Abstract
Insulin plays an essential role in metabolic homeostasis in mammals, and many of the underlying biochemical pathways are regulated via the canonical phosphatidylinositol 3-kinase/AKT pathway. To identify novel metabolic actions of insulin, we conducted a quantitative proteomics analysis of insulin-regulated 14-3-3-binding proteins in muscle cells. These studies revealed a novel role for insulin in the post-transcriptional regulation of mRNA expression. EDC3, a component of the mRNA decay and translation repression pathway associated with mRNA processing bodies, was shown to be phosphorylated by AKT downstream of insulin signaling. The major insulin-regulated site was mapped to Ser-161, and phosphorylation at this site led to increased 14-3-3 binding. Functional studies indicated that induction of 14-3-3 binding to EDC3 causes morphological changes in processing body structures, inhibition of microRNA-mediated mRNA post-transcriptional regulation, and alterations in the protein- protein interactions of EDC3. These data highlight an important new arm of the insulin signaling cascade in the regulation of mRNA utilization.
Insulin plays an essential role in regulating fuel homeostasis, cellular differentiation, and growth (1). One of the primary signaling cascades activated by insulin is the PI3K1/AKT pathway. This pathway mediates many of the metabolic actions of insulin via phosphorylation of key metabolic substrates such as glycogen synthase kinase-3 for glycogen synthesis (2), FOXO1 for transcription (3), and TSC2 for protein synthesis (4). Intriguingly, insulin also plays a major role in mRNA decay and translation repression; however, the signaling molecules linking insulin with these pathways have not been fully elucidated (5–9). Several important mechanisms exist by which cells target mRNAs for post-transcriptional regulation including AU-rich element-binding proteins and through the actions of microRNAs (miRNAs) (10–12). The targeted mRNAs are then regulated by degradation or by inert reversible storage in cytoplasmic protein complexes called processing bodies (p-bodies) (13, 14). Such stored complexes may serve as a readily releasable reservoir of mRNA that could theoretically be rapidly mobilized for translation under anabolic conditions such as insulin stimulation. Enhancer of mRNA decapping 3 (EDC3) is a p-body protein that has previously been shown to have a role in regulating the removal of the mRNA 5′ cap structure (15). To achieve this function, EDC3 interacts with a number of other proteins through its three domains (16, 17). The N-terminal Lsm domain is responsible for p-body targeting of EDC3 and can mediate interactions with DCP1A/B (16, 17). The central FDF domain has no known function but enables the interaction of EDC3 with the RNA helicase DDX6 and the decapping enzyme DCP2, which functions in conjunction with DCP1A/B to remove the 5′ cap structure from mRNA (16, 17). The C-terminal Yjef homology domain is responsible for the homodimerization of EDC3 that may be important for its function (17, 18). Once the 5′ cap structure is removed from the mRNA, it is quickly degraded by exonucleases such as XRN1, which is also present in p-bodies (19).
The insulin-regulated AKT substrate repertoire identified to date reflects the diversity of the actions of insulin in relevant target cells (20). Many AKT substrates undergo 14-3-3 binding upon phosphorylation, and so these two processes may be linked in modulating protein function (21). In the present study, we took advantage of the high affinity of 14-3-3 for AKT substrates combined with quantitative mass spectrometry methods based on stable isotope labeling with amino acids in cell culture (SILAC) (22, 23) to search for novel actions of insulin. Here, we report the identification of a novel insulin-regulated phosphoprotein, EDC3, a component of p-bodies (13, 16), thus revealing a novel action of insulin on mRNA post-transcriptional regulation.
EXPERIMENTAL PROCEDURES
Materials
NIH-3T3 murine fibroblasts, L6 fibroblasts, and Chinese hamster ovary (CHO) cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Dulbecco's modified Eagle's medium (DMEM) was obtained from Invitrogen, Myoclone-Plus FCS was from Trace Scientific (Melbourne, Australia), and antibiotics were from Invitrogen. Paraformaldehyde was from ProSciTech (Thuringowa Central, Australia). Insulin was obtained from Calbiochem, and BSA was from USB Corp. (Cleveland, OH). Bicinchoninic acid (BCA) reagent, Supersignal West Pico chemiluminescent substrate, and protein G-agarose beads were from Pierce. Lipofectamine LTX and Plus Reagent were from Invitrogen. PVDF membrane was from Millipore (Billerica, MA). Trypsin was from Promega (Madison, WI). C18 cartridges were from Michrom Bioresources, Inc. (Auburn, CA). Magic C18 material was from (Alltech, Deerfield, IL). Complete protease inhibitor mixture tablets were from Roche Applied Science. All other materials were obtained from Sigma. Antibodies were kindly provided by Dr. Jens Lykke-Andersen (EDC3 and DCP1A; University of Colorado). Antibodies were purchased from Novus Biologicals (EDC3; Littleton, CO), MBL International (DDX6; Woburn, MA), Santa Cruz Biotechnology (Hedls, HMGA-2, and pan-14-3-3; Santa Cruz Biotechnology), Cell Signaling Technology (phospho-AKT substrate, AKT, AKT Ser(P)-473, and GST; Danvers, MA), and Sigma (FLAG M2). Antibodies against AS160 have been described previously (24). Cy5- and Cy3-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Alexa Fluor 680-conjugated secondary antibodies were obtained from Invitrogen. IrDye 800-conjugated secondary antibodies were obtained from Rockland Immunochemicals (Gilbertsville, PA). HRP-conjugated secondary antibodies were from GE Healthcare. Active AKT2 for the in vitro kinase assay was from Cell Signaling Technology.
Production of Rabbit EDC3 Ser(P)-161 Antibody
The antibody specific against EDC3 Ser(P)-161 was produced by 21st Century Biochemicals. Briefly, rabbits were inoculated with EDC3 Ser(P)-161 peptides encoding amino acids 154–169 of mouse EDC3 conjugated to keyhole limpet hemocyanin carrier protein five times over a 72-day period. Subsequently, the phosphospecific antibody was affinity-purified by removal of antibodies that bind the non-phosphorylated peptide and then affinity purification using the phosphorylated peptide.
Cell Culture
NIH-3T3 fibroblasts (25), L6 cells (26), HEK 293T, and CHO IR/IRS-1 cells (27) were cultured as described previously. Briefly, NIH-3T3 cells were grown in DMEM supplemented with 10% FCS, 2 mm l-glutamine, 100 units/liter penicillin, and 100 μg/liter streptomycin at 37 °C in 10% CO2 and passaged at ∼60% confluence. To establish basal conditions before use, cells were incubated in serum-free DMEM for 16 h at 37 °C in 10% CO2. L6 myoblasts were cultured in minimal essential medium α supplemented with 10% FCS, 2 mm l-glutamine, 100 units/liter penicillin, and 100 μg/liter streptomycin at 37 °C in 10% CO2 and passaged at ∼60% confluence. For differentiation into myotubes, cells were cultured in minimal essential medium α supplemented with 2% FCS, 2 mm l-glutamine, 100 units/liter penicillin, and 100 μg/liter streptomycin at 37 °C in 10% CO2 and were maintained in this medium postdifferentiation. Myotubes were used for experiments 5–7 days after differentiation. For SILAC labeling of L6 and HEK 293T cells, arginine- and lysine-free Dulbecco's minimal essential medium or minimal essential medium α, respectively, was used and supplemented with stable isotope-labeled arginine and lysine in addition to dialyzed FCS as described previously (22). CHO IR/IRS-1 cells were cultured in F-12 medium containing 10% FCS, 800 μg/ml G418, 2 mm l-glutamine, 100 units/liter penicillin, and 100 μg/liter streptomycin at 37 °C in 10% CO2. CHO IR/IRS-1 cells, NIH-3T3, and HEK 293T cells were transiently transfected with DNA constructs using Lipofectamine LTX according to the manufacturer's instructions. For specific inhibition of PI3K in cells, 100 nm wortmannin was added to the basal cell culture medium 30 min prior to stimulation of cells with insulin.
