (Grants 90919001, 30700138, 30900835, 30830036, 30721002, 30871236, and 90913016), Chinese Academy of Sciences (Grants KSCX1-YW-R65, KSCX2-YW-R-139, and INFO-115-C01-SDB4-36), and National Science Foundation for Postdoctoral Scientists (Grant 20080430100).
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- SNP
- single nucleotide polymorphism
- phosSNP
- phosphorylation-related SNP
- nsSNP
- non-synonymous SNP
- PTM
- post-translational modification
- PK
- protein kinase
- cSNP
- SNP in the coding region
- PTC
- premature termination codon
- NMD
- nonsense-mediated mRNA decay
- PSP
- phosphorylation site peptide
- MAPK
- mitogen-activated protein kinase
- PKC
- protein kinase C
- nt
- nucleotides
- CDK
- cyclin-dependent kinase
- VEGFR
- vascular endothelial growth factor receptor
- TK
- tyrosine kinase
- IKK
- IκB kinase complex
- PIKK
- phosphoinositol 3-kinase-related protein kinase
- DNAPK
- DNA-dependent protein kinase
- PKA
- protein kinase A
- DMPK
- myotonic dystrophy protein kinase
- ROCK
- Rho kinase
- CFH
- complement factor H
- CFHR1
- complement factor H-related 1
- PSEN1
- Presenilin-1
- HTR2A
- 5-HT2A serotonin receptor.
REFERENCES
- 1.Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C. R., Lim E. P., Kalyanaraman N., Nemesh J., Ziaugra L., Friedland L., Rolfe A., Warrington J., Lipshutz R., Daley G. Q., Lander E. S. (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet 22, 231–238 [DOI] [PubMed] [Google Scholar]
- 2.Collins F. S., Brooks L. D., Chakravarti A. (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8, 1229–1231 [DOI] [PubMed] [Google Scholar]
- 3.Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., Stuve L. L., Gibbs R. A., Belmont J. W., Boudreau A., Hardenbol P., Leal S. M., Pasternak S., Wheeler D. A., Willis T. D., Yu F., Yang H., Zeng C., Gao Y., Hu H., Hu W., Li C., Lin W., Liu S., Pan H., Tang X., Wang J., Wang W., Yu J., Zhang B., Zhang Q., Zhao H., Zhao H., Zhou J., Gabriel S. B., Barry R., Blumenstiel B., Camargo A., Defelice M., Faggart M., Goyette M., Gupta S., Moore J., Nguyen H., Onofrio R. C., Parkin M., Roy J., Stahl E., Winchester E., Ziaugra L., Altshuler D., Shen Y., Yao Z., Huang W., Chu X., He Y., Jin L., Liu Y., Shen Y., Sun W., Wang H., Wang Y., Wang Y., Xiong X., Xu L., Waye M. M., Tsui S. K., Xue H., Wong J. T., Galver L. M., Fan J. B., Gunderson K., Murray S. S., Oliphant A. R., Chee M. S., Montpetit A., Chagnon F., Ferretti V., Leboeuf M., Olivier J. F., Phillips M. S., Roumy S., Sallée C., Verner A., Hudson T. J., Kwok P. Y., Cai D., Koboldt D. C., Miller R. D., Pawlikowska L., Taillon-Miller P., Xiao M., Tsui L. C., Mak W., Song Y. Q., Tam P. K., Nakamura Y., Kawaguchi T., Kitamoto T., Morizono T., Nagashima A., Ohnishi Y., Sekine A., Tanaka T., Tsunoda T., Deloukas P., Bird C. P., Delgado M., Dermitzakis E. T., Gwilliam R., Hunt S., Morrison J., Powell D., Stranger B. E., Whittaker P., Bentley D. R., Daly M. J., de Bakker P. I., Barrett J., Chretien Y. R., Maller J., McCarroll S., Patterson N., Pe'er I., Price A., Purcell S., Richter D. J., Sabeti P., Saxena R., Schaffner S. F., Sham P. C., Varilly P., Altshuler D., Stein L. D., Krishnan L., Smith A. V., Tello-Ruiz M. K., Thorisson G. A., Chakravarti A., Chen P. E., Cutler D. J., Kashuk C. S., Lin S., Abecasis G. R., Guan W., Li Y., Munro H. M., Qin Z. S., Thomas D. J., McVean G., Auton A., Bottolo L., Cardin N., Eyheramendy S., Freeman C., Marchini J., Myers S., Spencer C., Stephens M., Donnelly P., Cardon L. R., Clarke G., Evans D. M., Morris A. P., Weir B. S., Tsunoda T., Mullikin J. C., Sherry S. T., Feolo M., Skol A., Zhang H., Zeng C., Zhao H., Matsuda I., Fukushima Y., Macer D. R., Suda E., Rotimi C. N., Adebamowo C. A., Ajayi I., Aniagwu T., Marshall P. A., Nkwodimmah C., Royal C. D., Leppert