Abstract
Although gemcitabine monotherapy is the standard treatment for advanced pancreatic cancer, patient outcome varies significantly, and a considerable number do not benefit adequately. We therefore searched for new biomarkers predictive of overall patient survival. Using LC-MS, we compared the base-line plasma proteome between 29 representative patients with advanced pancreatic cancer who died within 100 days and 31 patients who survived for more than 400 days after receiving at least two cycles of the same gemcitabine monotherapy. Identified biomarker candidates were then challenged in a larger cohort of 304 patients treated with the same protocol using reverse-phase protein microarray. Among a total of 45,277 peptide peaks, we identified 637 peaks whose intensities differed significantly between the two groups (p < 0.001, Welch's t test). Two MS peaks with the highest statistical significance (p = 2.6 × 10−4 and p = 5.0 × 10−4) were revealed to be derived from α1-antitrypsin and α1-antichymotrypsin, respectively. The levels of α1-antitrypsin (p = 8.9 × 10−8) and α1-antichymotrypsin (p = 0.001) were significantly correlated with the overall survival of the 304 patients. We selected α1-antitrypsin (p = 0.0001), leukocyte count (p = 0.066), alkaline phosphatase (p = 8.3 × 10−8), and performance status (p = 0.003) using multivariate Cox regression analysis and constructed a scoring system (nomogram) that was able to identify a group of high risk patients having a short median survival time of 150 days (95% confidence interval, 123–187 days; p = 2.0 × 10−15, log rank test). The accuracy of this model for prognostication was internally validated and showed good calibration and discrimination with a bootstrap-corrected concordance index of 0.672. In conclusion, an increased level of α1-antitrypsin is a biomarker that predicts short overall survival of patients with advanced pancreatic cancer receiving gemcitabine monotherapy. Although an external validation study will be necessary, the current model may be useful for identifying patients unsuitable for the standardized therapy.
Invasive ductal adenocarcinoma of the pancreas is one of the most aggressive and lethal malignancies (1). It is the fifth leading cause of cancer-related death in Japan and the fourth in the United States, accounting for an estimated >23,000 deaths per year in Japan and >33,000 in the United States (2, 3). Because the majority of patients have distant metastases even at their first presentation (4, 5), the main therapeutic modality for pancreatic cancer is systemic chemotherapy, and gemcitabine monotherapy is the current standard (6). Gemcitabine treatment has significantly improved the median survival time of patients with advanced pancreatic cancer (7). However, the outcome of the treatment varies significantly among individuals, and a considerable portion of patients do not appear to benefit significantly from it. It therefore seems necessary to assess the efficacy and adverse effects of the drug before administration and tailor the treatment accordingly for each person.
We previously identified a predictive biomarker for hematologic toxicity, which is one of the most frequent and potentially life-threatening adverse effects associated with gemcitabine monotherapy (8). As a next step, we performed a large scale proteome analysis in this study to identify biomarkers predictive of patient survival after gemcitabine monotherapy. Several factors and their combinations have been reported to correlate significantly with outcome in patients with advanced pancreatic cancer receiving gemcitabine, such as performance status, metastases, serum albumin, alkaline phosphatase, and peripheral leukocyte count (9–11). Unfortunately, however, the accuracy of survival prediction based on these conventional prognostic factors seems unsatisfactory (9).
In recent years, there has been considerable interest in applying advanced proteomics technologies to the discovery of predictive biomarkers (12, 13). We and others have successfully applied MALDI MS-based protein profiling techniques for predicting the efficacy of chemoradiotherapy and molecular targeting therapy (14, 15). Two-dimensional image converted analysis of liquid chromatography and mass spectrometry (2DICAL)1 is a new LC-MS-based proteomics platform that was developed in our laboratory (16). 2DICAL can quantify protein content accurately across a theoretically unlimited number of samples without isotope labeling and thus has considerable advantages over conventional LC-MS-based methods for clinical studies (17). The predictive biomarker protein for hematologic toxicity described above was identified using 2DICAL (8).
It has been generally accepted that tumor responses do not always correlate with the outcome of patients (10, 18, 19). The rates of complete and partial responses (Response Evaluation Criteria in Solid Tumors guideline) to gemcitabine monotherapy are limited to ∼10% (20–22), and the majority of pancreatic cancers do not show significant tumor regression. Given that the ultimate goal of gemcitabine therapy for pancreatic cancer is to achieve prolonged survival, it would be desirable to stratify patients according to survival rather than tumor response (9). In the present study, using 2DICAL, we compared the base-line plasma proteome of two extreme populations of patients who had shown distinct clinical courses after identical gemcitabine treatment.
EXPERIMENTAL PROCEDURES
Patients
Samples were collected from a total of 304 patients who had all been included in our previous study (8). All patients had metastatic (stage IVb; n = 285) or locally advanced (stage IVa; n = 19) (23) histologically or cytologically proven pancreatic ductal adenocarcinoma and had received at least two cycles of gemcitabine monotherapy (1,000 mg/m2 intravenously over 30 min on days 1, 8, and 15 of a 28-day cycle). Two hundred eighty-one patients (92%) received gemcitabine as a first line therapy (supplemental Table S1).
Two hundred sixty-two patients (86%) were treated consecutively at the National Cancer Center (NCC) Hospital (Tokyo, Japan) between September 2002 and June 2007, and 42 patients (14%) were treated consecutively at the NCC Hospital East (Kashiwa, Japan) between September 2002 and July 2004. Survival times were determined as of May 2008. During this period, 248 patients (82%) died, and 56 patients (18%) were censored. Tumor response was evaluated after the first two cycles of gemcitabine using the Response Evaluation Criteria in Solid Tumors guideline.