Production of Thrombin-cleaved Human 14-3-3β in E. coli for 14-3-3 Pulldown Experiments
GST-14-3-3β was prepared as described previously (28) and when bound to glutathione beads was incubated with 1000 units/ml thrombin in 50 mm Tris-HCl, pH 8, and 1 mm CaCl2 overnight at room temperature. The supernatant was buffer-exchanged three times into bead coupling buffer using Amicon 10-kDa molecular mass cutoff ultrafiltration devices (Millipore). The concentrated 14-3-3 protein was then coupled to cyanogen bromide-activated Sepharose 4B (GE Healthcare) according to the manufacturer's instructions at a concentration of 5 mg of 14-3-3 coupled per 1 ml of packed beads. After extensive washing with TBS, the beads were stored at 4 °C as a 50% slurry in TBS containing 0.02% sodium azide.
14-3-3 Pulldown from SILAC-labeled L6 Cells or Mouse Quadriceps
Frozen mouse quadriceps (isolated under the approval of the Garvan/St. Vincent's animal ethics committee) were lysed using a Polytron mixer (Kinematica, Lucerne, Switzerland) for 30 s in IP buffer (1% Nonidet P-40, 50 mm Tris-Cl, pH 7.4, 10% glycerol, 150 mm NaCl, Roche Applied Science Complete protease inhibitor mixture, 2 mm sodium orthovanadate, 1 mm sodium pyrophosphate, 1 mm ammonium molybdate, and 10 mm sodium fluoride) and incubated for 30 min at 4 °C. L6 myotubes were lysed in IP buffer by scraping down the cells on ice and syringing the lysate six times with a 22-gauge needle followed by three times with a 27-gauge needle. The lysate was centrifuged for 30 min at 18,000 × g at 4 °C. Equal protein amounts of each sample were then incubated with 14-3-3 beads that had been washed once in IP buffer (50 μg of 14-3-3/1 mg of lysate) and incubated overnight at 4 °C. Beads were then washed once with IP buffer, once with high salt (0.5 m NaCl) IP buffer, and finally once more in IP buffer. Beads were transferred to a spin column (Pierce) and centrifuged dry at 500 × g for 1 min. SDS-PAGE sample buffer that had been preheated to 65 °C was then added and incubated at 65 °C for 5 min. The eluate was collected by centrifugation at 500 × g for 1 min at room temperature.
SDS-PAGE and Immunoblotting
All samples were subjected to SDS-PAGE analysis on 10% resolving gels according to Laemmli (29) but with the addition of 50 mm tris(carboxyethyl)phosphine (Bond-Breaker TCEP neutral pH solution, Pierce) in the sample buffer. Equal amounts of protein were loaded for each sample in a single experiment with 10 μg/lane. For mass spectrometric identification, SYPRO Ruby staining was performed according to the manufacturer's instructions. For Western blotting, separated proteins were electrophoretically transferred to PVDF membrane, blocked with 5% nonfat skim milk in 0.1% Tween 20 in TBS (TBST), and incubated with primary antibody in 5% BSA in TBST overnight at 4 °C. After incubation, membranes were washed three times in TBST and incubated with HRP-labeled or Alexa Fluor 680/IrDye 800-labeled secondary antibodies in 5% nonfat skim milk in TBST. Proteins were visualized using Supersignal West Pico chemiluminescent substrate and imaged with x-ray film (Fuji) for HRP-labeled secondary antibodies or a Licor Odyssey imager for Alexa Fluor 680/IrDye 800-labeled secondary antibodies.
Production of Human GST-14-3-3β in E. coli and 14-3-3 Overlay Assay
GST-14-3-3β was prepared as described previously (28) and stored at 1 mg/ml in PBS containing 0.02% sodium azide. For the 14-3-3 overlay assay, proteins were transferred to PVDF membranes as described above for immunoblotting. Membranes were blocked in 5% skim milk TBS and 0.1% Tween 20 for 1 h at room temperature. Membranes were then washed twice in TBS and 0.1% Tween 20 and incubated overnight at 4 °C in 5% BSA TBS and 0.1% Tween 20 containing 1 μg/ml GST-14-3-3β. Membranes were then detected as described above using a GST-specific primary antibody.
Immunoprecipitation of FLAG-tagged EDC3 from CHO IR/IRS-1 Cells
CHO IR/IRS-1 cells transiently transfected with TagRFP alone or TagRFP-FLAG-EDC3 were lysed after 24 h in IP buffer and centrifuged at 18,000 × g for 30 min at 4 °C. Anti-FLAG antibody and protein G beads were added to the supernatant and incubated for 1 h at 4 °C with mixing. The beads were then washed three times with IP buffer, and FLAG-EDC3 complexes were eluted by incubation of beads in 0.2 mg/ml 3× FLAG peptide (Sigma) in TBS for 1 h at 4 °C.
Immunoprecipitation of SILAC-labeled FLAG-tagged EDC3 from HEK 293T Cells
SILAC-labeled HEK 293T cells were transiently transfected with either a TagRFP-only negative control (light label), TagRFP-FLAG-EDC3 S161A-R18 (medium label), or TagRFP-FLAG-EDC3 S161A-R18-KK (heavy label) (supplemental Fig. S5). After 24 h, cells were lysed in IP buffer and centrifuged at 18,000 × g for 30 min at 4 °C. Anti-FLAG antibody and protein G beads were added to the supernatant and incubated for 1 h at 4 °C with mixing. The beads were then washed three times with IP buffer, and FLAG-EDC3 complexes were eluted by incubation of beads in 0.2 mg/ml 3× FLAG peptide (Sigma) in TBS for 1 h at 4 °C. As each lysate is mixed prior to immunoprecipitation, low affinity interactions will allow exchange of the stable isotope labels and induce a decrease in the medium over light (M/L) and heavy over light (H/L) SILAC ratios for those low affinity binding proteins. To avoid disregarding these low affinity binders, we used a low stringency (M/L or H/L SILAC ratio >1) cutoff in our data analysis with MaxQuant in the elimination of contaminant nonspecific binding proteins. This yields excellent removal of environmental contaminants (keratins, trypsin, antibody, etc.) that do not incorporate stable isotopes and therefore have a very low M/L or H/L SILAC ratio. But this method will not remove some experimental contaminants such as STK38, which may have M/L and H/L SILAC ratios close to 1.
Dual-Luciferase Assay for Analysis of let-7-mediated mRNA Repression
The Dual-Luciferase assay was performed according to the method of Clancy et al. (30). Briefly, NIH-3T3 cells stably expressing FLAG-tagged EDC3 constructs or vector alone were transiently transfected with firefly and Renilla luciferase plasmids as described (30). After transfection, cells were incubated in medium containing 10% serum for 16 h and then assayed for luciferase expression as described (30).
Immunoprecipitation of EDC3 Phosphorylated at Ser-161
NIH-3T3 cells were lysed in IP buffer and centrifuged at 18,000 × g for 30 min at 4 °C. Anti-EDC3 Ser(P)-161 antibodies were coupled to cyanogen bromide-activated Sepharose 4B Beads (GE Healthcare) according to the manufacturer's instructions. The cell lysate was incubated with the antibody-coupled beads in the presence of a 10-fold molar excess of either Ser(P)-161 phosphopeptide or Ser-161 non-phosphopeptide overnight at 4 °C. Beads were washed three times in IP buffer and boiled in SDS-PAGE sample buffer.
Retroviral Transfection of NIH-3T3 Cells for Generation of Stable Cells
NIH-3T3 cells were grown to 20% confluency, and then pMXSC-IRES-GFP (pMIG) retrovirus constructs produced in Platinum-E cells were added in the presence of 4 μg/ml Polybrene (Sigma) for 16 h at 37 °C. Low level enhanced green fluorescent protein-expressing cells (bottom 10%) were then selected by fluorescence-activated cell sorting as described previously (31).