M. F., Dixon M., Peiffer A., Qiu R., Kent A., Kato K., Niikawa N., Adewole I. F., Knoppers B. M., Foster M. W., Clayton E. W., Watkin J., Gibbs R. A., Belmont J. W., Muzny D., Nazareth L., Sodergren E., Weinstock G. M., Wheeler D. A., Yakub I., Gabriel S. B., Onofrio R. C., Richter D. J., Ziaugra L., Birren B. W., Daly M. J., Altshuler D., Wilson R. K., Fulton L. L., Rogers J., Burton J., Carter N. P., Clee C. M., Griffiths M., Jones M. C., McLay K., Plumb R. W., Ross M. T., Sims S. K., Willey D. L., Chen Z., Han H., Kang L., Godbout M., Wallenburg J. C., L'Archevêque P., Bellemare G., Saeki K., Wang H., An D., Fu H., Li Q., Wang Z., Wang R., Holden A. L., Brooks L. D., McEwen J. E., Guyer M. S., Wang V. O., Peterson J. L., Shi M., Spiegel J., Sung L. M., Zacharia L. F., Collins F. S., Kennedy K., Jamieson R., Stewart J. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., Andrews T. D., Fiegler H., Shapero M. H., Carson A. R., Chen W., Cho E. K., Dallaire S., Freeman J. L., González J. R., Gratacòs M., Huang J., Kalaitzopoulos D., Komura D., MacDonald J. R., Marshall C. R., Mei R., Montgomery L., Nishimura K., Okamura K., Shen F., Somerville M. J., Tchinda J., Valsesia A., Woodwark C., Yang F., Zhang J., Zerjal T., Zhang J., Armengol L., Conrad D. F., Estivill X., Tyler-Smith C., Carter N. P., Aburatani H., Lee C., Jones K. W., Scherer S. W., Hurles M. E. (2006) Global variation in copy number in the human genome. Nature 444, 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinds D. A., Stuve L. L., Nilsen G. B., Halperin E., Eskin E., Ballinger D. G., Frazer K. A., Cox D. R. (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 [DOI] [PubMed] [Google Scholar]
- 6.Stenson P. D., Ball E. V., Mort M., Phillips A. D., Shiel J. A., Thomas N. S., Abeysinghe S., Krawczak M., Cooper D. N. (2003) Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat 21, 577–581 [DOI] [PubMed] [Google Scholar]
- 7.Reumers J., Maurer-Stroh S., Schymkowitz J., Rousseau F. (2006) SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics 22, 2183–2185 [DOI] [PubMed] [Google Scholar]
- 8.Reumers J., Schymkowitz J., Ferkinghoff-Borg J., Stricher F., Serrano L., Rousseau F. (2005) SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res 33, D527–D532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packer B. R., Yeager M., Burdett L., Welch R., Beerman M., Qi L., Sicotte H., Staats B., Acharya M., Crenshaw A., Eckert A., Puri V., Gerhard D. S., Chanock S. J. (2006) SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res 34, D617–D621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jegga A. G., Gowrisankar S., Chen J., Aronow B. J. (2007) PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res 35, D700–D706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J. O., Hwang S., Oh J., Bhak J., Sohn T. K. (2008) An integrated database-pipeline system for studying single nucleotide polymorphisms and diseases. BMC Bioinformatics 9, Suppl. 12, S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue P., Moult J. (2006) Identification and analysis of deleterious human SNPs. J. Mol. Biol 356, 1263–1274 [DOI] [PubMed] [Google Scholar]
- 13.Stitziel N. O., Binkowski T. A., Tseng Y. Y., Kasif S., Liang J. (2004) topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res 32, D520–D522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uzun A., Leslin C. M., Abyzov A., Ilyin V. (2007) Structure SNP (StSNP): a web server for mapping and modeling nsSNPs on protein structures with linkage to metabolic pathways. Nucleic Acids Res 35, W384–W392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kono H., Yuasa T., Nishiue S., Yura K. (2008) coliSNP database server mapping nsSNPs on protein structures. Nucleic Acids Res 36, D409–D413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savas S., Ozcelik H. (2005) Phosphorylation states of cell cycle and DNA repair proteins can be altered by the nsSNPs. BMC Cancer 5, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C. Y., Chang C. H., Yu Y. L., Lin T. C., Lee S. A., Yen C. C., Yang J. M., Lai J. M., Hong Y. R., Tseng T. L., Chao K. M., Huang C. Y. (2008) PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics 24, i14–20 [DOI] [PubMed] [Google Scholar]