Sample Preparation
Blood was collected before the first administration of gemcitabine. Plasma or serum was separated by centrifugation at 1,050 × g for 10 min at 4 °C and frozen until analysis as reported previously (8, 24). Macroscopically hemolyzed samples were excluded from the current analysis. Two hundred fifty-two plasma samples (83%) were collected from the NCC Hospital and Hospital East, and 52 serum samples (17%) were collected from the NCC Hospital. Written informed consent was obtained from all patients before blood sampling. The protocol of this retrospective study was reviewed and approved by the institutional ethics committee boards of the NCC (Tokyo, Japan) and the National Institute of Health Sciences (Tokyo, Japan).
LC-MS
Samples were blinded, randomized, and passed through an IgY-12 High Capacity Spin Column (Beckman Coulter, Fullerton, CA) in accordance with the manufacturer's instructions. The flow-through portion was digested with sequencing grade modified trypsin (Promega, Madison, WI) and analyzed in triplicate using a nanoflow high performance LC system (NanoFrontier nLC, Hitachi High Technologies, Tokyo, Japan) connected to an electrospray ionization quadrupole time-of-flight mass spectrometer (Q-Tof Ultima, Waters, Milford, MA). LC-MS run order was also randomized to eliminate any potential bias.
MS peaks were detected, normalized, and quantified using the in-house 2DICAL software package as described previously (16). A serial identification (ID) number was applied to each of the MS peaks detected (1–45,277). The stability of LC-MS was monitored by calculating the correlation coefficient (CC) and coefficient of variance (CV) of every triplicate measurement. The mean CC and CV ± S.D. for all 45,277 peaks observed in the 60 triplicate runs were as high as 0.970 ± 0.022 and as low as 0.056 ± 0.017, respectively.
Protein Identification by MS/MS
Peak lists were generated using the Mass Navigator software package (version 1.2) (Mitsui Knowledge Industry, Tokyo, Japan) and searched against the NCBInr database (downloaded on May 20, 2008) using the Mascot software package (version 2.2.1) (Matrix Science, London, UK). The search parameters used were as follows. A database of human proteins was selected. Trypsin was designated as the enzyme, and up to one missed cleavage was allowed. Mass tolerances for precursor and fragment ions were ±2.0 and ±0.8 Da, respectively. The score threshold was set to p < 0.05 based on the size of the database used in the search. If a peptide matched to multiple proteins, the protein name with the highest Mascot score was selected.
Western Blot Analysis
Primary antibodies used were rabbit polyclonal antibody against human α1-antitrypsin (Dako, Glostrup, Denmark), rabbit polyclonal antibody against human α1-antichymotrypsin (Dako), and mouse monoclonal antibody against human complement C3b-α (Progen, Heidelberg, Germany). Ten microliters of partitioned sample was separated by SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. The membrane was then incubated with the primary antibody and subsequently with the relevant horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG as described previously (25, 26). Blots were developed using an ECL detection system (GE Healthcare).
Reverse-phase Protein Microarray
Samples were serially diluted 1:500, 1:1,000, 1:2,000, and 1:4,000 using a Biomek 2000 Laboratory Automation Robot (Beckman Coulter) and randomly plotted onto ProteoChip® glass slides (Proteogen, Seoul, Korea) in quadruplicate in a 6,144-spot/slide format using a Protein Microarrayer Robot (Kaken Geneqs Inc., Matsudo, Japan). The spotted slides were incubated overnight with the same primary antibodies as those used in Western blotting. The slides were incubated with biotinylated anti-rabbit IgG (Vector Laboratories, Burlingame, CA) and subsequently with streptavidin-horseradish peroxidase conjugate (GE Healthcare). The peroxidase activity was detected using the Tyramide Signal Amplification (TSA®) Cyanine 5 System (PerkinElmer Life Sciences). The slides were counterstained with Alexa Fluor® 546-labeled goat anti-human IgG (Invitrogen) (spotting control).
The stained slides were scanned on a microarray scanner (InnoScan® 700AL, Innopsys, Carbonne, France). Fluorescence intensity, determined as the mean net value of quadruplicate samples, was determined using the Mapix® software package (Innopsys). All determined intensity values were transformed into logarithmic variables.
The reproducibility of reverse-phase protein microarray assay was revealed by repeating the same experiment. A plasma sample was serially diluted within a range of 1,024–16,384-fold. Each diluted sample was spotted in quadruplicate onto glass slides and blotted with anti-α1-antitrypsin antibody. In a representative quality control experiment, the CC value was 0.977 between days, and the median CV was 0.026 among quadruplicate samples.
Statistical Analysis
Overall survival time was defined as the period from the date of starting gemcitabine monotherapy until the date of death from any cause or until the date of the last follow-up at which point the data were censored. We used the Kaplan-Meier method to plot overall survival curves. Statistical significance of intergroup differences was assessed with Welch's t test, Wilcoxon test, χ2 test, or log rank test as appropriate. The maximally selected statistics (27) using the fitness of univariate Cox model (log likelihood) was used to determine which level (optimal cutoff point) of each factor best segregated patients in terms of survival.
Multivariate regression analysis was performed using ordinal Cox regression modeling. Factors included in the prediction model were selected with a forward stepwise selection procedure using Akaike's information criterion (AIC), and the result was confirmed using a backward stepwise procedure. The significance of differences between models with and without α1-antitrypsin was assessed with the likelihood ratio test. The survival prediction model was internally validated by measuring both discrimination and calibration (28). Discrimination was evaluated using the concordance index, which is similar in concept to the area under the receiver operating characteristic curve. Calibration was evaluated with a calibration curve whereby patients are categorized by predicted survival and then plotted as actual versus predicted survival. Both discrimination and calibration were evaluated for the whole study cohort using 200 cycles of bootstrap resampling. Statistical analyses were performed using the open source statistical language R (version 2.7.0) with the optional module Design package.