In-gel Protein Tryptic Digestion, LC-MS/MS, Identification, and Quantitation
Protein bands of interest were excised and destained in 1 ml of 50% acetonitrile and 250 mm ammonium bicarbonate at room temperature for 45 min with shaking. The gel slice was dehydrated by incubation in 1 ml of 100% acetonitrile for 10 min at room temperature. All solution was carefully removed prior to the addition of modified trypsin (12.5 ng/μl) in 100 mm NH4HCO3 and incubation overnight at 37 °C. Peptides were extracted by the addition of 0.1 ml of 5% formic acid and incubation at 37 °C for 1 h. Peptides were further extracted by the addition of 0.1 ml of 100% acetonitrile and incubation at 37 °C for 1 h. The gel slice was completely dehydrated by the addition of 0.5 ml of 100% acetonitrile and incubation at 37 °C for 10 min. The entire supernatant was then vacuum-dried. The peptides were redissolved in 20 μl of 5% formic acid for LC-MS/MS analysis. Peptide solutions (5 μl) were subsequently resolved on a 100-mm × 75-μm C18 Magic reverse phase analytical column using a Dionex Ultimate 3000 liquid chromatography system over a 1-h organic gradient with a flow rate of 250 nl/min. Peptides were ionized by nanoelectrospray ionization at 1.8 kV from the end of the column, which was pulled to an internal diameter of 5 μm by a P-2000 laser puller (Sutter Instrument Co.). Tandem mass spectrometry analysis was carried out on a Thermo Scientific (San Jose, CA) LTQ-FT Ultra mass spectrometer. The data-dependent acquisition method used was the FT10 protocol as described previously (32). Data were processed, searched, and quantified using the MaxQuant software version 1.0.13.13 package as described previously (33) using the default settings, and searches where performed using Mascot server version 2.2 and against the entire MaxQuant mouse database version 3.52 (with 111,130 sequences searched) or human database version 3.52 (with 148,380 sequences searched). The settings used for the Mascot search were as follows: two missed cleavages were allowed; enzyme was trypsin cleaving after arginine and lysine; variable modifications were methionine oxidation, propionamide cysteine, and phosphorylation of serine, threonine, or tyrosine; no fixed modifications were used; a mass tolerance of 7 ppm was used for precursor ions; and a tolerance of 0.5 Da was used for fragment ions. Using the default MaxQuant settings, a maximum false positive rate of 1% was allowed for both peptide and protein identification, and this was used as a cutoff score for accepting individual spectra as well as whole proteins in the combined search and quantitation output. This threshold has been shown previously to be a rigorous method of identifying true positive matches (33). Protein quantitation data were always derived from two or more peptides per protein. For the 14-3-3 SILAC screen, proteins were included in Table I if their ratio had a significance (B) <0.05 as described previously (33) and were detected in both experiments.
Table I. Novel insulin-responsive proteins identified by combined SILAC and 14-3-3 affinity analysis.
| UniProt accession no. | Protein name | Protein description | Average (insulin/basal)a | S.D. (insulin/basal) | Average (wort + insulin/basal)a | S.D. (wort + insulin/basal) |
|---|---|---|---|---|---|---|
| Q8K2D3 | EDC3 | Enhancer of mRNA-decapping protein 3 | 5.96 | 0.80 | 1.99 | 0.52 |
| O88380 | TRIP12 | Thyroid hormone receptor interactor 12 | 5.35 | 0.19 | 0.88 | 0.15 |
| Q9WVH4 | FOXO3 | Forkhead box O3a | 4.83 | 0.96 | 1.22 | 0.59 |
| Q9D1F4 | PRAS | Proline-rich AKT1 substrate 1 | 4.41 | 0.40 | 1.25 | 0.04 |
| Q62433 | NDRG1 | Protein NDRG1 | 4.20 | 0.07 | 1.23 | 0.16 |
| Q8BL80 | ARHGAP22 | Rho GTPase-activating protein 22 | 3.33 | 0.20 | 0.69 | 0.07 |
| Q9EP53 | TSC1 | Tuberous sclerosis 1 | 2.93 | 0.56 | 1.32 | 0.15 |
| Q8C570 | MRNP41 | mRNA export factor | 2.83 | 0.32 | 1.52 | 0.15 |
| Q99MS7 | EHBP1L1 | EH domain-binding protein 1-like protein 1 | 2.80 | 0.29 | 0.97 | 0.04 |
| P23949 | BRF2 | Butyrate response factor 2 | 2.77 | 0.35 | 2.03 | 0.58 |
| Q9Z1S8 | GAB2 | Growth factor receptor-bound protein 2-associated protein 2 | 2.63 | 1.08 | 1.02 | 0.11 |
| P23950 | BRF1 | Butyrate response factor 1 | 2.55 | 0.38 | 2.08 | 0.58 |
| Q61037 | TSC2 | Tuberous sclerosis 2 | 2.54 | 0.52 | 1.10 | 0.18 |
| Q7TPH6 | MYCBP2 | Probable E3 ubiquitin-protein ligase MYCBP2 | 2.49 | 0.00 | 1.62 | 0.23 |
| O08908 | PIK3R2 | Phosphatidylinositol 3-kinase regulatory subunit β | 2.43 | 0.20 | 2.32 | 0.30 |
| Q99J03 | BECN1 | Beclin 1 | 2.35 | 0.12 | 1.17 | 0.10 |
| Q148W8 | DUSP27 | Inactive dual specificity phosphatase 27 | 2.21 | 0.08 | 1.38 | 0.02 |
| Q9WTX5 | SKP1 | S phase kinase-associated protein 1 | 2.16 | 0.58 | 1.52 | 0.41 |
| Q6PDH0 | PHLDB1 | Pleckstrin homology-like domain family B member 1 | 2.10 | 0.08 | 1.03 | 0.25 |
| Q6ZQ58 | LARP1 | La-related protein | 1.98 | 0.08 | 1.03 | 0.05 |
| A0T1J8 | LMO7 | LIM domain only 7 | 1.83 | 0.28 | 1.46 | 0.13 |
| Q8BKC8 | PI4KB | Phosphatidylinositol 4-kinase β | 0.61 | 0.01 | 0.86 | 0.16 |
| A0JLR7 | AHNAK | AHNAK nucleoprotein isoform 1 | 0.59 | 0.14 | 0.80 | 0.08 |
| Q6P9S0 | MTSS1L | MTSS1-like protein | 0.58 | 0.04 | 0.58 | 0.11 |
| Q9JM13 | RABGEF1 | Rab5 GDP/GTP exchange factor | 0.56 | 0.13 | 0.83 | 0.18 |
| Q6AXD2 | ABI2 | Abl interactor 2 | 0.54 | 0.02 | 0.59 | 0.02 |
| O35063 | KIF13B | Kinesin family member 13B | 0.54 | 0.17 | 1.09 | 0.10 |
| Q8CI08 | SLAIN2 | SLAIN motif-containing protein 2 | 0.52 | 0.02 | 0.54 | 0.07 |
| P42128 | FOXK1 | Forkhead box protein K1 | 0.38 | 0.01 | 0.81 | 0.05 |
a Average ratio of -fold change in 14-3-3 binding with insulin stimulation or wortmannin (wort) and insulin stimulation compared with basal.
Immunofluorescence Microscopy
NIH-3T3 fibroblasts were cultured as described above on glass coverslips. The cells were serum-depleted for 16 h at 37 °C after which they were incubated in the absence or presence of 1 μm insulin for 30 min. Cells were then fixed with 3% paraformaldehyde in PBS. Fixed cells were washed with PBS, and free aldehyde groups were quenched with 50 mm glycine in PBS. The cells were then processed for immunolabeling by permeabilization and labeling in PBS containing 0.1% saponin and 2% BSA using standard procedures. Primary antibodies were detected with Alexa Fluor 488- or Cy3-conjugated secondary antibodies. Optical sections were analyzed by epifluorescence microscopy on a Zeiss Observer inverted microscope. Images were generated by the maximum projection of a z-stack taken from the top to the bottom of the cell monolayer. The contrast was adjusted for all images with the same settings. Quantitation of p-body intensity was performed in ImageJ software using a peak threshold setting, which was applied equally across all images, and calculation of the integrated intensity by the Analyze Particles function.
