Abstract
To enable the first physiologically relevant peptidomic survey of gastrointestinal tissue, we have developed a primary mouse colonic crypt model enriched for enteroendocrine L-cells. The cells in this model were phenotypically profiled using PCR-based techniques and showed peptide hormone and secretory and processing marker expression at mRNA levels that were increased relative to the parent tissue. Co-localization of glucagon-like peptide-1 and peptide YY, a characteristic feature of L-cells, was demonstrated by double label immunocytochemistry. The L-cells displayed regulated hormone secretion in response to physiological and pharmacological stimuli as measured by immunoassay. Using a high resolution mass spectrometry-based platform, more than 50 endogenous peptides (<16 kDa), including all known major hormones, were identified a priori. The influence of culture conditions on peptide relative abundance and post-translational modification was characterized. The relative abundance of secreted peptides in the presence/absence of the stimulant forskolin was measured by label-free quantification. All peptides exhibiting a statistically significant increase in relative concentration in the culture media were derived from prohormones, consistent with a cAMP-coupled response. The only peptides that exhibited a statistically significant decrease in secretion on forskolin stimulation were derived from annexin A1 and calcyclin. Biophysical interactions between annexin A1 and calcyclin have been reported very recently and may have functional consequences. This work represents the first step in characterizing physiologically relevant peptidomic secretion of gastrointestinally derived primary cells and will aid in elucidating new endocrine function.
The gut is one of the largest endocrine systems, secreting more than 30 peptide hormones that serve vital roles in glucose metabolism and energy homeostasis. At least 15 endocrine cell types can be found in the gut, yet they represent less than 1% of the total cellular population (1, 2). The colon is known to be one of the richest sections of the gut for PYY1/GLP-1-producing L-cells (3, 4). These peptide hormones play important roles in the modulation of feeding behavior, satiety, and body weight, and GLP-1 analogs and mimics have emerged as an important drug class for the treatment of diabetes (5). Until recently, the elucidation of nutrient-sensing mechanisms involved in regulated peptide hormone secretion has been hindered by the lack of availability of suitable in vitro models. Existing cell lines developed from fetal tissue or intestinal tumors display a poorly differentiated phenotype (6) or dysregulated prohormone gene expression (7). As the endocrine content of the gut changes significantly during development, primary cultures derived from fetal tissues may not accurately reflect the physiology of adult colon (8).
Recent studies have significantly advanced our understanding of factors influencing tissue growth and differentiation in normal intestinal epithelia. In separate studies, two laboratories recently manipulated Wnt signaling of crypt proliferation to produce robust long term cultures (9, 10). In addition, conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice were recently established (11). The caveat to these studies is that the characterization of the endocrine content of these cultures is limited to immunocytochemical methods.
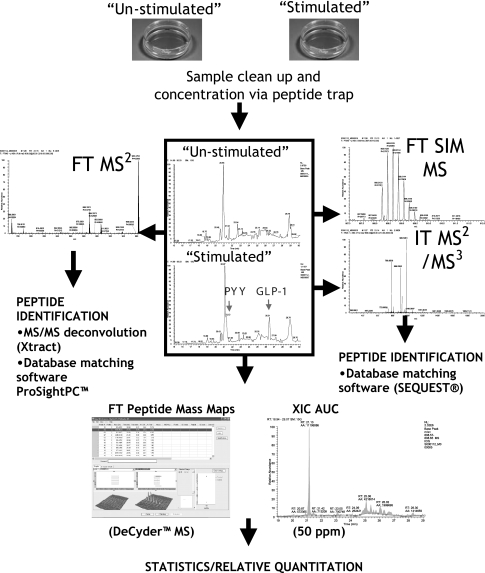
Our focus is on identification of secreted products from colonic endocrine cells in response to physiological and pharmacological stimulants. To enable these studies, we have developed and characterized a primary in vitro culture system enriched in endocrine cells representative of normal adult colonic epithelia. Our approach involves analysis of cell media samples by mass spectrometry with no cellular lysis, resulting in cleaner samples with reduced background from intracellular proteins (Scheme 1) (12). Secreted products were desalted, concentrated, and directly analyzed by high resolution LC/MS. Minimal sample manipulation significantly reduced adsorptive loss of low abundance peptides. Peptide identity was established through a variety of gas phase fragmentation techniques from which amino acid sequence was derived by MS informatics. The use of mass maps for relative quantification allowed us to see differences in peptide profiles reflective of cellular state (e.g. stimulated or unstimulated). The methodology contrasts with laborious “grind and find” approaches where large amounts of source material are processed for the identification of bioactive peptides. By adopting a “peptidomic” (13)/“top-down” (14) approach rather than proteomic methods that use trypsin digestion (15), we capture and characterize full-length endogenous peptides (<16 kDa) rather than proteolytic fragments for identification (12, 16). In addition, our platform captures the influence of culture conditions on peptide post-translational modifications that are critical for biological function.
Scheme 1.
Work flow for analysis of secreted peptides by nanoflow chromatography coupled to hybrid ion trap-Fourier transform mass spectrometer (modified from Ref. 12). XIC, extracted ion chromatogram; AUC, area under the curve.
EXPERIMENTAL PROCEDURES
Crypt Cell Isolation and Culture
All animal procedures were approved by the Institutional Animal Care and Use Committee at Amylin Pharmaceuticals, Inc. in accordance with Animal Welfare Act guidelines. Colonic crypts were isolated by sequential enzymatic and mechanical dispersion using a modification of the method of Booth and O'Shea (17). Colons were rapidly removed from 1-month-old C57BL/6 mice, washed in Ca2+- and Mg2+-free Hanks' balanced salt solution and then rinsed in DMEM supplemented with 5% fetal calf serum and antibiotics. The tissue was digested in 15 ml of DMEM with a mixture of enzymes consisting of type 1 collagenase (790 units/ml), hyaluronidase (800 units/mg), and DNase-I (5 units/mg) for 15 min at 37 °C in 5%CO2. The remaining tissue was further digested in the enzyme mixture consisting of type IV collagenase (107 units/ml) and dispase neutral protease (8 units/mg) for two 15-min intervals. Following mechanical disruption, the tissue was allowed to sediment in DMEM containing 2% sorbitol, and the supernatant containing the purified crypt fraction was collected by centrifugation at 250 × g at room temperature. The crypts were then plated on collagen 1-coated plates (BD Biosciences) in the growth media consisting of 50% DMEM/F-12 supplemented with 1 μg/ml hydrocortisone, 8 μg/ml insulin, 100 μg/ml gentamicin, and 10% FCS and 50% specialized bronchial epithelial growth medium (Lonza, Walkersville, MD) and incubated at 37 °C in CO2 (5%). In some cases, this medium was also supplemented with 35 or 350 μm ascorbate (Sigma).
Gene Profiling of Colon Tissue, Isolated Crypts, and Cultures by Real Time RT-PCR Low Density Array
Control tissue was harvested and then placed in RNAlater® (Ambion/Applied Biosystems) according to the manufacturer's protocol and stored at 4 °C. RNA was then isolated from tissue using TRI Reagent® according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). RNA was also prepared from isolated crypt cells that were cultured for 2 days in a 6-well tissue culture plate. Cells were lysed in 350 μl of RNeasy Lysis Buffer containing 143 mm β-mercaptoethanol. Lysates were passed over a QIAshredder column, and then total RNA was purified using RNeasy® mini columns according to the manufacturer's protocol (Qiagen Inc., Valencia, CA). Reverse transcription was carried out on 1 μg of RNA using a High Capacity cDNA Archive kit (Applied Biosystems) according to the manufacturer's protocol. PCRs were then prepared by adding 2× TaqMan Universal PCR Master Mix (Applied Biosystems) to each cDNA reaction and loaded onto a custom TaqMan® low density array (LDA) microfluidic card (Applied Biosystems). PCR amplification was performed using an ABI Prism® 7900HT Fast Real-Time PCR System (Applied Biosystems). Colon was used as the calibrator sample, 18 S was used as the endogenous control gene, and each primer set was present in duplicate on the LDA. Quantification of gene expression relative to the calibrator sample was then done using the comparative CT method as described in the manufacturer's manual.
Immunofluorescence Staining and Quantification of Double Labeled Enteroendocrine Cells
Cells were fixed in 10% buffered formalin and stored at 4 °C before staining and analysis. Primary antibodies used were rabbit anti-chromogranin A and B (Covance), rabbit anti-GLP-1 (Peninsula Laboratories, Inc.), and rabbit anti-PYY (Peninsula Laboratories, Inc.), and nuclei were stained with 4′,6-diamidino-2-phenylindole, dilactate (Molecular Probes, Carlsbad, CA). The secondary antibody was a donkey anti-rabbit rhodamine red X-conjugated (Jackson ImmunoResearch Laboratories, West Grove, PA). PYY was detected by direct immunofluorescence using primary antibody labeled using the Zenon® One Alexa Fluor® 488 kit (Molecular Probes).
Analysis of GLP-1 and PYY Secretion by Immunoassays
Colonic crypt cells were isolated and cultured for 48 h as described above. They were then washed four times with release/stimulation medium consisting of RPMI 1640 medium supplemented with 17.5 mm glucose, 55 mg/liter sodium pyruvate, and 15 mm HEPES. For secretion studies, the cultures were either stimulated with 10 μm forskolin or other stimuli as detailed under “Results” and incubated at 37 °C at 5% CO2 for different time periods. Media samples were then collected into tubes with dipeptidyl peptidase IV and protease inhibitors for immunoassay. GLP-1 concentrations were measured using a GLP-1 (active) ELISA (Millipore/LINCO Research, St. Charles, MO). PYY was measured by enzyme immunoassay (Bachem, Peninsula Laboratories).