RESULTS
The median survival estimate for the present study was 236 days (95% CI, 216–254 days), which is comparable to those of previous large scale studies (10, 22). To identify a prognostic factor in patients with advanced pancreatic cancer, we compared the base-line plasma proteome between 29 patients showing short term survival (<100 days) and 31 patients showing long term survival (>400 days) using 2DICAL. There was no significant difference in age, sex, body surface area, prior therapy, clinical stage, or gemcitabine pharmacokinetics (24) (Table I) between the two groups, but the patients with short term survival had significantly poorer base-line conditions such as liver function and Eastern Cooperative Oncology Group (ECOG) performance status than those with long term survival (Table I).
Table I. Clinical and laboratory data of patients with short term or long term survival.
Survival time was calculated from the date of starting gemcitabine therapy until the date of death from cancer. Wilcoxon test was applied to assess differences in values. 5-FU, 5-fluorouracil; LAPC, locally advanced pancreatic cancer; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; Cmax, peak concentration; AUC, area under the curve.
| Short term survivor (<100 days) | Long term survivor (>400 days) | p | |
|---|---|---|---|
| No. of patients | 29 | 31 | |
| Sex (no. of patients) | 0.361a | ||
| Male | 21 | 19 | |
| Female | 8 | 12 | |
| Age, mean (S.D.) (years) | 63 (7) | 67 (8) | 0.123 |
| ECOG performance status (no. of patients) | 0.008a | ||
| 0 | 8 | 20 | |
| 1 | 18 | 11 | |
| 2 | 3 | 0 | |
| Body surface area, mean (S.D.) (m2) | 1.59 (0.17) | 1.54 (0.15) | 0.333 |
| Prior therapy | 0.438a | ||
| None | 27 | 27 | |
| Chemoradiotherapy using 5-FU for LAPC | 2 | 4 | |
| Clinical stageb | 0.697a | ||
| IVa | 2 | 3 | |
| IVb | 27 | 28 | |
| Subsequent line chemotherapy after gemcitabine | 0.045a | ||
| None | 29 | 27 | |
| Yes | 0 | 4 | |
| Leukocytes, mean (S.D.) (×103/mm3) | 7.6 (3.6) | 5.2 (1.3) | 0.002 |
| Platelets, mean (S.D.) (×104/mm3) | 24.5 (7.6) | 20.2 (4.6) | 0.020 |
| Hemoglobin, mean (S.D.) (g/dl) | 11.7 (1.6) | 11.7 (1.5) | 0.491 |
| Albumin, mean (S.D.) (g/dl) | 3.4 (0.4) | 3.7 (0.3) | 0.014 |
| Creatinine, mean (S.D.) (mg/dl) | 0.70 (0.23) | 0.68 (0.23) | 0.726 |
| AST, mean (S.D.) (IU/liter) | 40 (25) | 26 (15) | 0.010 |
| ALT, mean (S.D.) (IU/liter) | 51 (44) | 27 (19) | 0.037 |
| ALP, mean (S.D.) (units/liter) | 728 (632) | 337 (160) | 0.026 |
| Pharmacokinetic parameters of gemcitabine | |||
| Cmax, mean (S.D.) (μg/ml) | 24.02 (7.52) | 24.91 (6.22) | 0.610 |
| AUC, mean (S.D.) (h·μg/ml) | 10.24 (2.83) | 10.75 (2.32) | 0.270 |
| α1-Antitrypsin,c mean (S.D.) | 64.6 (66.8) | 16.9 (7.9) | 0.0003 |
| α1-Antichymotrypsin,c mean (S.D.) | 706.4 (416.0) | 389.0 (216.5) | 0.0005 |
| Tumor responsed | <0.0001a | ||
| Complete response | 0 | 0 | |
| Partial response | 0 | 1 | |
| Stable disease | 2 | 22 | |
| Progressive disease | 24 | 0 | |
| Not evaluable | 3 | 8 |
a Calculated by χ2 test.
b According to Ref. 23.
c Intensity of the corresponding peak measured by quantitative mass spectrometry.
d Evaluated after the first two cycles of gemcitabine monotherapy.
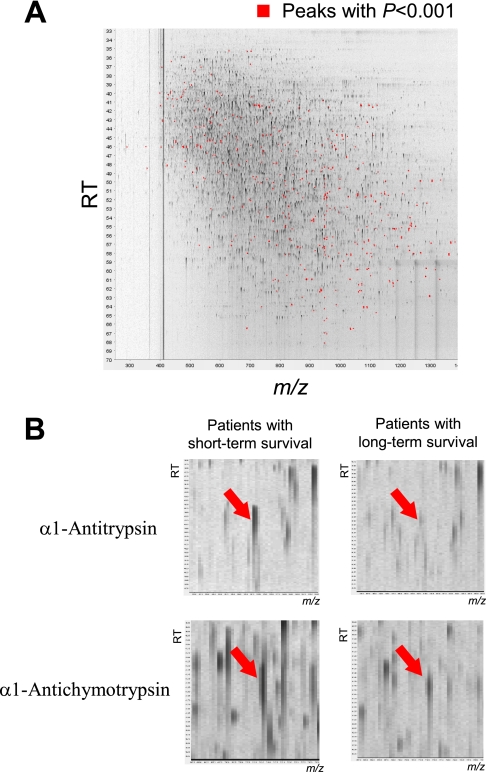
Among a total of 45,277 independent MS peaks detected within the range 250–1,600 m/z and within the time range of 20–70 min, we found that the mean intensity of triplicates differed significantly for 637 peaks (p < 0.001, Welch's t test). Fig. 1A is a representative two-dimensional view of all the MS peaks displayed with m/z along the x axis and the retention time of LC along the y axis. The 637 MS peaks whose expression differed significantly between patients with short term and long term survival are highlighted in red.