In Vitro AKT2 Kinase Assay Using [γ-32P]ATP
TagRFP-FLAG-EDC3 wild type or S161A mutant were expressed by transient transfection in CHO IR/IRS-1 cells, and FLAG was immunoprecipitated as described above. Eluted TagRFP-FLAG-EDC3 proteins were incubated with 1 μCi/ml [γ-32P]ATP, 1 mm cold ATP, 50 mM Tris-HCl, pH 7.4, and 150 mm NaCl and in the presence or absence of 10 ng/ml active AKT2 (Cell Signaling Technology) for 10 or 100 min at room temperature.
RESULTS
Identification of Insulin-responsive 14-3-3-binding Proteins Using SILAC and Mass Spectrometry
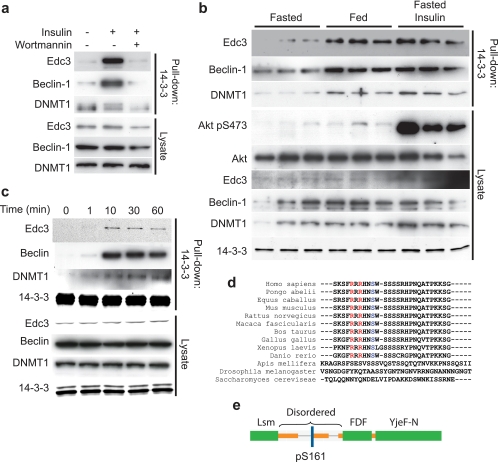
In the present study, we devised a strategy to identify novel insulin-regulated AKT substrates in insulin-responsive L6 myotubes (supplemental Fig. S1). By combining the high specificity for AKT substrates provided by 14-3-3 affinity chromatography and the inherent quantitative capability of SILAC (22), we identified from among the large cohort of intracellular 14-3-3-binding proteins those that do so in an insulin- and PI3K-dependent manner, thus providing a unique glimpse of the phosphoproteome. Analysis of these experiments using MaxQuant (33) software yielded >400 identified proteins in L6 myotubes of which 28 showed a significant (significance (B) <0.05) change in 14-3-3 binding after insulin stimulation (Table I and supplemental Table S1). Of these, we identified five previously reported AKT substrates (FOXO3A (34), PRAS40 (35), GAB2 (36), BRF1 (37), and TSC2 (4)), and we observed six previously unknown insulin-responsive proteins on which we chose to focus our study (supplemental Table S2). The majority of novel insulin-responsive proteins were wortmannin-sensitive, indicating that they were downstream of PI3K (Table I). Immunoblotting using specific antibodies was used to verify several of the novel insulin-responsive proteins. Beclin, EDC3, and DNA methyltransferase 1 underwent rapid insulin-dependent phosphorylation as demarcated by 14-3-3 binding in a wortmannin-sensitive manner (Fig. 1, a and c). Moreover, these proteins were also found to undergo increased 14-3-3 binding in mouse quadriceps in vivo under physiological conditions such as the fasted/fed transition (Fig. 1b). Bioinformatics analysis using Scansite (38) and Eukaryotic Linear Motifs (ELM) (39) software packages revealed that EDC3, TRIP12, and MYCBP2 contained consensus AKT phosphorylation motifs (supplemental Table S2).
Fig. 1.
Analysis of novel insulin-responsive proteins. a, immunoblotting of 14-3-3 pulldowns from L6 myotubes that were either left unstimulated, stimulated with 100 nm insulin for 30 min, or treated with the PI3K inhibitor wortmannin prior to insulin stimulation. b, immunoblotting of 14-3-3 pulldowns from quadriceps of 20-week-old male C57BL/6 mice. Mice were either fasted overnight followed by a mock intraperitoneal injection, ad libitum fed overnight followed by a mock intraperitoneal injection, or fasted overnight followed by an intraperitoneal injection of 1 unit/kg insulin. Mice were sacrificed 10 min after injection, and quadriceps were excised. c, immunoblotting of 14-3-3 pulldowns from L6 myotubes that were stimulated with 100 nm insulin for the indicated times. d, sequence conservation throughout evolution of amino acids surrounding Ser(P)-161 (blue). Critical arginines at −5 and −3 required for AKT phosphorylation are shown in red. e, domain structure of EDC3 showing the location of the putative AKT phosphorylation and 14-3-3 binding site Ser(P)-161. Immunoblots are from a representative experiment (n = 3).
The identification of these proteins potentially unveils important actions of insulin in a variety of novel pathways. Based on the key role of EDC3 in mRNA repression and the fact that an effect of insulin on this pathway has not been described previously, we prioritized this protein for further analysis and characterized the role of insulin-dependent phosphorylation of this protein in a range of model systems. Although all of the proteins identified in our screen contain 14-3-3 binding and AKT phosphorylation consensus sites (supplemental Table S2), EDC3 contains one Scansite high stringency AKT site at Ser-161 (in the top 0.008% of all sites in the Swiss-Prot vertebrate database) and two medium stringency AKT sites at Ser-262 and Ser-231. ClustalX alignment showed that the sequence surrounding Ser-161 is highly conserved in vertebrates (Fig. 1d). Strikingly, those residues that make EDC3 Ser-161 a high scoring AKT site in the Scansite algorithm were preserved throughout vertebrate evolution. Phosphorylation and 14-3-3 binding have been shown previously to be strongly associated with disordered regions of proteins (40). Using the GlobPlot algorithm (41), we analyzed the disorder around the putative 14-3-3/AKT sites in our subset of proteins (supplemental Table S2). These data revealed that EDC3 Ser-161 lies in the middle of a disordered region of the protein between its N-terminal Lsm-like domain and its central FDF domain (16) (Fig. 1e).
Identification of EDC3 Phosphorylation Sites and Interacting Partners
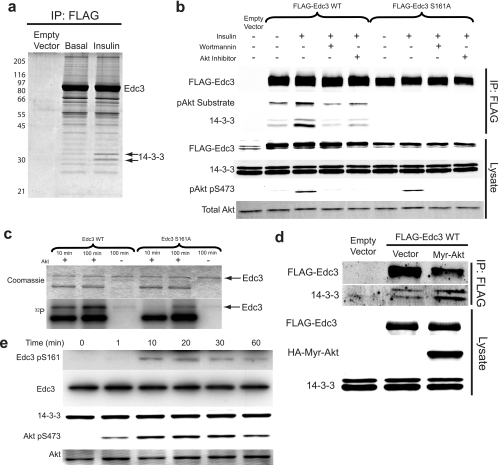
To identify phosphorylated 14-3-3 binding sites within EDC3, we expressed either TagRFP alone or a TagRFP-FLAG-tagged human EDC3 construct by transient transfection of CHO cells that stably express the insulin receptor and IRS-1 (CHO IR/IRS-1). The TagRFP-FLAG-EDC3 or vector control was FLAG-immunoprecipitated and subjected to SDS-PAGE and SYPRO Ruby staining. The most abundant band migrated at a position corresponding to the predicted molecular mass of TagRFP-FLAG-EDC3 (Fig. 2a), and several other bands with molecular masses of ∼55 and 68 kDa were also evident (Fig. 2a). Of note, two bands with an approximate molecular mass of 30 kDa showed increased abundance in the immunoprecipitate from insulin-stimulated cells and were not present in the vector control. Bands from each lane were excised and subjected to tryptic digestion and analysis by LC-MS/MS. These data revealed that the most abundant protein corresponded to human EDC3 containing five phosphopeptides, consistent with phosphorylation of EDC3 at Ser-161, Ser-131, Thr-173, Ser-109, and Tyr-225 (supplemental Table S3 and Fig. S2 for MS/MS spectra of phosphopeptides). Analysis of the entire gel lane from both the vector control and the TagRFP-FLAG-EDC3 immunoprecipitation revealed the presence of DCP1A/1B and DDX6, previously described EDC3 interactors, as well as six 14-3-3 isoforms within the 30-kDa bands from the EDC3 immunoprecipitates (supplemental Table S3). Together these data show that recombinant EDC3 co-immunoprecipitates with endogenous 14-3-3 isoforms in response to insulin in vivo and is phosphorylated at the putative 14-3-3 binding and AKT phosphorylation site Ser-161.
Fig. 2.