- 18.Ryu G. M., Song P., Kim K. W., Oh K. S., Park K. J., Kim J. H. (2009) Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucleic Acids Res 37, 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diella F., Gould C. M., Chica C., Via A., Gibson T. J. (2008) Phospho.ELM: a database of phosphorylation sites—update 2008. Nucleic Acids Res 36, D240–D244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller M. L., Blom N. (2009) Kinase-specific prediction of protein phosphorylation sites. Methods Mol. Biol 527, 299–310, x [DOI] [PubMed] [Google Scholar]
- 22.Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hjerrild M., Gammeltoft S. (2006) Phosphoproteomics toolbox: computational biology, protein chemistry and mass spectrometry. FEBS Lett 580, 4764–4770 [DOI] [PubMed] [Google Scholar]
- 24.Kobe B., Kampmann T., Forwood J. K., Listwan P., Brinkworth R. I. (2005) Substrate specificity of protein kinases and computational prediction of substrates. Biochim. Biophys. Acta 1754, 200–209 [DOI] [PubMed] [Google Scholar]
- 25.Li H., Xing X., Ding G., Li Q., Wang C., Xie L., Zeng R., Li Y. (2009) SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol. Cell. Proteomics 8, 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan C. S., Bodenmiller B., Pasculescu A., Jovanovic M., Hengartner M. O., Jørgensen C., Bader G. D., Aebersold R., Pawson T., Linding R. (2009) Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal 2, ra39. [DOI] [PubMed] [Google Scholar]
- 27.Luna L., Rolseth V., Hildrestrand G. A., Otterlei M., Dantzer F., Bjørås M., Seeberg E. (2005) Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res 33, 1813–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Dumont P., Della Pietra A., Shetler C., Murphy M. E. (2005) The codon 47 polymorphism in p53 is functionally significant. J. Biol. Chem 280, 24245–24251 [DOI] [PubMed] [Google Scholar]
- 29.Oh Y. T., Chun K. H., Park B. D., Choi J. S., Lee S. K. (2007) Regulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1 by protein kinase Cdelta-mediated phosphorylation. Apoptosis 12, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 30.Gentile S., Martin N., Scappini E., Williams J., Erxleben C., Armstrong D. L. (2008) The human ERG1 channel polymorphism, K897T, creates a phosphorylation site that inhibits channel activity. Proc. Natl. Acad. Sci. U.S.A 105, 14704–14708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., Sirotkin K. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29, 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruitt K. D., Tatusova T., Maglott D. R. (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35, D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maquat L. E. (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol 5, 89–99 [DOI] [PubMed] [Google Scholar]
- 35.Nagy E., Maquat L. E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci 23, 198–199 [DOI] [PubMed] [Google Scholar]
- 36.Han A., Kim W. Y., Park S. M. (2007) SNP2NMD: a database of human single nucleotide polymorphisms causing nonsense-mediated mRNA decay. Bioinformatics 23, 397–399 [DOI] [PubMed] [Google Scholar]
- 37.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 38.Pinna L. A., Ruzzene M. (1996) How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1314, 191–225 [DOI] [PubMed] [Google Scholar]