Sample Collection and Preparation for MS Analysis
After 48 h in culture, colonic crypt cells were washed four times with the release/stimulation medium described above. Cells were stimulated by addition of 10 μm forskolin and incubation at 37 °C at 5% CO2 for different time periods as described under “Results.” To reduce stimulant background, in a subset of samples, cells were treated in a “pulse-chase” format with 10 μm forskolin or 0.5 μm phorbol 12-myristate 13-acetate (PMA) stimulation for 5 min and fresh release medium (no stimulation factors) replacement, and cell media were collected after incubation for 90, 120, or 180 min at 37 °C in 5% CO2 (16). Media samples were collected as 900-μl aliquots in prewashed Lo-Bind tubes (Eppendorf) and diluted with ACN, formic acid, and TFA in water to a final concentration of 25% acetonitrile, 2.5% formic acid, and 0.1% TFA, respectively. Concentration and desalting of 1–2-ml aliquots of colonic crypt media/ACN/TFA/formic acid samples by solid phase extraction was performed using a peptide MicroTrap (Thermo Fisher Scientific Inc.) as described previously (16).
MALDI-TOF MS Analysis
MALDI-TOF MS was performed using a Voyager-DE STR instrument (Applied Biosystems) in positive ion linear mode. Colonic crypt samples were applied to a 100-well stainless steel MALDI target plate. For spotting on the target, 0.5 μl of sample (post-solid phase extraction, predilution) was mixed with 0.5 μl of a 1:1 mixture of α-cyano-4-hydroxycinnamic acid and 2,5-dihydroxybenzoic acid matrix solution, both dissolved at 20 mg/ml, directly on the target. Instrument settings were as follows: positive ion linear mode, manual control, accelerating voltage of 25,000, grid percentage of 92.5, delay time of 700 ns, 500 shots per spectrum, and mass range of 500–20,000 Da with a low mass gate of 500 Da. For reflectron mode, the settings were as follows: accelerating voltage of 20,000, grid percentage of 66.0, delay time of 250 ns, 500 shots per spectrum, and mass range of 1000–7001 Da with a low mass gate of 800 Da. Postacquisition data analysis was performed using Data Explorer, v.4.0 (Applied Biosystems). MALDI spectra were base line-corrected and noise-filtered/smoothed using a correlation factor of 0.5–0.7.
LC/MS Analysis
Samples for DeCyderMS Analysis
The samples (basal versus forskolin-stimulated cell culture media) for relative quantification were resolved by reversed phase chromatography nano-LC instrument (Paradigm MS4B, Michrom Bioresources Inc.) coupled to a 7-tesla LTQ-FTTM (Thermo Fisher Scientific Inc.) hybrid mass spectrometer operated using the Xcalibur 1.4 SR1/Tune Plus 1.1 operating system. The electrospray source was fitted with a NanoTrap interface (Michrom Bioresources Inc.). As previously described in detail (12, 16), 50 μl of sample in 30% acetonitrile, 0.1% TFA was loaded via an Endurance autosampler (Michrom Bioresources Inc.) onto the Michrom Peptide NanoTrap at 50 μl/min for 5 min and eluted with a linear gradient from 5% acetonitrile, 0.1% formic acid to 60% acetonitrile, 0.1% formic acid over 40 min at 500 nl/min from a 16-cm, 75-μm-inner diameter PicoFrit column (New Objective) packed with Magic C8 (Michrom Bioresources Inc.).
MS analysis was performed using a 7-Tesla LTQ-FT hybrid linear ion trap-ion cyclotron resonance mass spectrometer (Thermo Fisher Scientific Inc.). Instrument settings were as follows: capillary temperature was 175 °C, and source voltage was 2.3 kV. IT MS/MS spectra were collected in centroid mode, and full-scan FT spectra and MS/MS spectra were collected in profile mode. A data-dependent selected ion monitoring (SIM) duty cycle was used (18) supplemented with IT MS3 and FT MS2 scans with dynamic exclusion enabled (16). Automatic gain control was 5,000,000 for the FT MS full scan (400–1800 m/z range), 100,000 for the FT SIM scans (m/z window = 5), and 500,000 for FT MSn scans (200–1800 m/z range). The duty cycle consisted of a full-scan FT MS spectrum at 50,000 resolution, an FT SIM scan at 50,000 resolution (largest peak in the FT MS spectrum), an IT MS2 spectrum (largest peak in the FT spectrum), an IT MS3 spectrum (largest peak in the IT MS2), an FT SIM scan at 50,000 resolution (second largest peak in the FT MS spectrum), an IT MS2 spectrum (second largest peak in the FT spectrum), an IT MS3 spectrum (largest peak in the second IT MS2), and an FT MS2 spectrum (largest peak in the FT MS spectrum) at 100,000 resolution. The minimum signal intensity was 1, MS2 isolation width m/z window was 2, MS normalized collision energy was 35%, charge state 1 was rejected, exclusion and reject mass widths were 10 ppm, the repeat count was 1 with a 30-s duration, the exclusion duration was 15 s, the FT master scan was previewed, and the monoisotopic toggle was enabled.
Methods for Expanding Peptide Identification
Variations on the above methods to obtain more peptide identifications from these samples included (a) substitution of the column stationary phase to Jupiter C5 (Phenomenex) material, (b) starting the gradient at 10% mobile phase, (c) increasing the automatic gain control for SIM scans to 500,000 and FT MS2 scans to 1,000,000, (d) increasing the MSn isolation width m/z window to 5, and (e) disabling the monoisotopic toggle and increasing the exclusion window settings to 0.8 Da low and 2.2 Da high. Data acquired with these modified LC/MS methods were not included in the statistical analysis.
Peptide Identification
Ion trap and FT fragmentation experiments were analyzed by SEQUEST v.28 (revision 12) running on an eight-node Linux cluster with a Bioworks 3.3.1 SP1 (Thermo Fisher Scientific Inc.) interface. Searches were performed specifying monoisotopic precursor and fragment masses. Peak lists were generated using extract_msn version 4.0 specifying a group scan of 1, minimum ion threshold of 5, and intensity threshold of 100. Depending on whether the monoisotopic toggle feature was enabled, peptide mass tolerances were set at either 2 ppm on the precursor (enabled) or 2.5 Da (disabled) and 1.0 Da on the fragments. Pyroglutamate (−17.0266 Da on Gln), methionine oxidation (+15.9949 Da), and N-terminal acetylation (+42.0106 Da) were specified as variable modifications as was amidation (−0.9840 Da) (monoisotopic toggle enabled) or b ion fragmentation (−18.0106 Da) (monoisotopic toggle disabled) on the C terminus for MS3 data. The March 24, 2009 release of the GenBankTM “non-redundant” protein database was downloaded in FASTA format from the National Center for Biotechnology Information (NCBI) ftp site. An in-house Perl script was used to select sequences with mouse annotations from this file and, additionally, to remove all protein sequences containing nonspecific “X” residues. The edited FASTA-formatted file contained a total of 130,122 sequences. No enzyme was specified in searching this database for 600–7500-Da peptides (monoisotopic toggle enabled) or 600–7000-Da peptides (monoisotopic toggle disabled). Candidate peptides satisfying both criteria of Xcorr >1.5 (1+), >2.0 (2+), or >2.9 (≥3+) and peptide probability p < 1e−4 (rigorous searches) were automatically accepted as reverse database searches (19) on the same files indicated a 0% false discovery rate using these parameters. Similarly, peptides and gas phase fragments from MS3 experiments were accepted if they satisfied both the same Xcorr criteria and p < 1e−5 (tolerant searches) after a 0% false discovery rate was found using these parameters on the same data files generated with the monoisotopic toggle disabled and analyzed with a reverse database.
FT fragmentation experiments were also analyzed using ProSightPC v2.0 RC 0.9.1 (Thermo Fisher Scientific Inc.) operated in high throughput mode (20). Deconvolution of FT MS2 data and generation of monoisotopic mass lists were achieved using the Xtract algorithm. The precursor selection criteria was based on the peak of highest intensity, and multiple precursors were allowed with a relative precursor threshold of 10%. Other options were default, including a minimum signal/noise threshold of 3 and maximum charge of 40 on the precursor and fragments, respectively. Minimum fit and remainder thresholds were 40 and 20 for the precursor and 10 and 10 for the fragments, respectively. The minimum base peak fragmentation intensity was set at 1000 with the option to add the remainder afterward enabled. Deconvoluted FT MS2 experiments were filtered for a minimum intact mass of 1000 Da and 10 fragments and subject to a search tree involving (a) an absolute mass search and (b) a biomarker search against the same non-redundant database used in the SEQUEST searches (reformatted for ProSight). For absolute mass searches, the precursor mass and fragment masses were specified as monoisotopic with a 10-kDa precursor window, a fragment tolerance of 10 ppm in ΔM mode, and the high priority PTM acetylation specified as a variable modification. Preliminary hit filtering involved a maximum of 25 sequences with a minimum of three matching fragments being returned. Searches resulting in an E <1e−4 were deemed a preliminary success and were automatically loaded into a peptide repository. Searches not satisfying this criterion were further analyzed in biomarker mode. Conditions were the same as the absolute mass mode except that ΔM mode was disabled, and a 10-ppm tolerance was set for the precursor. All raw files were searched in this way using forward and reverse databases. It was found that an E <1e−7 corresponded to a 0% false discovery rate. In absolute mass mode, preliminary peptide sequence identifications had to be reconciled with their intact masses because this mode of searching detects uniform mass shifts between calculated and observed mass resulting from extensions, truncations, substitutions, or modifications. This was done through subsequent individual searches in biomarker, biomarker ΔM mode, or single protein searches with a proposed sequence. Peptides identified above the 0% false discovery rate thresholds for either SEQUEST or ProSight searches were defined as satisfying Criterion 1.