Analysis of EDC3 phosphorylation and 14-3-3 binding at Ser(P)-161. a, SDS-PAGE and SYPRO Ruby staining of immunoprecipitated (IP) TagRFP-FLAG-EDC3 WT or TagRFP vector alone transiently transfected into CHO IR/IRS-1 cells. Arrows indicate 14-3-3 identified by mass spectrometry; Edc3 marks the TagRFP-FLAG-EDC3 protein. b, immunoblotting of immunoprecipitated FLAG-EDC3 constructs from transiently transfected CHO IR/IRS-1 cells. Cells were either left unstimulated, stimulated with 100 nm insulin for 30 min, treated with the PI3K inhibitor wortmannin prior to insulin stimulation, or treated with AKTi prior to insulin stimulation. c, analysis of in vitro AKT phosphorylation of TagRFP-FLAG-EDC3 constructs in the presence of [γ-32P]ATP and active AKT2 kinase for the indicated times. Arrows indicate molecular weight of TagRFP-FLAG-EDC3. d, immunoblotting of immunoprecipitated FLAG-EDC3 WT from CHO IR/IRS-1 cells transiently transfected with the indicated constructs and serum-starved prior to analysis. e, immunoblotting of total cell lysates from NIH-3T3 fibroblasts that were stimulated with 1 μm insulin for the indicated times. Immunoblots and images are from a representative experiment (n = 3). pAkt, phospho-Akt; Myr, myristoylated.
EDC3 Ser(P)-161 Is a 14-3-3 Binding Site and Is Phosphorylated by Akt
To determine whether EDC3 Ser(P)-161 is indeed the 14-3-3 binding site, we generated an Ser-161 alanine mutant (S161A) in FLAG-tagged EDC3 and again analyzed FLAG immunoprecipitates from CHO IR/IRS-1 cells expressing either vector, FLAG-EDC3 WT, or FLAG-EDC3 S161A by transient transfection. Cells expressing these EDC3 constructs were either left unstimulated or stimulated with insulin. Additionally, some cells were pretreated with the PI3K inhibitor wortmannin or the specific AKT inhibitor 1/2 (AKTi). Subsequent immunoblotting of these immunoprecipitates revealed a number of important facets of EDC3 biochemistry (Fig. 2b). First, FLAG-EDC3 WT reacted with the phospho-AKT substrate antibody in an insulin-responsive and AKTi-sensitive manner, whereas the S161A mutant was not recognized by this antibody. Second, insulin increased the association between EDC3 and 14-3-3, and this effect was inhibited by wortmannin and AKTi, whereas no interaction was observed in the case of the EDC3 S161A mutant. Collectively, these data suggest that Ser-161 in EDC3 is phosphorylated in response to insulin principally by AKT and that this subsequently triggers 14-3-3 binding. To confirm that AKT phosphorylates EDC3 at Ser-161, we performed in vitro kinase assays using FLAG-EDC3 WT or the S161A mutant, constitutively active AKT2, and [γ-32P]ATP (Fig. 2c). Quantitation of 32P incorporation into EDC3 over two separate incubation time points revealed that AKT phosphorylated WT EDC3, but phosphorylation of the Ser-161 mutant was markedly impaired. Furthermore, we co-expressed EDC3 with either constitutively active myristoylated AKT or vector alone in CHO IR/IRS-1 cells and showed that EDC3 from myristoylated AKT-expressing cells displayed increased 14-3-3 binding in the unstimulated state (Fig. 2d). We next generated a phosphospecific antibody that recognized EDC3 Ser(P)-161 (supplemental Fig. S3). This antibody demonstrated that endogenous EDC3 was phosphorylated in vivo at Ser-161 and also showed a time-dependent increase in EDC3 Ser(P)-161 phosphorylation after insulin stimulation of NIH-3T3 cells (Fig. 2e).
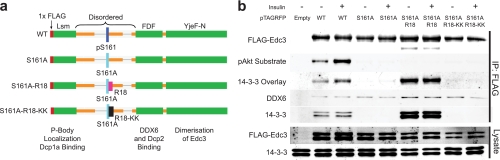
Expression of EDC3 Constructs with Altered 14-3-3 Binding Alters mRNA Processing Body Structure and Function
We next wanted to test the functional consequences of EDC3 phosphorylation and 14-3-3 binding. Previous studies have identified a small peptide motif (R18) that binds 14-3-3 strongly without requiring phosphorylation (42). We exploited this motif by cloning it into the EDC3 protein sequence three amino acids downstream of the S161A mutation (Fig. 3a). This generated a construct (S161A-R18) that could not be phosphorylated at Ser-161 but displayed constitutive 14-3-3 binding (Fig. 3b). We also made an additional construct that has a mutated R18 sequence that could not bind 14-3-3 (S161A-R18-KK) (27). To show that EDC3 WT binds 14-3-3 directly and that the S161A-R18 construct displays constitutive 14-3-3 binding, we used a bacterially produced GST-14-3-3β protein in a far-Western (overlay) analysis (Fig. 3b). These data showed that the S161A and S161A-R18-KK constructs did not bind 14-3-3, whereas the R18 construct bound ∼20–30 times more 14-3-3 than the insulin-treated WT construct. Furthermore, each EDC3 construct displayed 14-3-3 co-immunoprecipitation to the same degree as the 14-3-3 far-Western (overlay) analysis (Fig. 3b).
Fig. 3.
Generation of EDC3 constructs that constitutively bind 14-3-3. a, domain structure of FLAG-EDC3 constructs showing the location of the AKT phosphorylation and 14-3-3 binding site Ser(P)-161, the R18 or R18-KK peptide insertion site, and the known functions of EDC3 protein domains. b, immunoblotting of immunoprecipitated FLAG-EDC3 constructs from transiently transfected CHO IR/IRS-1 cells. Cells were either left unstimulated or stimulated with 100 nm insulin for 30 min. Immunoblots and images are from a representative experiment (n = 3). pAkt, phospho-Akt.
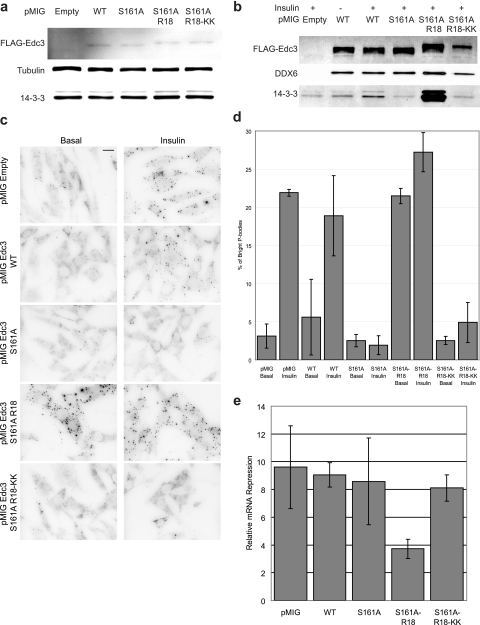
14-3-3 proteins often play roles in altering protein subcellular distribution and protein-protein interactions (43, 44). To qualitatively investigate whether phosphorylation of Ser-161 and subsequent 14-3-3 binding had an effect on p-body morphology, we transiently transfected NIH-3T3 cells with TagRFP-tagged constructs that expressed vector alone, EDC3 WT, S161A, S161A-R18, or S161A-R18-KK. The morphology of p-bodies within these cells was examined by immunofluorescence microscopy using antibodies specific for the endogenous p-body markers DDX6 or Hedls. Consistent with previous studies, p-bodies were evident as small puncta scattered randomly throughout the cytoplasm (supplemental Fig. S4). Insulin increased apparent p-body size/brightness and number substantially in both vector and EDC3 WT cells. This was not observed in cells expressing EDC3 S161A or S161A-R18-KK. Strikingly, cells expressing EDC3 S161A-R18 showed numerous large/bright p-body structures regardless of insulin stimulation. These data clearly indicate that insulin treatment and 14-3-3 binding to EDC3 modulate p-body morphology.