- 39.Blom N., Gammeltoft S., Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 40.Kreegipuu A., Blom N., Brunak S. (1999) PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res 27, 237–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreegipuu A., Blom N., Brunak S., Järv J. (1998) Statistical analysis of protein kinase specificity determinants. FEBS Lett 430, 45–50 [DOI] [PubMed] [Google Scholar]
- 42.Songyang Z., Lu K. P., Kwon Y. T., Tsai L. H., Filhol O., Cochet C., Brickey D. A., Soderling T. R., Bartleson C., Graves D. J., DeMaggio A. J., Hoekstra M. F., Blenis J., Hunter T., Cantley L. C. (1996) A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol 16, 6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19, 271–273 [DOI] [PubMed] [Google Scholar]
- 44.Milting H., Lukas N., Klauke B., Körfer R., Perrot A., Osterziel K. J., Vogt J., Peters S., Thieleczek R., Varsányi M. (2006) Composite polymorphisms in the ryanodine receptor 2 gene associated with arrhythmogenic right ventricular cardiomyopathy. Cardiovasc. Res 71, 496–505 [DOI] [PubMed] [Google Scholar]
- 45.Gray J. A., Compton-Toth B. A., Roth B. L. (2003) Identification of two serine residues essential for agonist-induced 5-HT2A receptor desensitization. Biochemistry 42, 10853–10862 [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Houle A. M., Letendre J., Richter A. (2008) RET Gly691Ser mutation is associated with primary vesicoureteral reflux in the French-Canadian population from Quebec. Hum. Mutat 29, 695–702 [DOI] [PubMed] [Google Scholar]
- 47.Erxleben C., Liao Y., Gentile S., Chin D., Gomez-Alegria C., Mori Y., Birnbaumer L., Armstrong D. L. (2006) Cyclosporin and Timothy syndrome increase mode 2 gating of CaV1.2 calcium channels through aberrant phosphorylation of S6 helices. Proc. Natl. Acad. Sci. U.S.A 103, 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamasaki C., Murakami K., Fujii Y., Sato Y., Harada E., Takeda J., Taniya T., Sakate R., Kikugawa S., Shimada M., Tanino M., Koyanagi K. O., Barrero R. A., Gough C., Chun H. W., Habara T., Hanaoka H., Hayakawa Y., Hilton P. B., Kaneko Y., Kanno M., Kawahara Y., Kawamura T., Matsuya A., Nagata N., Nishikata K., Noda A. O., Nurimoto S., Saichi N., Sakai H., Sanbonmatsu R., Shiba R., Suzuki M., Takabayashi K., Takahashi A., Tamura T., Tanaka M., Tanaka S., Todokoro F., Yamaguchi K., Yamamoto N., Okido T., Mashima J., Hashizume A., Jin L., Lee K. B., Lin Y. C., Nozaki A., Sakai K., Tada M., Miyazaki S., Makino T., Ohyanagi H., Osato N., Tanaka N., Suzuki Y., Ikeo K., Saitou N., Sugawara H., O'Donovan C., Kulikova T., Whitfield E., Halligan B., Shimoyama M., Twigger S., Yura K., Kimura K., Yasuda T., Nishikawa T., Akiyama Y., Motono C., Mukai Y., Nagasaki H., Suwa M., Horton P., Kikuno R., Ohara O., Lancet D., Eveno E., Graudens E., Imbeaud S., Debily M. A., Hayashizaki Y., Amid C., Han M., Osanger A., Endo T., Thomas M. A., Hirakawa M., Makalowski W., Nakao M., Kim N. S., Yoo H. S., De Souza S. J., Bonaldo Mde F., Niimura Y., Kuryshev V., Schupp I., Wiemann S., Bellgard M., Shionyu M., Jia L., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Zhang Q., Go M., Minoshima S., Ohtsubo M., Hanada K., Tonellato P., Isogai T., Zhang J., Lenhard B., Kim S., Chen Z., Hinz U., Estreicher A., Nakai K., Makalowska I., Hide W., Tiffin N., Wilming L., Chakraborty R., Soares M. B., Chiusano M. L., Suzuki Y., Auffray C., Yamaguchi-Kabata Y., Itoh T., Hishiki T., Fukuchi S., Nishikawa K., Sugano S., Nomura N., Tateno Y., Imanishi T., Gojobori T. (2008) The H-Invitational Database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res 36, D793–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent W. J. (2002) BLAT—the BLAST-like alignment tool. Genome Res 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.