Peptides not satisfying the false discovery rate thresholds for SEQUEST but with otherwise compelling evidence for a correct identification were also accepted (Criterion 2) with supporting information supplied in the supplemental material. This evidence included (a) precursor mass accuracy <2 ppm combined with (b) high resolution/accuracy FT MS2 peaks assigned to consecutive b or y ion fragments or IT MS3 fragmentation confirming the sequence of a gas phase fragment. Finally, peptide sequences were subjected to protein BLAST searches against UniProt and NCBI databases (21), and redundant Swiss-Prot and single GenBank accession numbers are provided in the supplemental material for those peptides with 100% homology. The GenBank accession number cross-referenced to the Swiss Prot accession number was preferentially chosen.
Relative Quantification of Peptides
Detection, intensity map comparison, and quantification were performed using DeCyderMS v1.0 (GE Healthcare) differential analysis software (22). FT full-scan precursor mass spectra were analyzed in fully automatic mode. Settings for peak detection were as follows: a uniform model; automatic background-subtracted quantification; LC peak shape tolerance at 20%; m/z shift tolerance at 0; m/z shape tolerance at 5%; remove overlapping peptides; time range, 0–50.0 min; and m/z range, 400–1800. The typical peak elution time was set from 0.65 min with an m/z tolerance set at 0.05 Da, and resolution settings were 50,000 at m/z 400 and 15,000 at m/z 1400. The LC/MS profiles corresponding to media collected from 14 basal and 20 stimulated colonic crypts run over a period of about 5 weeks on a Magic C8 (Michrom Bioresources Inc.) column were compared. Matching of signals across profiles was facilitated by a low mass tolerance (0.05 Da) combined with the high resolution of the isotopic envelopes afforded by the FT MS for accurate charge state determination. Statistics, including group to group comparisons, were generated using the DeCyderMS statistical toolset. Intensity information for the various charge states is expressed as log2 (analogous to gene expression arrays), and a two-tailed Student's t test was applied to measure statistical differences between the control and forskolin-stimulated group of LC/MS runs. Average ratios (forskolin/basal secretion) were calculated from actual intensities without taking the logarithms (see supplemental information). No normalization of peak intensities was performed as described previously (12). Only known peptides matched in three or more profiles or unknown masses matched in six or more profiles were included (see supplemental information). Other disparate masses were excluded as they corresponded to incorrectly assigned monoisotopic masses or charge states (e.g. overlapping isotope envelopes) or infrequently observed features. Forskolin-related features were also excluded from the analysis as described previously (12). All masses were manually inspected for goodness of fit to the calculated isotopic distribution pattern. As DeCyderMS v1.0 has a charge state limitation of 15+, several masses were misassigned and corrected after deconvolution using Xtract in the Xcalibur software suite (Thermo Fisher Scientific Inc.). The technical reproducibility of this label-free method has been assessed in previous publications (12, 22).
RESULTS
Characterization of Colonic Crypt Cultures
Viable colonic crypts were successfully isolated from adult mice using a combination of enzymatic digestion and mechanical dispersion. Crypts rapidly attached to collagen (type 1)-coated plates, and after 3 h in culture, epithelial cells began to migrate outward, forming colonies of polygonal cells arranged in a typical cobblestone pattern. To prevent fibroblast overgrowth and induce endocrine differentiation, the medium was changed to serum-free bronchial epithelial growth medium. To test whether endocrine enrichment was achieved, 2-day cultures were analyzed by real time RT-PCR and immunocytochemical staining. Freshly isolated crypts showed an increase in relative mRNA expression of the glucagon gene compared with whole tissue. Cultured crypts were further enriched in mRNA levels of neuroendocrine peptides and peptide processing and secretory machinery (Table I).
Table I. Enrichment of endocrine expression profile in colonic crypt cultures.
mRNA expression of neuroendocrine peptides and proteins involved in peptide processing and secretion was measured by real time PCR using a custom TaqMan LDA microfluidic card (Applied Biosystems). GNAS, Guanine Nucleotide Binding Protein, alpha stimulating; PAM, peptidyl-alpha-amidating monooxygenase; CART, cocaine-amphetamine regulated transcript; NPY, neuropeptide Y; NMU, neuromedin U; AgRP, agouti-related protein; PC, prohormone convertase; SNAP25, synaptosomal-associated protein 25.
| PCR-based TaqMan LDA |
Relative quantity |
||||
|---|---|---|---|---|---|
| Gene name | Gene symbol | Assay ID | Colon tissue | Isolated crypts | Crypt cultures |
| Secretory machinery | |||||
| Chromogranin A | Chga | Mm00514341_m1 | 1 | 1.5 | 3.4 |
| Chromogranin B | Chgb | Mm00483287_m1 | 1 | 1.3 | 4.9 |
| GNAS | Gnas | Mm00456660_m1 | 1 | 1.1 | 4.5 |
| Secretogranin II | Scg2 | Mm00843883_s1 | 1 | 0.8 | 2.6 |
| Secretogranin III | Scg3 | Mm00485961_m1 | 1 | 1.0 | 1.8 |
| SNAP-25 | Snap25 | Mm00456921_m1 | 1 | 0.7 | 1.3 |
| Syntaxin 1A | Stx1a | Mm00444008_m1 | 1 | 0.7 | 7.8 |
| Synaptophysin | Syp | Mm00436850_m1 | 1 | 0.8 | 1.4 |
| Peptide processing | |||||
| Furin | Furin | Mm00440646_m1 | 1 | 0.7 | 3.7 |
| PAM | Pam | Mm00449686_m1 | 1 | 0.6 | 1.4 |
| PC 1/3 | Pcsk1 | Mm00479023_m1 | 1 | 1.1 | 2.3 |
| PC 2 | Pcsk2 | Mm00500981_m1 | 1 | 0.6 | 1.6 |
| PC 4 | Pcsk4 | Mm00801899_m1 | 1 | 0.3 | 0.0 |
| PC 7 | Pcsk7 | Mm00476621_m1 | 1 | 0.8 | 0.6 |
| PC 9 | Pcsk9 | Mm00463738_m1 | 1 | 11.0 | 12.0 |
| Pro-SAAS | Pcsk1n | Mm00457410_m1 | 1 | 0.9 | 2.3 |
| Neuroendocrine peptides | |||||
| AgRP | Agrp | Mm00475829_g1 | 1 | 2.3 | 3.1 |
| CART | Cart | Mm00489086_m1 | 1 | 3.0 | 26.9 |
| Glucagon | Gcg | Mm00801712_m1 | 1 | 2.0 | 26.4 |
| NPY | Npy | Mm00445771_m1 | 1 | 1.4 | 5.8 |
| NMU | Nmu | Mm00479868_m1 | 1 | 0.8 | 1.1 |
| Resp18 | Resp18 | Mm00485697_m1 | 1 | 0.9 | 1.7 |
| Somatostatin | Sst | Mm00436671_m1 | 1 | 1.9 | 2.6 |
| VIP | Vip | Mm00660234_m1 | 1 | 0.6 | 1.4 |
A granular pattern of chromogranin (A and B), PYY, and GLP-1 expression was observed by immunofluorescence (Fig. 1, a, b, and c), consistent with their localization in the secretory granules. Morphometric analysis of chromogranin (A and B)-immunoreactive cells showed that enteroendocrine cells comprise ∼7% of the total cell population. GLP-1 and PYY immunoreactivity was primarily co-localized within a subpopulation of chromogranin (A and B)-immunoreactive cells consistent with L-cell expression (23) (Fig. 1).
Fig. 1.
Representative images (40× objective) of chromogranin (Chg) AB, PYY, and GLP-1 immunoreactivities in colonic crypt cultures. a–c, granular pattern of ChgAB (a), GLP-1 (b), and PYY (c) immunoreactivities. d–f, double immunofluorescent staining for PYY (green) and ChgAB (red) showing partial co-localization: d, ChgAB-positive cells; e, two PYY-positive cells in the same field of view; f, multiple exposure photomicrograph revealing two yellow doubly labeled ChgAB/PYY cells. g–i, double immunofluorescent staining for PYY (green) and GLP-1 (red) showing co-localization: g, GLP-1-positive cells; h, PYY-positive cells in the same field of view; i, multiple exposure photomicrograph revealing all doubly labeled GLP-1/PYY cells except one. To perform double labeling, indirect staining with GLP-1 or chromogranin (A and B) antibodies and secondary antibody conjugated to rhodamine red (red) was followed by PYY detection using Zenon Alexa Fluor 488-labeled rabbit anti-PYY antibody (green) and counterstained for 4′,6-diamidino-2-phenylindole, dilactate (blue, nucleus). The scale bar for a, b, and c is 10 μm, and for d–i, the scale bar is 20 μm.