Fig. 1.
A, two-dimensional display of all (>45,000) the MS peaks. The 637 MS peaks whose mean intensity differed significantly between patients with short term and long term survival (p < 0.001, Welch's t test) are highlighted in red. B, two MS peaks with the smallest p values (upper, p = 2.57 × 10−4; bottom, p = 5.03 × 10−4) in representative patients with short term (left) and long term (right) survival. RT, retention time.
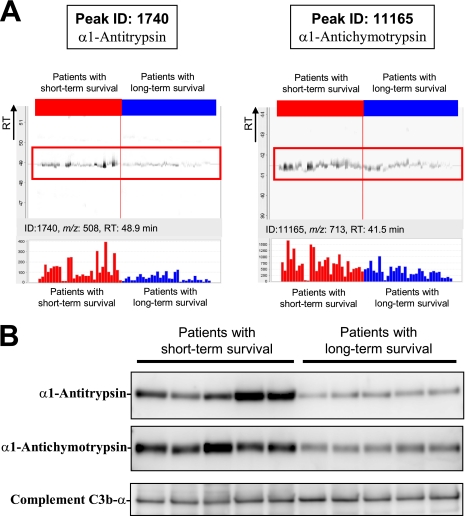
MS peaks that were increased in patients with short term survival with the highest statistical significance (p = 2.57 × 10−4) (Fig. 1B) matched the amino acid sequences of the α1-antitrypsin (AAT) gene product (supplemental Fig. S1A). The MS peak with the second highest statistical significance (p = 5.03 × 10−4) was revealed to be derived from the α1-antichymotrypsin (AACT) gene product (supplemental Fig. S1B). We calculated the false discovery rate (FDR) (29) and confirmed the significance of these MS peaks (FDR = 0.0327 for α1-antitrypsin and FDR = 0.0428 for α1-antichymotrypsin). Fig. 2A shows the distribution of the two peaks (ID 1740 (at 508 m/z and 48.9 min; α1-antitrypsin) and ID 11165 (at 713 m/z and 41.5 min; α1-antichymotrypsin)) in patients with short term (red) and long term survival (blue). The differential expression and identification of α1-antitrypsin and α1-antichymotrypsin were confirmed by denaturing SDS-PAGE and immunoblotting (Fig. 2B).
Fig. 2.
A, representative MS peaks in 60 triplicate LC-MS runs (29 with short term survival (red) and 31 with long term survival (blue)) aligned along the retention time (RT) of LC. Columns represent the mean intensity of triplicates (bottom). B, detection of α1-antitrypsin, α1-antichymotrypsin, and complement C3b-α (loading control) by immunoblotting.
Correlation of α1-Antitrypsin and α1-Antichymotrypsin with Overall Survival
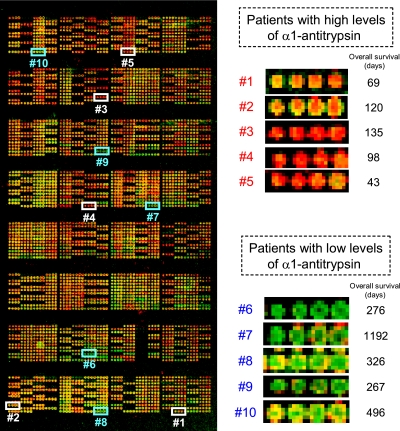
The relative levels of α1-antitrypsin and α1-antichymotrypsin in plasma or serum samples obtained from 304 patients with advanced pancreatic cancer prior to gemcitabine treatment (including 60 patients used in 2DICAL) were measured using reverse-phase protein microarrays (Fig. 3). Quadruplicate spots for representative patients with high and low levels of α1-antitrypsin are shown in Fig. 3. There were no differences between plasma (n = 252) and serum (n = 52) with regard to the levels of α1-antitrypsin and α1-antichymotrypsin (plasma versus serum (mean ± S.D.): α1-antitrypsin, 2.10 ± 0.19 versus 2.16 ± 0.16, p = 0.06; α1-antichymotrypsin, 4.44 ± 0.10 versus 4.45 ± 0.08, p = 0.67).
Fig. 3.
Left, representative reverse-phase protein microarray slide stained with anti-α1-antitrypsin antibody. Right, samples were randomly assigned, and quadruplicate spots of representative patients with high and low levels of α1-antitrypsin were extracted.