To quantify the functional consequences of 14-3-3 binding to EDC3, we generated a series of NIH-3T3 stable cell lines infected with retroviral vectors expressing FLAG-tagged EDC3 WT, S161A, S161A-R18, or S161A-R18-KK linked to IRES-GFP or an empty vector as control. These cell lines were sorted for GFP by fluorescence-activated cell sorting, enabling us to normalize for the expression of the different constructs (Fig. 4a), and displayed stable expression of these retroviral constructs for more than 10 passages. Analysis of FLAG immunoprecipitates from these cells showed that as expected the S161A and S161A-R18-KK constructs did not bind 14-3-3 and the R18 construct bound significantly more 14-3-3 than the insulin-treated WT construct (Fig. 4b). These data also showed that DDX6 did not change in its association with each of these constructs, indicating that 14-3-3 binding does not control the EDC3-DDX6 interaction. The morphology of p-bodies within each of these cell lines was examined by immunofluorescence microscopy (Fig. 4c) and found to be similar to that observed in transiently transfected cells. Quantitation of the intensity of the p-bodies in these images emphasized the marked increase in DDX6 staining intensity after insulin stimulation of vector and EDC3 WT stable cells (Fig. 4d). It also showed the total lack of response in EDC3 S161A and EDC3 S161A-R18-KK stable cells. In contrast, EDC3 S161A-R18 cells showed high intensity DDX6 p-body staining regardless of insulin stimulation.
Fig. 4.
Constitutive 14-3-3 binding to EDC3 alters p-body morphology and function. a, immunoblotting of total cell lysates showing FLAG-EDC3 constructs stably expressed in NIH-3T3 cells. b, immunoblotting of the immunoprecipitated FLAG-EDC3 constructs indicated stably expressed in NIH-3T3 cells. Cells were either left unstimulated or stimulated with 1 μm insulin for 30 min. c, immunofluorescence microscopy of endogenous DDX6 in NIH-3T3 cells stably expressing the indicated FLAG-EDC3 constructs. Cells were either left unstimulated or stimulated with 1 μm insulin for 30 min. The black scale bar in the top right panel indicates 10 μm. d, quantitation of staining intensity shown in c using the ImageJ Analyze Particles function. e, Dual-Luciferase assay for analysis of mRNA repression by the endogenous let-7 miRNA in NIH-3T3 cells stably expressing the indicated FLAG-EDC3 constructs. Values (arbitrary) indicate relative abilities of the let-7 miRNA to perform mRNA repression. Cells were incubated in the presence of serum after transfection and for 16 h prior to the Dual-Luciferase assay. Immunoblots and images are from a representative experiment (n = 3). Error bars indicate standard deviation.
To investigate whether the altered p-body morphology present in these cells correlated with altered miRNA function we used a Dual-Luciferase reporter assay to analyze repression by the let-7 miRNA (30, 45). Previous studies have shown that the let-7 miRNA is expressed in most cell types (46), and we exploited this by transiently transfecting plasmids expressing Renilla luciferase with either three WT or three mutant let-7 binding sites in its 3′-untranslated region. Only the mRNA containing the WT let-7 binding sites is a target for the let-7 miRNA. Each of these constructs was co-transfected with a firefly luciferase plasmid, which acted as a transfection control. By calculating the ratio of the firefly-corrected Renilla luciferase activity in cells transfected with the wild type let-7 binding plasmid or the mutant let-7 binding plasmid, we could evaluate the level of post-transcriptional regulation by let-7 in these cells. We found that the EDC3 S161A-R18 cells displayed an inhibition of this post-transcriptional regulation compared with the other stable cell lines tested (Fig. 4e). These data indicate that constitutive 14-3-3 binding to EDC3 is inhibitory for its role in miRNA function.
EDC3 Bound to 14-3-3 Displays a Distinct Protein Interaction Repertoire
To determine the mechanism by which 14-3-3 binding to EDC3 may alter its function, we comprehensively analyzed the interaction partners of EDC3 constructs either constitutively bound to 14-3-3 (S161A-R18) or deficient in 14-3-3 binding (S161A-R18-KK). This analysis was performed using SILAC (47) technology, which enabled quantitative analysis of regulated EDC3-interacting partners (supplemental Table S4 and Fig. S5). This analysis was performed by transient transfection into SILAC-labeled cells of either a vector-only negative control (light label), TagRFP-FLAG-EDC3 S161A-R18 (medium label), or TagRFP-FLAG-EDC3 S161A-R18-KK (heavy label) (supplemental Fig. S5). These studies revealed that EDC3 deficient in 14-3-3 binding co-immunoprecipitated ∼10-fold more poly(A)-binding protein 1 (PABP1), 7-fold more Y-box-binding protein 1, and ∼5-fold more IGF-2 mRNA-binding protein 1 compared with EDC3 with 14-3-3 constitutively bound (Table II and supplemental Table S4). Some of these interactions may be indirectly bridged by mRNA, and previous studies have shown that PABP or PABP-mRNA complexes may associate with various p-body components (48, 49). These observations are exciting because they may indicate that the interaction of EDC3 with 14-3-3 decreases the mRNA binding of the EDC3 complex. As expected the EDC3 S161A-R18 construct was able to co-immunoprecipitate 10-fold more 14-3-3 compared with the EDC3 S161A-R18-KK construct. The interaction between EDC3 and previously identified interaction partners such as DDX6 and DCP1A was not markedly altered between the two EDC3 constructs.
Table II. Differential protein-protein interactions of the EDC3 and 14-3-3 protein complex.
| UniProt accession no. | Protein name | Protein description | Average (KK/R18)a | S.D. (KK/R18) |
|---|---|---|---|---|
| P11940 | PABPC1 | Polyadenylate-binding protein 1 | 9.37 | 4.53 |
| P67809 | YBX1 | Nuclease-sensitive element-binding protein 1 | 6.55 | 2.62 |
| Q9NZI8 | IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 4.99 | 1.83 |
| P08107 | HSPA1A | Heat shock 70-kDa protein 1 | 2.56 | 0.37 |
| P11142 | HSPA8 | Heat shock cognate 71-kDa protein | 2.40 | 0.22 |
| P23396 | RPS3 | 40 S ribosomal protein S3 | 2.75 | 0.24 |
| P07900 | HSP90AA1 | Heat shock protein 90-kDa α | 2.38 | 0.39 |
| P08238 | HSP90AB1 | Heat shock protein HSP 90-β | 2.48 | 0.59 |
| P07437 | TUBB | Tubulin β chain | 1.67 | 0.20 |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | 1.49 | 0.27 |
| P26196 | DDX6 | Probable ATP-dependent RNA helicase DDX6 | 1.45 | 0.21 |
| Q9NPI6 | DCP1A | mRNA-decapping enzyme 1A | 1.54 | 0.48 |
| A8K674 | UBC | ubiquitin C | 0.94 | 0.36 |
| Q8IZD4 | DCP1B | mRNA-decapping enzyme 1B | 1.40 | 0.49 |
| P62258 | YWHAE | 14-3-3 protein ϵ | 0.13 | 0.12 |
| P31946 | YWHAB | 14-3-3 protein β/α | 0.13 | 0.13 |
| P61981 | YWHAG | 14-3-3 protein γ | 0.13 | 0.12 |
| Q04917 | YWHAH | 14-3-3 protein η | 0.12 | 0.12 |
| P27348 | YWHAQ | 14-3-3 protein θ | 0.11 | 0.11 |
| P63104 | YWHAZ | 14-3-3 protein ζ/δ | 0.12 | 0.12 |
a Average ratio of protein-protein interaction with EDC3 S161A-R18-KK (no 14-3-3 binding) compared with EDC3 S161A-R18 (14-3-3 constitutively bound).
DISCUSSION
In the present study, we used quantitative mass spectrometry methods based on SILAC to delineate additional elements of the insulin signaling pathway downstream of PI3K. In addition to well known AKT substrates, we identified several novel insulin-regulated proteins including EDC3, DNA methyltransferase 1, TRIP12, and Beclin. The identification of these phosphoproteins points to novel actions of insulin and other growth factors that activate PI3K/AKT, including regulation of autophagy (50), DNA methylation (51), and protein ubiquitylation (52). In this study, we further interrogated EDC3 and unveiled a novel action of insulin on mRNA post-transcriptional regulation, a pathway that plays an essential role in controlling protein expression in eukaryotic cells most notably via miRNAs (53–57). This discovery opens up new avenues of research in trying to identify the cellular basis of metabolic disease.