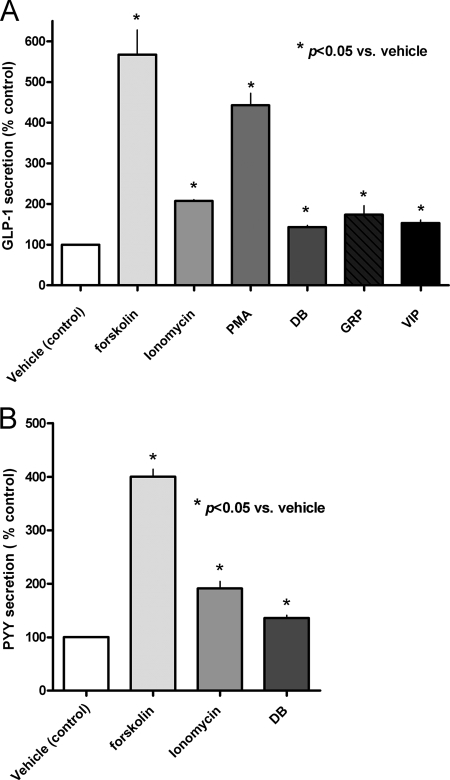
To further characterize endocrine cell function in the 2-day colonic crypt cultures, we measured GLP-1 concentrations in media by immunoassay. The responsiveness of the cells to pharmacological and physiological stimuli was evaluated. GLP-1 secretion was stimulated by forskolin, an activator of adenylate cyclase. Increased GLP-1 secretion was also observed following stimulation with PMA, an activator of protein kinase C signaling, and ionomycin, a calcium ionophore (Fig. 2A). GLP-1 secretion was also increased by stimulation with agonists of physiologically relevant G-protein-coupled receptors such as VIP, gastrin-releasing peptide, and denatonium benzoate (DB) (Fig. 2A). Incubation of cells with forskolin, ionomycin, and DB resulted in a similar stimulation of PYY secretion (Fig. 2B).
Fig. 2.
GLP-1 and PYY secretion from colon crypt cultures in response to variety of pharmacological and physiological stimuli. A, stimulation of GLP-1 secretion in response to 1-h incubation with medium alone (vehicle control), 10 μm forskolin (n = 3), 1 μm ionomycin (n = 3), or 1 μm PMA (n = 3). One-hour incubation with 40 nm VIP (n = 3) or 100 nm gastrin-releasing peptide (GRP) (n = 4) or 2-h incubation with 5 mm DB (n = 3) resulted in about 50% stimulation of GLP-1 secretion. GLP-1 secretion is expressed as a percentage of the control. *, p < 0.05 versus control. B, PYY secretion in response to 10 μm forskolin (n = 3), 1 μm ionomycin (n = 4), and 5 mM DB (n = 3). PYY secretion is expressed as a percentage of the vehicle control (p < 0.05). Error bars represent standard errors of the means.
Optimization of Media Sample Collection for LC/MS
To optimize sample collection for MS analysis, we evaluated a time course of forskolin stimulation to identify conditions for maximal GLP-1 secretion and an increased ratio of stimulated versus basal secretion to enable differential peptide identification. Stimulation with 10 μm forskolin resulted in cumulative levels of released GLP-1 approaching 400 pm in the cell culture media after 120 min (Fig. 3). Pulse-chase experiments (16) with forskolin resulted in GLP-1 secretion and accumulation similar to that for continuous incubation.
Fig. 3.
Dynamics of forskolin-stimulated GLP-1 secretion from primary colon crypt cultures. GLP-1 concentrations were measured in samples of media collected 5, 15, 30, 60, 90, and 120 min after addition of 10 μm forskolin (n = 3).
Peptidomic Profiling of Media Samples from Colonic Crypt Cultures
Peptidomic profiling of media samples from colonic crypt cultures consisted of a preliminary low resolution survey of MALDI-TOF profiles of the concentrated/desalted peptides followed by high resolution separation by nano-LC FT/IT/MSn experiments for peptide identification and relative quantification. We used a hybrid linear ion trap ion cyclotron resonance mass spectrometer (LTQ-FT) and capitalized on the multiple scanning options available to this instrument to capture peptides with a wide range of molecular weights. We operated the instrument in a modification of the SIM mode described by Andersen et al. (18) with IT MS2 and MS3 scan events as well an FT MS2 scan event. In this mode, the FT ICR cell is overfilled with ions in the initial FT MS scan over the full m/z range (400–1800), thus sacrificing mass accuracy (∼10 ppm) through space charging effects in favor of sensitivity. This is compensated by a data-dependent SIM scan over a small scan range (m/z window = 5) with fewer ions allowed to enter this ICR cell. Consequently, a higher mass accuracy (<2 ppm) is attained for the SIM scan that is used by the informatics for accurate precursor ion determination (Scheme 1). We operated the instrument in two modes with either the monoisotopic toggle feature of the Xcalibur software enabled or disabled. Having the feature enabled allowed the peak finding program extract_msn to accurately determine monoisotopic masses from the SIM scans such that very rigorous SEQUEST searches (2-ppm mass accuracy on the precursor) could be performed on LC/MS runs acquired in this mode. However, many peaks that were either of low intensity or high charge state did not trigger a sequence identifying event. Apparently, this was a consequence of over-rigorous requirements for isotopic envelopes being recognized as bona fide signals (24). Disabling the monoisotopic toggle resulted in previously ignored low abundance/high molecular weight peptides triggering MSn sequencing events; however, extract_msn could not determine accurate monoisotopic masses postacquisition. Consequently, SEQUEST searches had to be performed with high error tolerances (2.5 Da) to capture large peptides where the monoisotopic peak tends to be small. Bona fide MS2 hits were usually characterized by incremental 1.0- or 2.0-Da mass errors in the SEQUEST output, corresponding to the larger peaks in the isotopic envelope. Subsequent examination of the raw data indicated high mass accuracy matches to the true monoisotopic masses. Currently, a new SEQUEST interface called “Proteome Discover” (Thermo Fisher Scientific Inc.) is being developed that can accurately determine monoisotopic masses postacquisition without the strict real time requirements of the monoisotopic toggle. Unfortunately, the interface was not conducive to parallel processing on our Linux cluster at the time of writing, making the Proteome Discover impractical for long “no enzyme” searches for 600–7500-Da peptides. In contrast, accurate monoisotopic masses were captured when FT data were analyzed by the Xtract deconvolution algorithm (Thermo Fisher Scientific Inc.), resulting in peptide identification by ProSight in the recently established high throughput mode (20). Aside from the monoisotopic toggle, of the other LC/MS conditions varied, increasing the FT MS2 m/z isolation window from 2 to 5 seemed most important in attaining high quality high resolution fragmentation. Using SEQUEST and ProSight algorithms, we identified 53 peptides ranging from <1 to 16 kDa (see Tables II–V). Of these, 39 satisfied Criterion 1 (0% false discovery rates), and 14 satisfied Criterion 2 (high accuracy precursor combined with high accuracy fragment peaks or IT MS3 for sequence confirmation). The latter peptides predominantly came to our attention in “Multiconsensus” views in Bioworks with less stringent filters for gene products with peptides satisfying Criterion 1. Full details of the peptide identifications can be found in the supplemental information.
Table II. Peptides from prohormone precursors.
Peptide modifications are indicated as follows: [, amide; #, pyroglutamate. Bold, increase (p < 0.05) or present only on forskolin stimulation. SEQ, SEQUEST; PS, ProSight; CCK, cholecystokinin; var, variant; Gly-ex, Gly-extended.