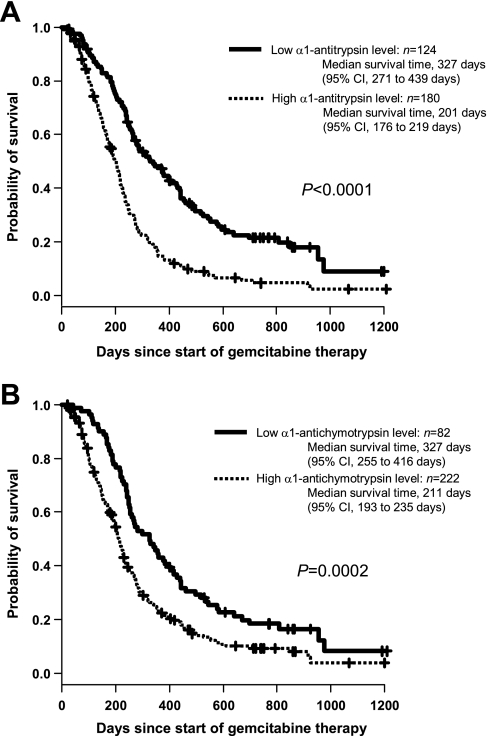
Although the levels of α1-antitrypsin and α1-antichymotrypsin were not mutually correlated (Pearson's r = 0.274), either level showed a significant correlation with overall survival (Table II). When the most optimal cutoff value was determined by maximally selected analysis, the median survival time of patients with high levels of α1-antitrypsin (>2.09 arbitrary units) was significantly shorter than that of patients with low levels (≤2.09) (201 days (95% CI, 176–219 days) versus 327 days (95% CI, 271–439 days), log rank p = 2.26 × 10−9; Fig. 4A). Similarly, the median survival time was significantly shorter in patients with α1-antichymotrypsin levels of >4.41 (211 days (95% CI, 193 to 235 days)) than in those with levels of ≤4.41 (327 days (95% CI, 255–416 days)) (p = 2.02 × 10−4; Fig. 4B). Even when the 60 patients used for 2DICAL were excluded, the differences in survival separated by α1-antitrypsin and α1-antichymotrypsin levels were still significant (supplemental Fig. S2, A and B). However, the level of either α1-antitrypsin or α1-antichymotrypsin was not associated with tumor response (Spearman's ρ = 0.090 and ρ = 0.017, respectively). The increased level of α1-antitrypsin in 58 patients who subsequently developed progressive diseases was statistically significant (p = 0.020; supplemental Fig. S3) but quite modest, confirming that it is not a predictive biomarker of tumor response.
Table II. Univariate and multivariate Cox regression analyses of overall survival since the start of gemcitabine therapy (n = 304).
Factors except sex are regarded as continuous variables. A forward stepwise selection based on Akaike's information criterion was used to select parameters for multivariate analysis. p values of <0.050 are shown in bold. AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase.
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| Hazard ratioa (95% CI) | p | Hazard ratioa (95% CI) | p | |
| Age (years) | 0.99 (0.98–1.01) | 0.380 | ||
| Female sex (vs. male) | 1.07 (0.83–1.38) | 0.610 | ||
| ECOG performance status | 1.49 (1.22–1.80) | <0.001 | 1.36 (1.11–1.67) | 0.003 |
| Body surface area (m2) | 0.70 (0.33–1.50) | 0.360 | ||
| Leukocytes | 1.08 (1.05–1.11) | <0.0001 | 1.04 (1.00–1.08) | 0.066 |
| Platelets | 1.07 (0.90–1.28) | 0.450 | ||
| Hemoglobin (g/dl) | 0.93 (0.85–1.01) | 0.098 | ||
| Albumin (g/dl) | 0.61 (0.45–0.82) | 0.001 | ||
| Creatinine (mg/dl) | 1.13 (0.60–2.14) | 0.700 | ||
| AST (IU/liter) | 1.01 (1.00–1.01) | <0.001 | ||
| ALT (IU/liter) | 1.00 (1.00–1.01) | 0.033 | ||
| ALP | 1.09 (1.06–1.11) | <0.0001 | 1.07 (1.05–1.10) | <0.0001 |
| α1-Antitrypsinb | 5.92 (3.09–11.37) | <0.0001 | 3.66 (1.89–7.11) | 0.0001 |
| α1-Antichymotrypsinb | 11.60 (2.69–50.01) | 0.001 | ||
| Clinical stage IVac (vs. IVb) | 1.10 (0.85–1.38) | 0.453 | ||
a Hazard ratios are per 1,000/mm3 increase for leukocytes, per 10 × 104/mm3 increase for platelets, and per 100 units/liter increase for ALP. Hazard ratios for other continuous variables are per 1 unit increase for each variable.
b Logarithmic variable determined by reverse-phase protein microarray.
c According to Ref. 23.
Fig. 4.
Kaplan-Meier plots of overall survival according to α1-antitrypsin (A) and α1-antichymotrypsin (B) levels.
Construction and Validation of Model Predicting Overall Survival Time
Univariate Cox regression analysis revealed that ECOG performance status and laboratory values including leukocyte count, albumin, aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase, α1-antitrypsin, and α1-antichymotrypsin were correlated with overall survival of the 304 patients (p < 0.05; Table II). Because none of the parameters were able to predict survival outcome satisfactorily when used individually (data not shown), we attempted to construct a multivariate predictive model for estimation of overall survival. We searched for parameters using a forward stepwise selection procedure by AIC from all the clinical and laboratory data listed in Table II (available for all 304 cases) and found that a combination of α1-antitrypsin, alkaline phosphatase, leukocyte count, and ECOG performance status provided the lowest AIC value. We also searched for parameters using a backward elimination algorithm and found that this identified the same combination of factors as that selected by a forward stepwise procedure. The base-line α1-antitrypsin level was the second most significant contributor to the model (Table II). The prediction model using this combination of parameters was significantly compromised when the level of α1-antitrypsin was excluded (Δχ2 = 14.12, df = 1, p = 0.0002, likelihood ratio test).