The canonical PI3K/AKT pathway plays an essential role in regulating numerous biological pathways downstream of tyrosine kinase growth factor receptors. In the context of insulin action, AKT has been shown to be critical in many if not most of the metabolic actions of insulin, including glucose transport and protein synthesis. Most of these pathways were mapped using a reverse discovery process whereby the metabolic actions of insulin were first identified and subsequently used as an end point to define the upstream biochemistry. Here, we describe a forward discovery process whereby identification of novel insulin-responsive 14-3-3-binding proteins can be used to define novel actions of insulin, and we predict that such an approach can be used to identify many more actions of this and other hormones. The approach takes advantage of the unusually high affinity of 14-3-3 proteins for AKT substrates and quantitative mass spectrometry using SILAC. The latter was a particularly important adaptation of the technique, allowing us to define from within the large repertoire of 14-3-3 proteins those that bind in an insulin- and PI3K-regulated manner. One of the important advantages of this technique compared with other techniques is that it overcomes the need to identify specific phosphopeptides in the mass spectrometer, which is limited by poor phosphopeptide separation, ionization, and fragmentation in LC-MS/MS (58, 59). To quantify proteins in this study, we were not dependent on the identification of specific peptides; rather any peptide from a target protein will be sufficient for generation of quantitative data for that protein. Furthermore, our technique can also allow the identification of insulin-responsive protein complexes where only one protein in the complex may bind to 14-3-3. Although the use of 14-3-3 preferentially selects for isolation of AGC family kinase substrates, we may overlook insulin-regulated phosphoproteins that do not interact with 14-3-3. Further studies using complementary technologies such as combined strong cation exchange/titanium dioxide chromatography or affinity chromatography using other phosphoprotein binding domains will be needed for a comprehensive analysis.
The observation that EDC3 is phosphorylated in response to insulin via the AGC kinase AKT is exciting because this reinforces the possibility that functions associated with p-bodies can be rapidly regulated (60). To control mRNA decay or translation repression, EDC3 binds to several core p-body components, and so it is well positioned as a key regulatory target (16–18, 62, 63). Intriguingly, other mRNA regulatory proteins such as BRF-1 (37, 64) and tristetraprolin (65) are also regulated by insulin and other growth factors via AKT-dependent phosphorylation, suggesting that this may represent a coordinated regulation of many components of the mRNA decay/repression process via AKT.
In this report, we show that AKT-dependent phosphorylation of EDC3 occurred within a disordered region between its Lsm and FDF domains. Phosphorylation at this site triggered 14-3-3 binding, and five observations suggest that this modulates EDC3 function. First, insulin treatment of cells significantly increased both the number and size of p-bodies. Second, the use of a 14-3-3 binding peptide (R18) engineered into EDC3 mediated constitutive 14-3-3 binding when expressed in cells, and this, like insulin, exaggerated the formation of large/bright p-bodies. Third, expression of an EDC3 mutant incapable of insulin-regulated phosphorylation and 14-3-3 binding in cells blocked this insulin effect. Fourth, 14-3-3 binding to EDC3 was associated with a significant inhibition of let-7 miRNA-induced mRNA repression. Finally, 14-3-3 binding to EDC3 resulted in a significant decrease in the binding of several key mRNA regulatory factors such as PABP and Y-box-binding protein 1 to EDC3. Collectively, these findings suggest that in the absence of insulin the mRNA decay/repression pathway is active and that this pathway corresponds with morphologically small p-bodies. Insulin may then inhibit this decay/repression pathway by phosphorylation of EDC3 and other proteins in the same pathway, including BRF-1, leading to 14-3-3 binding. Consistent with this, it has been shown that mRNA decay and miRNA-mediated mRNA repression can occur in the absence of visible p-body structures (66–70). It seems likely based on our data that 14-3-3 binding rather than phosphorylation per se is the key regulator of EDC3 function. We speculate based on the bivalent nature of 14-3-3 that the transition from active to inactive mRNA decay/repression is mediated by the displacement of EDC3 and probably other factors from functional p-bodies into larger aggregate structures (18, 61). This mechanism would suggest that insulin has the capacity to increase protein synthesis via multiple mechanisms: transcription, mRNA stability, mRNA accessibility to translation machinery, and translation efficiency.
Our studies have considerable implications for the regulation of protein synthesis in eukaryotic cells. In view of the tissue specificity of miRNA expression and growth factor regulation, one can envisage that this pathway might rapidly regulate the expression of a unique repertoire of proteins that play a unique role in a particular tissue. Thus, in the context of insulin action, this pathway may regulate the expression of proteins involved in the adipogenic program in adipocytes, whereas in other cell types, different growth factors may utilize this same pathway to regulate the expression of a unique subset of proteins via an alternate family of cell-specific miRNAs. This provides the basis for a combinatorial mechanism of cell-specific regulation involving this canonical AKT/p-body signaling pathway. Hence, it will be of interest to determine the impact of this pathway in disease states such as diabetes, cardiovascular disease, and cancer that at least in part are characterized by alterations in PI3K signaling.
Supplementary Material
Acknowledgments
We thank Jens Lykke Anderson for the EDC3 constructs and all those colleagues who provided antibodies used in this study. Mass spectrometric analysis for this work was carried out at the Bioanalytical Mass Spectrometry Facility, University of New South Wales.
* This work was supported in part by grants from the National Health and Medical Research Council of Australia and Diabetes Australia Research Trust (to D. E. J.). Mass spectrometric analysis was supported in part by grants from the Australian Government Systemic Infrastructure Initiative and Major National Research Facilities Program (University of New South Wales (UNSW) node of the Australian Proteome Analysis Facility) and by the UNSW Capital Grants Scheme.
 This article contains supplemental Tables S1–S4 and Figs. S1–S5.
This article contains supplemental Tables S1–S4 and Figs. S1–S5.
1 The abbreviations used are:
- PI3K
- phosphatidylinositol 3-kinase
- AGC
- cAMP-dependent protein kinase A/protein kinase G/protein kinase C
- AKTi
- AKT inhibitor 1/2
- CHO
- Chinese hamster ovary
- DMEM
- Dulbecco's modified Eagle's medium
- EDC3
- enhancer of mRNA decapping 3
- HRP
- horseradish peroxidase
- IR
- insulin receptor
- IRES
- internal ribosome entry site
- IRS-1
- insulin receptor substrate 1
- miRNA
- microRNA
- p-body
- processing body
- pMIG
- pMSCV-IRES-GFP
- GFP
- green fluorescent protein
- SILAC
- stable isotope labeling with amino acids in cell culture
- Trip12
- thyroid hormone receptor-interacting protein 12
- TSC2
- tuberous sclerosis protein 2
- WT
- wild type
- M
- medium
- L
- light
- H
- heavy
- FOXO
- Forkhead box protein O
- PABP
- poly(A)-binding protein.