| Gene and identified peptide | Named peptides | Algorithm | MH+ (mono) | ΔM | z |
|---|---|---|---|---|---|
| ppm | |||||
| Peptide YY | |||||
| YPAKPEAPGEDASPEELSRYYASLRHYLNLVTRQRYG | PYY(1–37) | SEQ/PS | 4,297.142 | 0.0 | 6 |
| YPAKPEAPGEDASPEELSRYYASLRHYLNLVTRQRY[ | PYY(1–36) amide | SEQ | 4,239.137 | −1.4 | 5 |
| AKPEAPGEDASPEELSRYYASLRHYLNLVTRQRYG | PYY(3–37) | SEQ/PS | 4,037.026 | −0.3 | 6 |
| AKPEAPGEDASPEELSRYYASLRHYLNLVTRQRY[ | PYY(3–36) amide | SEQ | 3,979.021 | −0.3 | 5 |
| PEELSRYYASLRHYLNLVTRQRY[ | PYY(14–36) amide | SEQ | 3,522.706 | −1.8 | 3 |
| DVPAALFSKLLFTDDSDSENLPFRPEGLDQW | Pro-PYY C terminus | SEQ/PS | 2,926.543 | 0.0 | 4 |
| DVPAALFSKLL | Pro-PYY C terminus | SEQ | 1,173.688 | 0.2 | 2 |
| PAALFSKLL | Pro-PYY C terminus | SEQ | 959.592 | 0.0 | 2 |
| Peptide YY, isoform CRA_a | |||||
| DVPAALFSKLLFTDDSDSENLPFRSRPEGLDQW | Pro-PYY (var) C terminus | SEQ | 3,765.840 | 13.5 | 4 |
| Glucagon | |||||
| HALQDTEENPRSFPASQTEAHEDPDEMNEDKRHSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIA | Glicentin | PS | 8,152.773 | 5.4 | 7 |
| HAEGTFTSDVSSYLEGQAAKEFIAWLVKGRGR | GLP-1(7–38) | SEQ MS3/PS | 3,510.776 | 0.2 | 5 |
| HAEGTFTSDVSSYLEGQAAKEFIAWLVKGRG | GLP-1(7–37) | SEQ/PS | 3,354.675 | 0.0 | 4 |
| HAEGTFTSDVSSYLEGQAAKEFIAWLVKGR[ | GLP-1(7–36) amide | SEQ | 3,296.670 | −0.8 | 4 |
| EGTFTSDVSSYLEGQAAKEFIAWLVKGRG | GLP-1(9–37) | SEQ | 3,146.579 | −0.7 | 3 |
| SSYLEGQAAKEFIAWLVKGRG | GLP-1(17–37) | SEQ | 2,310.224 | −0.2 | 3 |
| HSQGTFTSDYSKYLDSRRAQDFVQWLMNTKRNRNNIA | Oxyntomodulin | SEQ/PS | 4,448.170 | 0.0 | 6 |
| HADGSFSDEMSTILDNLATRDFINWLIQTKITD | GLP-2 | SEQ MS3 | 3,767.822 | 14.5 | 3 |
| Vasoactive intestinal polypeptide | |||||
| HADGVFTSDYSRLLGQISAKKYLESLIG | PHI-27 (Gly-ex) | SEQ | 3,068.609 | −1.3 | 4 |
| Cholecystokinin | |||||
| Q#PVVPAEATDPVEQRAQEAPRRQL | Pro-CCK N terminus | SEQ | 2,668.380 | 0.1 | 3 |
Table III. Peptides from granins and anti-inflammatory, cell stress, and cytoskeletal precursors.
Peptide modifications are indicated as follows: ], N-acetylation; @, half-cystine; *, oxidation. Underlined, decrease on forskolin stimulation (p < 0.05). SEQ, SEQUEST; PS, ProSight; ox, oxidized.
| Gene and identified peptide | Named peptide | Algorithm | MH+ (mono) | ΔM | z |
|---|---|---|---|---|---|
| ppm | |||||
| Granin | |||||
| Secretogranin II | |||||
| IPVGSLKNEDTPNRQYLDEDMLLKVLEYLNQEQAEQGREHLA | SEQ | 4,896.447 | 0.7 | 5 | |
| TNEIVEEQYTPQSLATLESVFQELGKLTGPSNQ | Secretoneurin | SEQ | 3,650.807 | 0.4 | 3 |
| Inflammation | |||||
| Annexin A1 | |||||
| A]MVSEFLKQARFLENQEQEYVQAVK | Murine Ac(2–26) | SEQ | 3,027.524 | 1.3 | 3 |
| A]MVSEFLKQARFLENQEQEYVQAVKSYK | Murine Ac(2–29) | SEQ | 3,405.715 | −0.8 | 4 |
| A]MVSEFLKQARFLENQEQEYVQAVKSY | Murine Ac(2–28) | SEQ | 3,277.620 | −0.6 | 3 |
| A]M*VSEFLKQARFLENQEQEYVQAVKSYK | Murine Ac(2–29) ox | SEQ | 3,421.710 | −1.1 | 4 |
| TYB4 | |||||
| S]DKPDMAEIEKFDKSKLKKTETQEKNPLPSKETIEQEKQAGES | Thymosin β 4 | SEQ | 4,961.494 | −0.4 | 6 |
| S]DKPDM*AEIEKFDKSKLKKTETQEKNPLPSKETIEQEKQAGES | Thymosin β 4 ox | SEQ | 4,977.489 | 1.2 | 5 |
| Cell stress | |||||
| Ubiquitin | |||||
| MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG | PS | 8,560.624 | 2.7 | 9 | |
| Heat shock protein 1 (chaperonin 10) | |||||
| A]GQAFRKFLPLFDRVLVERSAAETVTKGGIMLPEKSQGKVLQATVVAVGSGGKGKSGEIEPVSVKVGDKVLLPEYGGTKVVLDDKDYFLFRDSDILGKYVD | PS | 10,867.843 | −2.4 | 15 | |
| Superoxide dismutase 1, soluble | |||||
| A]MKAVCVLKGDGPVQGTIHFEQKASGEPVVLSGQITGLTEGQHGFHVHQYGDNTQGC@TSAGPHFNPHSKKHGGPADEERHVGDLGNVTAGKDGVANVSIEDRVISLSGEHSIIGRTMVVHEKQDDLGKGGNEESTKTGNAGSRLAC@GVIGIAQ | PS | 15,842.767 | 0.7 | 15 | |
| Cytoskeletal | |||||
| Keratin complex 2, basic, gene 8 | |||||
| SLNNKFASFIDKVRFLEQQNKMLETKWSLLQQQK | SEQ | 4,140.206 | 1.3 | 5 | |
| AVVVKKIETRDGKLVSESSDVVSK | SEQ | 2,573.451 | 1.6 | 4 | |
| Actin, β | |||||
| RGILTLKYPIEHGIVTNWEDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPK | SEQ | 6,174.206 | 0.0 | 7 |
Table IV. Peptides from histones and ribosomal precursors.
Peptide modifications are indicated as follows: ], N-acetylation; &, cysteinyl-cysteine; SEQ, SEQUEST; PS, ProSight; str., structure.
| Gene and identified peptide | Function | Informatics | MH+ (mono) | ΔM | z |
|---|---|---|---|---|---|
| ppm | |||||
| Histone cluster 1, H2bb | |||||
| LPGELAKHAVSEGTKAVTKYTSSK | Chromatin str. | SEQ | 2,502.356 | −0.7 | 4 |
| PGELAKHAVSEGTKAVTKYTSSK | Chromatin str. | SEQ | 2,389.272 | 1.4 | 4 |
| GELAKHAVSEGTKAVTKYTSSK | Chromatin str. | SEQ | 2,292.219 | 0.5 | 4 |
| ELAKHAVSEGTKAVTKYTSSK | Chromatin str. | SEQ | 2,235.198 | −0.8 | 4 |
| H2A1 | |||||
| S]GRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYSERVGAGAPVYLAAVLEYLTAEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNKLLGRVTIAQGGVLPNIQAVLLPKKTESHHKAKGK | Chromatin str. | PS | 14,038.927 | −4.3 | 14 |
| Histone H4-related | |||||
| S]GRGKGGKGLGKGGAKRHRKVLRDNIQGITKPVIRRLARRGGVKRISGLIYEETRGVLKVFLENVIRDAVTYTEHAKRKTVTAMDVVYALKRQGRTLYGFGG | Chromatin str. | PS | 11,300.390 | 3.3 | 11 |
| Ribosomal protein S19 | |||||
| PGVTVKDVNQQEFVRALAAFLKKSGKLKVPEWVDTVKLAK | Translation | SEQ | 4,437.538 | 0.3 | 7 |
| 40 S ribosomal protein S28 | |||||
| M]DTSRVQPIKLARVTKVLGRTGSQGQCTQVRVEFMDDTSRSIIRNVKGPVREGDVLTLLESEREARRLR | Translation | PS | 7,879.219 | −1.5 | 10 |
| M]DTSRVQPFKLARVTKVLGRTGSQGQC&TQVLVEFMDDTSRSIIRNVKGPVREGDVLTLLESEREARRLR | Translation | PS | 7,998.223 | −0.8 | 11 |
Table V. Peptides from outer (OMM) and inner (IMM) mitochondrial membrane protein precursors and others.
Peptide modifications are indicated as follows: ], N-acetylation; @, half-cystine; &, cysteinyl-cysteine. Underlined, decrease on forskolin stimulation (p < 0.05). SEQ, SEQUEST; PS, ProSight; Trans., transport; Immun., immunity; prolif., proliferation.