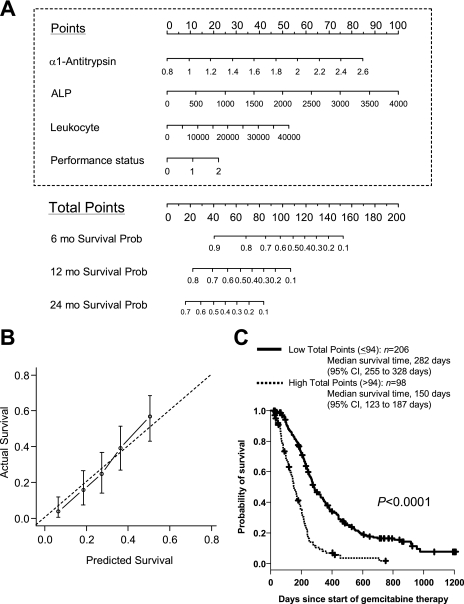
Based on the results of multivariate Cox regression analysis, we constructed a scoring system (nomogram) in which the values of the four parameters (α1-antitrypsin, alkaline phosphatase, leukocyte count, and ECOG performance status) were integrated into a single score (total point) to estimate the survival outcome (Fig. 5A). The accuracy of the nomogram for prognostication was internally validated. The bootstrap-corrected concordance index was 0.672, and the calibration curve demonstrated good agreement between the predicted and observed outcomes (Fig. 5B). It was possible to estimate high risk patients by calculating the total points using the nomogram. The median survival time was 150 days (95% CI, 123–187 days) for patients with a total point score of >94 (n = 98) and 282 days (95% CI, 255–328 days) for patients with a score of ≤94 (n = 206), and the difference was significant (p = 2.00 × 10−15, log rank test; Fig. 5C). Even when the 60 patients used for 2DICAL analyses were excluded from the total points calculation, the difference was still significant (p = 5.23 × 10−10; supplemental Fig. S2C). The median survival time was 171 days (95% CI, 147–205 days) for patients with a score of >92 (n = 83) and 270 days (95% CI, 243–299 days) for patients with a score of ≤92 (n = 161). The cutoff value that optimally segregated patients into subgroups with a poor and good prognosis was determined by using the maximally selected statistics.
Fig. 5.
A, nomogram for estimating the probability (Prob) of survival 6, 12, and 24 months (mo) after gemcitabine treatment. See supplemental Fig. S4 and its legend for details of usage. B, calibration curve demonstrating the correlation between predicted and actual survival at 12 months after gemcitabine treatment. Bars represent 95% CI. C, Kaplan-Meier plots of overall survival according to total points. ALP, alkaline phosphatase.
DISCUSSION
Currently, no diagnostic tool has been established for stratifying patients with advanced pancreatic cancer according to their likelihood of obtaining a survival benefit from gemcitabine treatment. Because some high risk patients may achieve prolonged survival through modification (or even withdrawal) of therapeutic protocols, a diagnostic method that can accurately identify such patients is necessary. We first compared the plasma proteome of two groups of patients who showed distinct clinical courses after receiving the same gemcitabine protocol (Fig. 1) and found that individuals who showed poor clinical courses had shown high base-line levels of plasma α1-antitrypsin and α1-antichymotrypsin (Figs. 1B and 2A). α1-Antitrypsin is an abundant plasma protein that usually cannot be measured by MS. However, antibody-based protein depletion (30) allowed us to accentuate the differences in α1-antitrypsin levels.
The results obtained by 2DICAL were then validated in a 5-fold larger cohort using a different methodology: high density reverse-phase protein microarray (Figs. 3 and 4 and Table II). Reverse-phase protein microarray is an emerging proteomics technology capable of validating new biomarkers because of its overwhelmingly high throughput (31, 32). Furthermore, reverse-phase protein microarrays require significantly smaller amounts of clinical samples for quantification than established clinical tests, such as ELISA. The prognostic significance of α1-antitrypsin was further supported by multivariate survival analysis with stepwise covariate selection. The level of α1-antitrypsin was selected as the second most significant factor following alkaline phosphatase (Table II), but α1-antichymotrypsin was not selected. To derive clinical applicability from the above findings, we constructed a model (nomogram) including α1-antitrypsin to estimate the survival period of pancreatic cancer patients (Fig. 5A), and its significance was internally validated (Fig. 5B). One previous study has demonstrated a correlation between an increased serum level of α1-antitrypsin and short survival in patients treated surgically for pancreatic cancer (33). Although the number of cases examined was small (n = 44), the results support our present findings.
α1-Antitrypsin and α1-antichymotrypsin are members of the serine protease inhibitor (serpin) superfamily that plays key roles in the regulation of inflammatory cascades (34, 35). α1-Antitrypsin and α1-antichymotrypsin interact mainly with neutrophil elastase and neutrophil cathepsin G, respectively, and inhibit their protease activities (36). A protease-to-protease inhibitor imbalance in patients with genetic α1-antitrypsin deficiency is reported to confer a higher risk of chronic pancreatitis (37). However, the serum level of α1-antitrypsin in patients with pancreatic cancer varied significantly from case to case, and its clinical significance has remained unclear. We showed that increased concentrations of α1-antitrypsin and α1-antichymotrypsin in plasma/serum correlated with poor survival, indicating that patients with poor outcomes have lower base-line protease activities than those with favorable outcomes. How such a protease imbalance affects the progression of pancreatic cancer awaits further clarification in future studies.
In conclusion, we identified a prognostic biomarker potentially useful for selecting high risk patients with advanced pancreatic cancer who are unlikely to gain adequate survival benefit from the standard treatment. This may be of great clinical importance, especially when an alternative therapeutic option becomes available for patients with advanced pancreatic cancer in the future. However, the level of α1-antitrypsin was not significantly correlated with the efficacy of gemcitabine, indicating that it may reflect the natural course of pancreatic cancer irrespective of treatment. Therefore, an independent prospective validation study will be definitely necessary to confirm the universality of the present findings. The absolute concentration of α1-antitrypsin can be measured by nephelometry, but this measurement requires a larger sample volume than reverse-phase microarrays and for this reason could not be performed in this study. While bearing all these limitations in mind, the present findings may not only help to stratify patients with pancreatic cancer but also provide novel insights into the molecular mechanisms behind the malignant progression of this neoplasm, possibly leading to the development of novel therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Ayako Igarashi, Tomoko Umaki, and Yuka Nakamura for technical assistance.
* This work was supported by the “Program for Promotion of Fundamental Studies in Health Sciences” conducted by the National Institute of Biomedical Innovation of Japan and the “Third-Term Comprehensive Control Research for Cancer” and “Research on Biological Markers for New Drug Development” conducted by the Ministry of Health and Labor of Japan.