REFERENCES
- 1.Cohen P. (2006) Timeline—the twentieth century struggle to decipher insulin signalling. Nat. Rev. Mol. Cell Biol 7, 867–873 [DOI] [PubMed] [Google Scholar]
- 2.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin-mediated by protein kinase B. Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 3.Rena G., Guo S., Cichy S. C., Unterman T. G., Cohen P. (1999) Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem 274, 17179–17183 [DOI] [PubMed] [Google Scholar]
- 4.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 5.Christ B., Nath A. (1993) The glucagon-insulin antagonism in the regulation of cytosolic protein binding to the 3′ end of phosphoenolpyruvate carboxykinase mRNA in cultured rat hepatocytes—possible involvement in the stabilization of the mRNA. Eur. J. Biochem 215, 541–547 [DOI] [PubMed] [Google Scholar]
- 6.Flores-Riveros J. R., McLenithan J. C., Ezaki O., Lane M. D. (1993) Insulin down-regulates expression of the insulin-responsive glucose transporter (Glut4) gene—effects on transcription and mRNA turnover. Proc. Natl. Acad. Sci. U.S.A 90, 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Brien R. M., Granner D. K. (1996) Regulation of gene expression by insulin. Physiol. Rev 76, 1109–1161 [DOI] [PubMed] [Google Scholar]
- 8.Okubo M., Villar-Palasi C., Nagasaka Y., Larner J., Larner A. C., Bai G., Lee E. Y. C. (1991) Long-term effects of insulin on the enzyme activity and messenger RNA of glycogen synthase in rat hepatoma H4 cells—an effect of insulin on glycogen synthase mRNA stability. Arch. Biochem. Biophys 288, 126–130 [DOI] [PubMed] [Google Scholar]
- 9.Woodcroft K. J., Hafner M. S., Novak R. F. (2002) Insulin signaling in the transcriptional and posttranscriptional regulation of CYP2E1 expression. Hepatology 35, 263–273 [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001) Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 [DOI] [PubMed] [Google Scholar]
- 11.Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 [DOI] [PubMed] [Google Scholar]
- 12.Lee R. C., Ambros V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 [DOI] [PubMed] [Google Scholar]
- 13.Franks T. M., Lykke-Andersen J. (2008) The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker R., Sheth U. (2007) P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 [DOI] [PubMed] [Google Scholar]
- 15.Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S. F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. (2007) Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21, 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J. (2005) Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20, 905–915 [DOI] [PubMed] [Google Scholar]
- 17.Tritschler F., Braun J. E., Eulalio A., Truffault V., Izaurralde E., Weichenrieder O. (2009) Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell 33, 661–668 [DOI] [PubMed] [Google Scholar]
- 18.Ling S. H., Decker C. J., Walsh M. A., She M., Parker R., Song H. (2008) Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol 28, 5965–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. (2005) A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning B. D., Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obata T., Yaffe M. B., Leparc G. G., Piro E. T., Maegawa H., Kashiwagi A., Kikkawa R., Cantley L. C. (2000) Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem 275, 36108–36115 [DOI] [PubMed] [Google Scholar]
- 22.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 23.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24.Larance M., Ramm G., Stöckli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. (2005) Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem 280, 37803–37813 [DOI] [PubMed] [Google Scholar]
- 25.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 26.Hoehn K. L., Hohnen-Behrens C., Cederberg A., Wu L. E., Turner N., Yuasa T., Ebina Y., James D. E. (2008) IRS1-independent defects define major nodes of insulin resistance. Cell Metab 7, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramm G., Larance M., Guilhaus M., James D. E. (2006) A role for 14-3-3 in insulin-stimulated GLUT4 translocation through its interaction with the RabGAP AS160. J. Biol. Chem 281, 29174–29180 [DOI] [PubMed] [Google Scholar]
- 28.Brummer T., Martin P., Herzog S., Misawa Y., Daly R. J., Reth M. (2006) Functional analysis of the regulatory requirements of B-Raf and the B-Raf(V600E) oncoprotein. Oncogene 25, 6262–6276 [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U. K. (1970) Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Clancy J. L., Nousch M., Humphreys D. T., Westman B. J., Beilharz T. H., Preiss T. (2007) Methods to analyze microRNA-mediated control of mRNA translation. Methods Enzymol 431, 83–111 [DOI] [PubMed] [Google Scholar]
- 31.Brummer T., Larance M., Herrera Abreu M. T., Lyons R. J., Timpson P., Emmerich C. H., Fleuren E. D., Lehrbach G. M., Schramek D., Guilhaus M., James D. E., Daly R. J. (2008) Phosphorylation-dependent binding of 14-3-3 terminates signalling by the Gab2 docking protein. EMBO J 27, 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villén J., Gygi S. P. (2006) Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 33.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 34.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 35.Kovacina K. S., Park G. Y., Bae S. S., Guzzetta A. W., Schaefer E., Birnbaum M. J., Roth R. A. (2003) Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem 278, 10189–10194 [DOI] [PubMed] [Google Scholar]
- 36.Lynch D. K., Daly R. J. (2002) PKB-mediated negative feedback tightly regulates mitogenic signalling via Gab2. EMBO J 21, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidlin M., Lu M., Leuenberger S. A., Stoecklin G., Mallaun M., Gross B., Gherzi R., Hess D., Hemmings B. A., Moroni C. (2004) The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J 23, 4760–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D. M., Ausiello G., Brannetti B., Costantini A., Ferrè F., Maselli V., Via A., Cesareni G., Diella F., Superti-Furga G., Wyrwicz L., Ramu C., McGuigan C., Gudavalli R., Letunic I., Bork P., Rychlewski L., Küster B., Helmer-Citterich M., Hunter W. N., Aasland R., Gibson T. J. (2003) ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 31, 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustos D. M., Iglesias A. A. (2006) Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins 63, 35–42 [DOI] [PubMed] [Google Scholar]
- 41.Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B., Yang H., Liu Y. C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999) Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 [DOI] [PubMed] [Google Scholar]
- 43.Mackintosh C. (2004) Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J 381, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaffe M. B. (2002) How do 14-3-3 proteins work?—gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett 513, 53–57 [DOI] [PubMed] [Google Scholar]
- 45.Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. (2005) Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 46.Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. (2006) Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol 172, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brengues M., Parker R. (2007) Accumulation of polyadenylated mRNA, Pab1, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P. (2007) Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol 179, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 51.Bestor T., Laudano A., Mattaliano R., Ingram V. (1988) Cloning and sequencing of a cDNA-encoding DNA methyltransferase of mouse cells— the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol 203, 971–983 [DOI] [PubMed] [Google Scholar]
- 52.Park Y., Yoon S. K., Yoon J. B. (2008) TRIP12 functions as an E3 ubiquitin ligase of APP-BP1. Biochem. Biophys. Res. Commun 374, 294–298 [DOI] [PubMed] [Google Scholar]
- 53.Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 55.Flynt A. S., Lai E. C. (2008) Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet 9, 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meister G. (2007) miRNAs get an early start on translational silencing. Cell 131, 25–28 [DOI] [PubMed] [Google Scholar]
- 57.Pillai R. S., Bhattacharyya S. N., Filipowicz W. (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends. Cell Biol 17, 118–126 [DOI] [PubMed] [Google Scholar]
- 58.Aebersold R., Goodlett D. R. (2001) Mass spectrometry in proteomics. Chem. Rev 101, 269–295 [DOI] [PubMed] [Google Scholar]
- 59.Mann M., Ong S. E., Grønborg M., Steen H., Jensen O. N., Pandey A. (2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 20, 261–268 [DOI] [PubMed] [Google Scholar]
- 60.Eulalio A., Behm-Ansmant I., Izaurralde E. (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol 8, 9–22 [DOI] [PubMed] [Google Scholar]
- 61.Aizer A., Brody Y., Ler L. W., Sonenberg N., Singer R. H., Shav-Tal Y. (2008) The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19, 4154–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tritschler F., Eulalio A., Helms S., Schmidt S., Coles M., Weichenrieder O., Izaurralde E., Truffault V. (2008) Similar modes of interaction enable trailer hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell. Biol 28, 6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tritschler F., Eulalio A., Truffault V., Hartmann M. D., Helms S., Schmidt S., Coles M., Izaurralde E., Weichenrieder O. (2007) A divergent Sm fold in EDC3 proteins mediates DCPI binding and P-body targeting. Mol. Cell. Biol 27, 8600–8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamin D., Schmidlin M., Min L., Gross B., Moroni C. (2006) BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol. Cell. Biol 26, 9497–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marderosian M., Sharma A., Funk A. P., Vartanian R., Masri J., Jo O. D., Gera J. F. (2006) Tristetraprolin regulates Cyclin D1 and c-myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene 25, 6277–6290 [DOI] [PubMed] [Google Scholar]
- 66.Chu C. Y., Rana T. M. (2006) Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4, e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stalder L., Mühlemann O. (2009) Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Decker C. J., Teixeira D., Parker R. (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol 179, 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. (2007) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol 27, 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoecklin G., Mayo T., Anderson P. (2006) ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep 7, 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.