| Gene and identified peptide | Location/function | Algorithm | MH+ (mono) | ΔM | z |
|---|---|---|---|---|---|
| ppm | |||||
| ATP synthase, H+-transporting, mitochondrial F1 complex, α subunit, isoform 1 | |||||
| TGAIVDVPVGEELLGRVVDALGNAIDGKGPIGSK | IMM/OXPHOS | SEQ MS3 | 3,316.811 | 2.6 | 3 |
| ATP synthase, H+-transporting, mitochondrial F1 complex, ϵ subunit | |||||
| VAYWRQAGLSYIRFSQICAKAVRDALKTEFKANAEKTSGSSIKIVKVSKKE | IMM/OXPHOS | PS | 5,704.116 | 0.2 | 8 |
| ATP synthase-coupling factor 6, mitochondrial (CF6) | |||||
| NKELDPVQKLFVDKIREYKSKRQASGGPVDIGPEYQQDLDRELYKLKQMYGKGEMDTFPTFKFDDPKFEVIDKPQS | IMM/OXPHOS | PS | 8,940.533 | −2.9 | 7 |
| Cytochrome c oxidase, subunit VIIa 2 | |||||
| FENKVPEKQKLFQEDNGMPVHLKGGASDALLYRATMALTLGGTAYAIYLLAMAAFPKKQN | IMM/OXPHOS | PS | 6,569.419 | 1.4 | 6 |
| Cytochrome c oxidase subunit VIb polypeptide 1 | |||||
| A]EDIKTKIKNYKTAPFDSRFPNQNQTKNC@WQNYLDFHRC@EKAMTAKGGDVSVC@EWYRRVYKSLC@PVSWVSAWDDRIAEGTFPGKI | IMM/OXPHOS | PS | 9,972.825 | 6.3 | 11 |
| Cytochrome c oxidase, subunit VIIc | |||||
| SHYEEGPGKNLPFSVENKWRLLAMMTVYFGSGFAAPFFIVRHQLLKK | IMM/OXPHOS | SEQ | 5,440.828 | 1.7 | 6 |
| Solute carrier family 25, member 5 | |||||
| T]DAAVSFAKDFLAGGVAAAISK | OMM/trans. | SEQ | 2,152.129 | −0.1 | 3 |
| Diazepam binding inhibitor isoform 2 | |||||
| S]QAEFDKAAEEVKRLKTQPTDEEMLFIYSHFKQATVGDVNTDRPGLLDLKGKAKWDSWNKLKGTSKESAMKTYVEKVDELKKKYGI | Trans. | PS | 9,906.120 | 6.8 | 12 |
| S100 calcium-binding protein A6 (calcyclin) | |||||
| A]C&PLDQAIGLLVAIFHKYSGKEGDKHTLSKKELKELIQKELTIGSKLQDAEIARLMDDLDRNKDQEVNFQEYVAFLGALALIYNEALK | Cell cycle/prolif. | PS | 10,075.270 | 0.7 | 14 |
| β2-Microglobulin | |||||
| IQKTPQIQVYSRHPPENGKPNILNC@YVTQFHPPHIEIQMLKNGKKIPKVEMSDMSFSKDWSFYILAHTEFTPTETDTYAC@RVKHASMAEPKTVYWDRDM | Immun. | PS | 11,634.702 | −2.1 | 8 |
| α-Globin (Mus musculus) | |||||
| AAWGKIGGHGAEYVAEALERMFASFPTTK | Trans. | SEQ | 3,095.541 | 0.7 | 4 |
Full-scan FT data were used to assemble two-dimensional mass maps for peptide relative quantification using the DeCyderMS algorithm. Peptide carryover is a challenge for relative quantification for intact peptide/protein analysis. In contrast to tryptic peptides, endogenous peptides and proteins tend to exhibit nonspecific adsorption to surfaces, and rather extreme washing conditions were required between injections to eliminate carryover as described previously (12). In our last published study, we performed technical replicates to assess the system reproducibility of our approach using pools of cell culture media from a readily available insulinoma cell line (RINm5f) (12). In the current study, we analyzed a range of culture and stimulation conditions using different crypt preparations from independent experiments to obtain a comprehensive survey of peptides secreted from colonic enteroendocrine cells. We found four peptides and two unknown masses in the stimulated samples that exhibited increased secretion with statistical significance (p < 0.05) and a further six peptides present only in stimulated samples (3–6 profiles). Although the two unknowns have not been unequivocally identified, it is interesting that their intact masses correspond to the singly and doubly oxidized forms of a novel PYY prohormone-derived splice variant with 12.2- and 11.2-ppm, respectively (consistent with high target value full scan mass errors). Three peptides exhibited statistically significant lower secretion on forskolin stimulation. Interestingly, they were derived from the protein precursors annexin A1 and calcyclin (S100A6), which are known to be expressed in enteroendocrine cell types (25, 26).
DISCUSSION
In the current investigation, we combined a primary endocrine cell culture system with a refined LC/MS platform for peptide hormone discovery. Secretion media from endocrine cell lines including pancreatic insulinomas and carcinomas (12, 16, 27–29), a pituitary corticotrophe-derived cell line (29) and a human medullary thyroid carcinoma (30) have previously been probed by peptidomic techniques. However, the current report on mature colonic epithelium is the first to our knowledge to probe secretion media from a complex adherent primary culture derived from any tissue. By enriching the endocrine population but preserving cellular heterogeneity, we have created a local microenvironment more reflective of tissue than transformed clonal cell lines primarily derived from endocrine tumors (7, 31).
Our approach complements proteomics (15, 32–35) and MALDI-TOF profiling reports (36, 37) on isolated colonic crypts that were primarily aimed at the identification of protein biomarkers of colon disease including carcinoma. It should be noted that none of these previous studies identified peptide hormones. In the case of the proteomics investigations that used one-dimensional and two-dimensional gel-based techniques, it may be simply that the peptide hormones were too small for electrophoretic resolution. However another possibility is that, by examining freshly isolated colon crypts or epithelial cells, these approaches favor abundant cytosolic and cytoskeletal components over relatively sparse endocrine cells. In the MALDI-TOF studies, only 10 crypt cells of unspecified phenotype were probed from different areas of the crypt, and the protein profiles were compared with those of an adenoma (36). Although we cannot make a direct comparison with these previous studies, we speculate that observation of peptides hormones in the current investigation was facilitated by the endocrine enrichment in the primary crypt cultures in addition to our non-gel-based, non-cytolytic peptidomics approach.
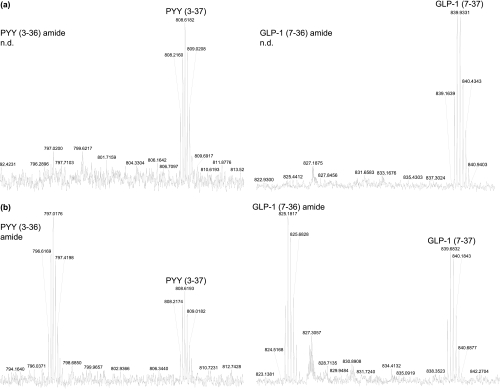
In the current study, the MALDI-TOF MS profile of the secreted media was dominated by L-cell components, predominantly glycine-extended precursors of GLP-1 and PYY (Fig. 4). This finding was initially surprising given that GLP-1(7–36) amide is the predominant form in the colon (38) and that the glycine-extended form of PYY has only been recently reported in extracts from dog ileum (39). By supplementing peptide-amidating enzymes with ascorbate in the growth media, we saw more efficient conversion of the glycine-extended precursors of these hormones to their corresponding amides (Fig. 5) (40), demonstrating that the state of post-translational modification was influenced by cell culture conditions.
Fig. 4.
MALDI-TOF profile of media from forskolin-stimulated cultured colonic crypts enriched in endocrine cells post-solid phase extraction reveal L-cell-dominant peptide profile. Bovine insulin is a remnant of the growth media. oxyn, oxyntomodulin.
Fig. 5.
Comparison of (MH4)4+ peaks for GLP-1(7–37) acid and GLP-1(7–36) amide and (MH5)5+ peaks for PYY(3–37) acid and PYY(3–36) amide cultured in absence (a) and presence (b) of 350 μm ascorbic acid. Spectra were signal-averaged over the LC/MS elution profiles of the acids and amides from forskolin-stimulated cell cultures. n.d., not detected.
Two-dimensional mass maps were compared for stimulated samples and basal secretions to identify any peptides that were differentially secreted (Scheme 1). Validation of our platform for the identification of neuroendocrine peptides came from the fact that all peptides that were either consistently present only in forskolin-stimulated samples or exhibited a statistically significant increased differential secretion were derived from prohormones. This is consistent with their release being mediated through a G-protein-coupled receptor response. During this label-free relative quantitation, peptides were simultaneously identified by database searching algorithms and partial de novo sequencing using high and low resolution data acquired on a linear hybrid ion trap-ion cyclotron resonance mass spectrometer (LTQ-FT, Thermo Fisher Scientific Inc.) (41–44). Over 50 distinct peptides, including 19 peptides from four different prohormones and one splice variant, were identified. Among these are peptides that comprise all known major peptide hormones derived from the glucagon and PYY prohormones at various stages of post-translational processing (Table II). Peptides were identified that appeared to be GLP-1 and PYY metabolites. Bioactive GLP-1(7–36) amide, PYY(1–36) amide, and PYY(3–36) amide are prone to degradation during sample processing procedures (45). Therefore, qualitatively, it was encouraging that the peak intensities of their metabolites were low relative to the bioactive/glycine-extended forms in the current investigation. Partially processed peptide hormone intermediates such as GLP-1(7–38) were also observed. The novel splice variant was initially flagged for identification by statistically significant differential secretion across multiple data sets. Initially, partial de novo sequencing (43) combined with a mass-sensitive BLAST search (44) revealed that it was a novel splice variant from the PYY remnant region that had an Arg-Ser insertion in the region homologous to the rat peptide (data not shown). The identity of the peptide was confirmed when the murine FASTA database was updated and the data were reanalyzed by SEQUEST. A novel remnant from cholecystokinin was also identified as well as the glycine-extended peptide histidine isoleucine, the dominant circulating form in the ileum (46). These data confirm that our approach is an engine for targeted discovery of prohormone-derived peptides.
Other peptides up to 16 kDa were identified, and their PTMs were characterized (see supplemental information). Two secretogranin-derived peptides from enteroendocrine cells were observed (Table III). The peptides were derived from precursors that undergo processing identical to that of peptide hormones via peptide hormone convertase activity and carboxypeptidase E processing at dibasic cleavage sites in the precursor sequence. Other peptides and small proteins possibly represent the products of cellular stress and damage including ubiquitin, heat shock proteins, superoxide dismutase, and peptides derived from cytoskeletal components (Table III). Overlap between peptides identified in the current study with previous proteomics investigations include heat shock protein 10, histone 2ba (15), and annexin A1. Thymosins and annexins have known anti-inflammatory roles (47, 48). Interestingly, they were the only peptides in this study exhibiting oxidized methionine residues. In the case of thymosin, this modification contributes to its anti-inflammatory activity (47).