 This article contains supplemental Figs. S1–S4 and Table S1.
This article contains supplemental Figs. S1–S4 and Table S1.
1 The abbreviations used are:
- 2DICAL
- two-dimensional image converted analysis of liquid chromatography and mass spectrometry
- AIC
- Akaike's information criterion
- CC
- correlation coefficient
- CI
- confidence interval
- CV
- coefficient of variance
- ECOG
- Eastern Cooperative Oncology Group
- NCC
- National Cancer Center
- ID
- identification
- FDR
- false discovery rate.
REFERENCES
- 1.Honda K., Hayashida Y., Umaki T., Okusaka T., Kosuge T., Kikuchi S., Endo M., Tsuchida A., Aoki T., Itoi T., Moriyasu F., Hirohashi S., Yamada T. (2005) Possible detection of pancreatic cancer by plasma protein profiling. Cancer Res 65, 10613–10622 [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society (2007) Cancer Facts and Figures 2007, pp. 16–17, American Cancer Society, Atlanta, GA [Google Scholar]
- 3.Ministry of Health, Labour and Welfare (2009) Japanese Government: Vital Statistics of Japan, Ministry of Health, Labour and Welfare, Tokyo [Google Scholar]
- 4.Rosewicz S., Wiedenmann B. (1997) Pancreatic carcinoma. Lancet 349, 485–489 [DOI] [PubMed] [Google Scholar]
- 5.Nieto J., Grossbard M. L., Kozuch P. (2008) Metastatic pancreatic cancer 2008: is the glass less empty? Oncologist 13, 562–576 [DOI] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network (2009) Clinical Practice Guidelines in Oncology: Pancreatic Adenocarcinoma, Vol. 1, National Comprehensive Cancer Network, Fort Washington, PA: [DOI] [PubMed] [Google Scholar]
- 7.Burris H. A., 3rd, Moore M. J., Andersen J., Green M. R., Rothenberg M. L., Modiano M. R., Cripps M. C., Portenoy R. K., Storniolo A. M., Tarassoff P., Nelson R., Dorr F. A., Stephens C. D., Von Hoff D. D. (1997) Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol 15, 2403–2413 [DOI] [PubMed] [Google Scholar]
- 8.Matsubara J., Ono M., Negishi A., Ueno H., Okusaka T., Furuse J., Furuta K., Sugiyama E., Saito Y., Kaniwa N., Sawada J., Honda K., Sakuma T., Chiba T., Saijo N., Hirohashi S., Yamada T. (2009) Identification of a predictive biomarker for hematologic toxicities of gemcitabine. J. Clin. Oncol 27, 2261–2268 [DOI] [PubMed] [Google Scholar]
- 9.Stocken D. D., Hassan A. B., Altman D. G., Billingham L. J., Bramhall S. R., Johnson P. J., Freemantle N. (2008) Modelling prognostic factors in advanced pancreatic cancer. Br. J. Cancer 99, 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louvet C., Labianca R., Hammel P., Lledo G., Zampino M. G., André T., Zaniboni A., Ducreux M., Aitini E., Taïeb J., Faroux R., Lepere C., de Gramont A. (2005) Gemcitabine in combination with oxaliplatin compared with gemcitabine alone in locally advanced or metastatic pancreatic cancer: results of a GERCOR and GISCAD phase III trial. J. Clin. Oncol 23, 3509–3516 [DOI] [PubMed] [Google Scholar]
- 11.Storniolo A. M., Enas N. H., Brown C. A., Voi M., Rothenberg M. L., Schilsky R. (1999) An investigational new drug treatment program for patients with gemcitabine: results for over 3000 patients with pancreatic carcinoma. Cancer 85, 1261–1268 [PubMed] [Google Scholar]
- 12.Hanash S. (2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi U., Nakayama R., Honda K., Ichikawa H., Hasegawa T., Shitashige M., Ono M., Shoji A., Sakuma T., Kuwabara H., Shimada Y., Sasako M., Shimoda T., Kawai A., Hirohashi S., Yamada T. (2008) Distinct gene expression-defined classes of gastrointestinal stromal tumor. J. Clin. Oncol 26, 4100–4108 [DOI] [PubMed] [Google Scholar]
- 14.Hayashida Y., Honda K., Osaka Y., Hara T., Umaki T., Tsuchida A., Aoki T., Hirohashi S., Yamada T. (2005) Possible prediction of chemoradiosensitivity of esophageal cancer by serum protein profiling. Clin. Cancer Res 11, 8042–8047 [DOI] [PubMed] [Google Scholar]
- 15.Taguchi F., Solomon B., Gregorc V., Roder H., Gray R., Kasahara K., Nishio M., Brahmer J., Spreafico A., Ludovini V., Massion P. P., Dziadziuszko R., Schiller J., Grigorieva J., Tsypin M., Hunsucker S. W., Caprioli R., Duncan M. W., Hirsch F. R., Bunn P. A., Jr., Carbone D. P. (2007) Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J. Natl. Cancer Inst 99, 838–846 [DOI] [PubMed] [Google Scholar]
- 16.Ono M., Shitashige M., Honda K., Isobe T., Kuwabara H., Matsuzuki H., Hirohashi S., Yamada T. (2006) Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 1338–1347 [DOI] [PubMed] [Google Scholar]
- 17.Negishi A., Ono M., Handa Y., Kato H., Yamashita K., Honda K., Shitashige M., Satow R., Sakuma T., Kuwabara H., Omura K., Hirohashi S., Yamada T. (2009) Large-scale quantitative clinical proteomics by label-free liquid chromatography and mass spectrometry. Cancer Sci 100, 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha Lima C. M., Green M. R., Rotche R., Miller W. H., Jr., Jeffrey G. M., Cisar L. A., Morganti A., Orlando N., Gruia G., Miller L. L. (2004) Irinotecan plus gemcitabine results in no survival advantage compared with gemcitabine monotherapy in patients with locally advanced or metastatic pancreatic cancer despite increased tumor response rate. J. Clin. Oncol 22, 3776–3783 [DOI] [PubMed] [Google Scholar]
- 19.Van Cutsem E., Vervenne W. L., Bennouna J., Humblet Y., Gill S., Van Laethem J. L., Verslype C., Scheithauer W., Shang A., Cosaert J., Moore M. J. (2009) Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J. Clin. Oncol 27, 2231–2237 [DOI] [PubMed] [Google Scholar]
- 20.Heinemann V., Quietzsch D., Gieseler F., Gonnermann M., Schönekäs H., Rost A., Neuhaus H., Haag C., Clemens M., Heinrich B., Vehling-Kaiser U., Fuchs M., Fleckenstein D., Gesierich W., Uthgenannt D., Einsele H., Holstege A., Hinke A., Schalhorn A., Wilkowski R. (2006) Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J. Clin. Oncol 24, 3946–3952 [DOI] [PubMed] [Google Scholar]
- 21.Moore M. J., Goldstein D., Hamm J., Figer A., Hecht J. R., Gallinger S., Au H. J., Murawa P., Walde D., Wolff R. A., Campos D., Lim R., Ding K., Clark G., Voskoglou-Nomikos T., Ptasynski M., Parulekar W. (2007) Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol 25, 1960–1966 [DOI] [PubMed] [Google Scholar]
- 22.Herrmann R., Bodoky G., Ruhstaller T., Glimelius B., Bajetta E., Schüller J., Saletti P., Bauer J., Figer A., Pestalozzi B., Köhne C. H., Mingrone W., Stemmer S. M., Tàmas K., Kornek G. V., Koeberle D., Cina S., Bernhard J., Dietrich D., Scheithauer W. (2007) Gemcitabine plus capecitabine compared with gemcitabine alone in advanced pancreatic cancer: a randomized, multicenter, phase III trial of the Swiss Group for Clinical Cancer Research and the Central European Cooperative Oncology Group. J. Clin. Oncol 25, 2212–2217 [DOI] [PubMed] [Google Scholar]
- 23.Japan Pancreas Society (ed) (2002) General Rules for the Study of Pancreatic Cancer, 5th Ed., Kanehara, Tokyo [Google Scholar]
- 24.Sugiyama E., Kaniwa N., Kim S. R., Kikura-Hanajiri R., Hasegawa R., Maekawa K., Saito Y., Ozawa S., Sawada J., Kamatani N., Furuse J., Ishii H., Yoshida T., Ueno H., Okusaka T., Saijo N. (2007) Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J. Clin. Oncol 25, 32–42 [DOI] [PubMed] [Google Scholar]
- 25.Honda K., Yamada T., Hayashida Y., Idogawa M., Sato S., Hasegawa F., Ino Y., Ono M., Hirohashi S. (2005) Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 128, 51–62 [DOI] [PubMed] [Google Scholar]
- 26.Idogawa M., Yamada T., Honda K., Sato S., Imai K., Hirohashi S. (2005) Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/beta-catenin complex. Gastroenterology 128, 1919–1936 [DOI] [PubMed] [Google Scholar]
- 27.Hothorn T., Zeileis A. (2008) Generalized maximally selected statistics. Biometrics 64, 1263–1269 [DOI] [PubMed] [Google Scholar]
- 28.Wang S. J., Fuller C. D., Kim J. S., Sittig D. F., Thomas C. R., Jr., Ravdin P. M. (2008) Prediction model for estimating the survival benefit of adjuvant radiotherapy for gallbladder cancer. J. Clin. Oncol 26, 2112–2117 [DOI] [PubMed] [Google Scholar]
- 29.Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L., Harvie G., Feitelson J. S., Gramatikoff K., Herold D. A., Allen D. L., Amunngama R., Hagler R. A., Pisano M. R., Zhang W. W., Fang X. (2005) Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics 5, 3314–3328 [DOI] [PubMed] [Google Scholar]
- 31.Grote T., Siwak D. R., Fritsche H. A., Joy C., Mills G. B., Simeone D., Whitcomb D. C., Logsdon C. D. (2008) Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19–9 levels in pancreatic cancer. Proteomics 8, 3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wulfkuhle J. D., Liotta L. A., Petricoin E. F. (2003) Proteomic applications for the early detection of cancer. Nat. Rev. Cancer 3, 267–275 [DOI] [PubMed] [Google Scholar]
- 33.Trichopoulos D., Tzonou A., Kalapothaki V., Sparos L., Kremastinou T., Skoutari M. (1990) Alpha 1-antitrypsin and survival in pancreatic cancer. Int. J. Cancer 45, 685–686 [DOI] [PubMed] [Google Scholar]
- 34.Lomas D. A., Mahadeva R. (2002) Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J. Clin. Investig 110, 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoller J. K., Aboussouan L. S. (2005) Alpha1-antitrypsin deficiency. Lancet 365, 2225–2236 [DOI] [PubMed] [Google Scholar]
- 36.Beatty K., Bieth J., Travis J. (1980) Kinetics of association of serine proteinases with native and oxidized alpha-1-proteinase inhibitor and alpha-1-antichymotrypsin. J. Biol. Chem 255, 3931–3934 [PubMed] [Google Scholar]
- 37.Rabassa A. A., Schwartz M. R., Ertan A. (1995) Alpha 1-antitrypsin deficiency and chronic pancreatitis. Dig. Dis. Sci 40, 1997–2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.