Interestingly, we identified three peptides that decreased in relative concentration in the culture media upon forskolin stimulation. These are derived from the proteins annexin A1 and calcyclin (S100A6) and belong to a family of Ca2+-binding proteins that function in response to changing calcium levels. Protein expression of both peptides has been shown in endocrine cell types within pancreatic islets and intestine (25, 26), and a role for calcyclin and annexin A1 in regulating insulin secretion has been proposed (26, 49, 50). Very recently, a specific Ca2+-dependent interaction was demonstrated between these two peptides using biophysical methods (51). Our report is the first to demonstrate that both peptides are differentially released into the medium from intestinal cell culture. Their function in intestinal epithelium is largely unknown, but it is interesting to speculate that they may play a physiological role in regulating enteroendocrine (GLP-1 and PYY) secretion. Alternatively, annexin-calcyclin interactions may be indicative of proliferation as observed in normal immune B-cells (52), and therefore, decreased secretion of these peptides may be consistent with a cell-specific inhibition of proliferation by forskolin (53).
The presence of peptides derived from histones/ribosomal precursors, including a peptide with a novel cysteinyl-cysteine PTM, was somewhat surprising in the secretion media (Table IV). However, precedents for secreted histones exist in the literature where they serve antimicrobial functions (54) in addition to their role in maintaining chromatin structure in the nucleus. Histones, ribosomal proteins, and mitochondrial proteins also dominated the alkaline proteins of the human colon crypt proteome studied by two-dimensional PAGE (15). The observation of secreted peptides and proteins of mitochondrial origin (Table V) may represent apoptotic events in the cell culture. Alternatively, their presence may indicate the shedding of extramitochondrial OXPHOS machinery from the plasma membrane. Interestingly, plasma membrane-expressed mitochondrial ATPase is the putative enterostatin receptor (55), and the secreted coupling factor 6 (CF6) that we characterized has been proposed to exhibit “hormone-like” behavior (56). It is interesting to speculate that we may be observing a physiologically relevant secreted form of CF6.
CONCLUSION
In summary, we report a peptidomics survey of adult mouse colonic epithelium using a physiologically relevant endocrine enriched culture model. All known L-cell-derived peptide hormones were secreted and identified at various stages of post-translation modification, which can in turn be influenced by the cell culture conditions. We also catalogued some novel remnants of known peptide hormones and other cellular components. These components include differentially secreted annexin A1-calcyclin peptides that may interact (51) to play a biological role in intestinal epithelia. We believe that this work represents a new approach for characterizing physiologically relevant peptide secretion throughout the gastrointestinal tract and elucidating new endocrine function.
Supplementary Material
Acknowledgments
We acknowledge Soumitra Ghosh, Mary Erickson, Michael Hanley, and Richard Pittner for useful discussions. We are grateful to Anders Kaplan (GE Healthcare), Jim Shofstahl, Collette Rudd, Thomas Möhring (Thermo Fisher Scientific Inc.), Kerry Nugent (Michrom Bioresources), Eoin Fahy (University of California, San Diego), and the Kelleher laboratory (University of Illinois) for early access to, and advice on, hardware and software used in this investigation.
 This article contains supplemental peptide identification and relative quantification details.
This article contains supplemental peptide identification and relative quantification details.
1 The abbreviations used are:
- PYY
- peptide YY
- DB
- denatonium benzoate
- DMEM
- Dulbecco's modified Eagle's medium
- GLP-1
- glucagon-like peptide-1
- IT
- ion trap
- PMA
- phorbol 12-myristate 13-acetate
- PTM
- post-translational modification
- SIM
- selected ion monitoring
- VIP
- vasoactive intestinal peptide
- BLAST
- Basic Local Alignment Search Tool
- CF6
- coupling factor 6
- Chg
- chromogranin
- LDA
- low density array
- OXPHOS
- oxidative phosphorylation.
REFERENCES
- 1.Roth K. A., Gordon J. I. (1990) Spatial differentiation of the intestinal epithelium: analysis of enteroendocrine cells containing immunoreactive serotonin, secretin, and substance P in normal and transgenic mice. Proc. Natl. Acad. Sci. U.S.A 87, 6408–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sjölund K., Sandén G., Håkanson R., Sundler F. (1983) Endocrine cells in human intestine: an immunocytochemical study. Gastroenterology 85, 1120–1130 [PubMed] [Google Scholar]
- 3.Drucker D. J. (2006) The biology of incretin hormones. Cell Metab 3, 153–165 [DOI] [PubMed] [Google Scholar]
- 4.Ballantyne G. H. (2006) Peptide YY(1–36) and peptide YY(3–36): part I. distribution, release and actions. Obes. Surg 16, 651–658 [DOI] [PubMed] [Google Scholar]
- 5.Stonehouse A., Okerson T., Kendall D., Maggs D. (2008) Emerging incretin based therapies for type 2 diabetes: incretin mimetics and DPP-4 inhibitors. Curr. Diabetes Rev 4, 101–109 [DOI] [PubMed] [Google Scholar]
- 6.Brubaker P. L., Schloos J., Drucker D. J. (1998) Regulation of glucagon-like peptide-1 synthesis and secretion in the GLUTag enteroendocrine cell line. Endocrinology 139, 4108–4114 [DOI] [PubMed] [Google Scholar]
- 7.Cao X., Flock G., Choi C., Irwin D. M., Drucker D. J. (2003) Aberrant regulation of human intestinal proglucagon gene expression in the NCI-H716 cell line. Endocrinology 144, 2025–2033 [DOI] [PubMed] [Google Scholar]
- 8.Dall'Olio F., Malagolini N., Di Stefano G., Ciambella M., Serafini-Cessi F. (1990) Postnatal development of rat colon epithelial cells is associated with changes in the expression of the beta 1,4-N-acetylgalactosaminyltransferase involved in the synthesis of Sda antigen of alpha 2,6-sialyltransferase activity towards N-acetyl-lactosamine. Biochem. J 270, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 [DOI] [PubMed] [Google Scholar]
- 10.Ootani A., Li X., Sangiorgi E., Ho Q. T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I. L., Capecchi M. R., Kuo C. J. (2009) Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med 15, 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead R. H., Robinson P. S. (2009) Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol 296, G455–G460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor S. W., Andon N. L., Bilakovics J. M., Lowe C., Hanley M. R., Pittner R., Ghosh S. S. (2006) Efficient high-throughput discovery of large peptidic hormones and biomarkers. J. Proteome Res 5, 1776–1784 [DOI] [PubMed] [Google Scholar]
- 13.Boonen K., Landuyt B., Baggerman G., Husson S. J., Huybrechts J., Schoofs L. (2008) Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J. Sep. Sci 31, 427–445 [DOI] [PubMed] [Google Scholar]
- 14.Breuker K., Jin M., Han X., Jiang H., McLafferty F. W. (2008) Top-down identification and characterization of biomolecules by mass spectrometry. J. Am. Soc. Mass Spectrom 19, 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X. M., Patel B. B., Blagoi E. L., Patterson M. D., Seeholzer S. H., Zhang T., Damle S., Gao Z., Boman B., Yeung A. T. (2004) Analyzing alkaline proteins in human colon crypt proteome. J. Proteome Res 3, 821–833 [DOI] [PubMed] [Google Scholar]
- 16.Taylor S. W., Sun C., Hsieh A., Andon N. L., Ghosh S. S. (2008) A sulfated, phosphorylated 7 kDa secreted peptide characterized by direct analysis of cell culture media. J. Proteome Res 7, 795–802 [DOI] [PubMed] [Google Scholar]
- 17.Booth C., O'Shea J. A. (2002) Isolation and culture of intestinal epithelial cells, in Culture of Epithelial Cells, Wiley-Liss, Inc., New York: 303–335 [Google Scholar]
- 18.Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 19.Moore R. E., Young M. K., Lee T. D. (2002) Qscore: an algorithm for evaluating SEQUEST database search results. J. Am. Soc. Mass Spectrom 13, 378–386 [DOI] [PubMed] [Google Scholar]
- 20.Boyne M. T., Garcia B. A., Li M., Zamdborg L., Wenger C. D., Babai S., Kelleher N. L. (2009) Tandem mass spectrometry with ultrahigh mass accuracy clarifies peptide identification by database retrieval. J. Proteome Res 8, 374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 22.Kaplan A., Söderström M., Fenyö D., Nilsson A., Fälth M., Sköld K., Svensson M., Pettersen H., Lindqvist S., Svenningsson P., Andrén P. E., Björkesten L. (2007) An automated method for scanning LC-MS data sets for significant peptides and proteins, including quantitative profiling and interactive confirmation. J. Proteome Res 6, 2888–2895 [DOI] [PubMed] [Google Scholar]
- 23.Nilsson O., Bilchik A. J., Goldenring J. R., Ballantyne G. H., Adrian T. E., Modlin I. M. (1991) Distribution and immunocytochemical colocalization of peptide YY and enteroglucagon in endocrine cells of the rabbit colon. Endocrinology 129, 139–148 [DOI] [PubMed] [Google Scholar]
- 24.Sutton J., Richmond T., Shi X., Athanas M., Ptak C., Gerszten R., Bonilla L. (2008) Performance characteristics of an FT MS-based workflow for label-free differential MS analysis of human plasma: standards, reproducibility, targeted feature investigation, and application to a model of controlled myocardial infarction. Proteomics Clin. Appl 2, 862–881 [DOI] [PubMed] [Google Scholar]
- 25.Timmons P. M., Chan C. T., Rigby P. W., Poirier F. (1993) The gene encoding the calcium binding protein calcyclin is expressed at sites of exocytosis in the mouse. J. Cell Sci 104, 187–196 [DOI] [PubMed] [Google Scholar]
- 26.Jägerbrink T., Lexander H., Palmberg C., Shafqat J., Sharoyko V., Berggren P. O., Efendic S., Zaitsev S., Jörnvall H. (2007) Differential protein expression in pancreatic islets after treatment with an imidazoline compound. Cell. Mol. Life Sci 64, 1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jost M. M., Budde P., Tammen H., Hess R., Kellmann M., Schulte I., Rose H. (2005) The concept of functional peptidomics for the discovery of bioactive peptides in cell culture models. Comb. Chem. High Throughput Screen 8, 767–773 [DOI] [PubMed] [Google Scholar]
- 28.Sasaki K., Sato K., Akiyama Y., Yanagihara K., Oka M., Yamaguchi K. (2002) Peptidomics-based approach reveals the secretion of the 29-residue COOH-terminal fragment of the putative tumor suppressor protein DMBT1 from pancreatic adenocarcinoma cell lines. Cancer Res 62, 4894–4898 [PubMed] [Google Scholar]
- 29.Che F. Y., Yuan Q., Kalinina E., Fricker L. D. (2004) Examination of the rate of peptide biosynthesis in neuroendocrine cell lines using a stable isotopic label and mass spectrometry. J. Neurochem 90, 585–594 [DOI] [PubMed] [Google Scholar]
- 30.Sasaki K., Satomi Y., Takao T., Minamino N. (2009) Snapshot peptidomics of the regulated secretory pathway. Mol. Cell. Proteomics 8, 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bruïne A. P., Dinjens W. N., van der Linden E. P., Pijls M. M., Moerkerk P. T., Bosman F. T. (1993) Extracellular matrix components induce endocrine differentiation in vitro in NCI-H716 cells. Am. J. Pathol 142, 773–782 [PMC free article] [PubMed] [Google Scholar]
- 32.Patel B. B., Li X. M., Dixon M. P., Blagoi E. L., Seeholzer S. H., Chen Y., Miller C. G., He Y. A., Tetruashvily M., Chaudhry A. H., Ke E., Xie J., Cooper H., Bellacosa A., Clapper M. L., Boman B. M., Zhang T., Litwin S., Ross E. A., Conrad P., Crowell J. A., Kopelovich L., Knudson A., Yeung A. T. (2007) Searchable high-resolution 2D gel proteome of the human colon crypt. J. Proteome Res 6, 2232–2238 [DOI] [PubMed] [Google Scholar]
- 33.Brouillard F., Bensalem N., Hinzpeter A., Tondelier D., Trudel S., Gruber A. D., Ollero M., Edelman A. (2005) Blue native/SDS-PAGE analysis reveals reduced expression of the mClCA3 protein in cystic fibrosis knock-out mice. Mol. Cell. Proteomics 4, 1762–1775 [DOI] [PubMed] [Google Scholar]
- 34.Yeung A. T., Patel B. B., Li X. M., Seeholzer S. H., Coudry R. A., Cooper H. S., Bellacosa A., Boman B. M., Zhang T., Litwin S., Ross E. A., Conrad P., Crowell J. A., Kopelovich L., Knudson A. (2008) One-hit effects in cancer: altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. Cancer Res 68, 7579–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanni P., Mezzanotte L., Roda G., Caponi A., Levander F., James P., Roda A. (2009) Differential proteomic analysis of HT29 Cl. 16E and intestinal epithelial cells by LC ESI/QTOF mass spectrometry. J. Proteomics 72, 865–873 [DOI] [PubMed] [Google Scholar]
- 36.Xu B. J., Caprioli R. M., Sanders M. E., Jensen R. A. (2002) Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J. Am. Soc. Mass. Spectrom 13, 1292–1297 [DOI] [PubMed] [Google Scholar]
- 37.Xu B. J., Li J., Beauchamp R. D., Shyr Y., Li M., Washington M. K., Yeatman T. J., Whitehead R. H., Coffey R. J., Caprioli R. M. (2009) Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol. Cell. Proteomics 8, 936–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deacon C. F., Johnsen A. H., Holst J. J. (1995) Human colon produces fully processed glucagon-like peptide-1 (7–36) amide. FEBS Lett 372, 269–272 [DOI] [PubMed] [Google Scholar]
- 39.Keire D. A., Whitelegge J. P., Bassilian S., Faull K. F., Wiggins B. W., Mehdizadeh O. B., Reidelberger R. D., Haver A. C., Sayegh A. I., Reeve J. R., Jr. (2008) A new endogenous form of PYY isolated from canine ileum: Gly-extended PYY(1–36). Regul. Pept 151, 61–70 [DOI] [PubMed] [Google Scholar]
- 40.Glembotski C. C. (1986) The characterization of the ascorbic acid-mediated alpha-amidation of alpha-melanotropin in cultured intermediate pituitary lobe cells. Endocrinology 118, 1461–1468 [DOI] [PubMed] [Google Scholar]
- 41.Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 42.Taylor G. K., Kim Y. B., Forbes A. J., Meng F., McCarthy R., Kelleher N. L. (2003) Web and database software for identification of intact proteins using “top down” mass spectrometry. Anal. Chem 75, 4081–4086 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z., McElvain J. S. (2000) De novo peptide sequencing by two-dimensional fragment correlation mass spectrometry. Anal. Chem 72, 2337–2350 [DOI] [PubMed] [Google Scholar]
- 44.Han Y., Ma B., Zhang K. (2005) SPIDER: software for protein identification from sequence tags with de novo sequencing error. J. Bioinform. Comput. Biol 3, 697–716 [DOI] [PubMed] [Google Scholar]
- 45.Stengel A., Keire D., Goebel M., Evilevitch L., Wiggins B., Taché Y., Reeve J. R., Jr. (2009) The RAPID method for blood processing yields new insight in plasma concentrations and molecular forms of circulating gut peptides. Endocrinology 150, 5113–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agoston D. V., Fahrenkrug J., Mikkelsen J. D., Whittaker V. P. (1989) A peptide with N-terminal histidine and C-terminal isoleucine amide (PHI) and vasoactive intestinal peptide (VIP) are copackaged in myenteric neurones of the guinea pig ileum. Peptides 10, 571–573 [DOI] [PubMed] [Google Scholar]
- 47.Young J. D., Lawrence A. J., MacLean A. G., Leung B. P., McInnes I. B., Canas B., Pappin D. J., Stevenson R. D. (1999) Thymosin beta 4 sulfoxide is an anti-inflammatory agent generated by monocytes in the presence of glucocorticoids. Nat. Med 5, 1424–1427 [DOI] [PubMed] [Google Scholar]
- 48.Hayhoe R. P., Kamal A. M., Solito E., Flower R. J., Cooper D., Perretti M. (2006) Annexin 1 and its bioactive peptide inhibit neutrophil-endothelium interactions under flow: indication of distinct receptor involvement. Blood 107, 2123–2130 [DOI] [PubMed] [Google Scholar]
- 49.Okazaki K., Niki I., Iino S., Kobayashi S., Hidaka H. (1994) A role of calcyclin, a Ca(2+)-binding protein, on the Ca(2+)-dependent insulin release from the pancreatic beta cell. J. Biol. Chem 269, 6149–6152 [PubMed] [Google Scholar]
- 50.Ohnishi M., Tokuda M., Masaki T., Fujimura T., Tai Y., Itano T., Matsui H., Ishida T., Konishi R., Takahara J. (1995) Involvement of annexin-I in glucose-induced insulin secretion in rat pancreatic islets. Endocrinology 136, 2421–2426 [DOI] [PubMed] [Google Scholar]
- 51.Streicher W. W., Lopez M. M., Makhatadze G. I. (2009) Annexin I and annexin II N-terminal peptides binding to S100 protein family members: specificity and thermodynamic characterization. Biochemistry 48, 2788–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francés R., Tumang J. R., Rothstein T. L. (2007) Extreme skewing of annexin II and S100A6 expression identified by proteomic analysis of peritoneal B-1 cells. Int. Immunol 19, 59–65 [DOI] [PubMed] [Google Scholar]
- 53.Stork P. J., Schmitt J. M. (2002) Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki H., Iwamuro S. (2008) Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets 8, 195–205 [DOI] [PubMed] [Google Scholar]
- 55.Park M., Lin L., Thomas S., Braymer H. D., Smith P. M., Harrison D. H., York D. A. (2004) The F1-ATPase beta-subunit is the putative enterostatin receptor. Peptides 25, 2127–2133 [DOI] [PubMed] [Google Scholar]
- 56.Osanai T., Tanaka M., Kamada T., Nakano T., Takahashi K., Okada S., Sirato K., Magota K., Kodama S., Okumura K. (2001) Mitochondrial coupling factor 6 as a potent endogenous vasoconstrictor. J. Clin. Investig 108, 1023–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.