Abstract
Genes involved in cancer susceptibility and progression can serve as templates for searching protein networks for novel cancer genes. To this end, we introduce a general network searching method, MaxLink, and apply it to find and rank cancer gene candidates by their connectivity to known cancer genes. Using a comprehensive protein interaction network, we searched for genes connected to known cancer genes. First, we compiled a new set of 812 genes involved in cancer, more than twice the number in the Cancer Gene Census. Their network neighbors were then extracted. This candidate list was refined by selecting genes with unexpectedly high levels of connectivity to cancer genes and without previous association to cancer. This produced a list of 1891 new cancer candidates with up to 55 connections to known cancer genes. We validated our method by cross-validation, Gene Ontology term bias, and differential expression in cancer versus normal tissue. An example novel cancer gene candidate is presented with detailed analysis of the local network and neighbor annotation. Our study provides a ranked list of high priority targets for further studies in cancer research. Supplemental material is included.
The function of a protein can be expressed in terms of its interactions with other molecules. All interactions between all proteins define the “protein interactome,” i.e. the complete interaction network of the proteins of an organism. These networks form the backbone of molecular pathways and cellular processes. Thus, the construction of interaction networks will shed light on many aspects of the dynamic and interactive function of human proteins.
Several efforts in reconstructing the human interactome are ongoing. Interactions may be measured directly with high throughput yeast two-hybrid or pulldown assays (1, 2). Experimental interactions have been collected from multiple sources to build large interaction networks (3–5). The network can be augmented considerably by inferred interactions either in the same or from other species (6–10). The largest predicted human interactome is currently provided by FunCoup (11), which uses eight types of evidence and transfers interactions extensively from model organism orthologs.
The development of new therapeutics and diagnostics rely on the understanding of disease mechanisms. Therefore, the identification of novel disease-associated genes is of great importance. Disease genes have traditionally been found by genetic linkage analysis or gene association studies, but this is very time-consuming and costly and often fails due to lack of data. Particularly for complex diseases involving many genes, these methods are unreliable (12).
Bioinformatics methods can be used to accelerate disease gene discovery either based on gene annotation and sequence features (13, 14) or based on network analysis (15–19). The network-based methods normally connect gene networks with phenotype networks to infer gene-disease relationships. These works, however, are limited to using only direct interaction data and/or were only applied to rank a short list of candidate genes in a genomic interval.
Here we describe a new generic network-based approach, MaxLink, for predicting novel candidate members to known biomolecular processes and pathways. A typical application is the identification of new disease genes based on a set of known disease genes. We applied MaxLink to the human interactome generated by FunCoup to screen for new cancer genes. To seed the screen, we compiled a list of 812 known cancer genes, 364 from the Cancer Gene Census (20) and 448 genes from text mining.
MaxLink assigns a score to every new candidate gene based on the number of links to a seed set. We show that the maxlink score is a useful indicator of candidate reliability by three types of validations: cross-validation, differential cancer expression, and GO1 term analysis. The screen resulted in nearly 2000 candidates of which nearly 200 are connected to over 10 known cancer genes. These genes have, to our knowledge, no clear former evidence supporting association with cancer. However, their network connection to cancer genes makes them worth particular focus when developing biomarkers or studying oncogenesis. As the candidate list is long, it makes sense to explore the top ranking genes first.
MATERIALS AND METHODS
Retrieval of Known Cancer Genes
The input data set of known cancer genes was collected from Swiss-Prot (21) and from the Cancer Gene Census (20). The Swiss-Prot genes were identified by searching annotations in the CC field, which represents curated annotations and includes a subcategory for annotations indicating disease involvement. The disease annotations of the CC field were matched against cancer-specific terms (see supplemental Table 4), and genes for which a match could be found were added to the set of known cancer genes. Genes and matching keywords are detailed in supplemental Table 1.
GO Analysis of Known Cancer Genes
The Gene Ontology functional term analysis was done using the amiGO web site. Enrichment analysis of terms in the major cluster (348 genes) versus UniProtKB (20,740 genes) resulted in a total of 231 terms with p < 10−2. This list was abbreviated by requiring p < 10−10 and enrichment >5, resulting in 34 GO terms (supplemental Table 2).
Network-based Identification of Candidate Genes
We used the human FunCoup protein network (11) to identify network neighbors to the previously retrieved input genes. Only links with a confidence value >0.75 were considered. Each candidate gene was assigned a maxlink score for ranking that equals the number of linked known cancer genes.
Annotation Filter
To identify genes with possible cancer annotations, the complete UniProt (22) DE, KW, CC, and FT fields as well as reference titles were searched for cancer-specific text terms (supplemental Table 4). Genes with a match were excluded from the candidates list. Additionally, genes with a gene identifier not found in the current version (version 51) of Ensembl (23) were also excluded.
Connectivity Filter
If the majority of a candidate's connections were to non-cancer genes, it was deemed of low cancer specificity and was rejected. For this analysis, we divided all genes into two sets: 1) the known cancer genes plus all genes with any cancer annotation (see “Annotation Filter” above) and 2) all other genes. The gene counts of these sets were 4953 and 12,198. Consequently, genes exhibiting over 2.46 times more links to genes not associated with cancer than to the known cancer genes were removed.
Differential Expression in Human Protein Atlas
We devised a score (differential expression score (DE)) for differential protein expression levels in 18 different cancer types relative to their normal tissue counterparts (see Table I) from the 3.0 version of the Human Protein Atlas. DE was calculated by subtracting the average expression in a normal tissue from the average expression in the corresponding cancer tissue for each gene and tissue. To avoid tissue-specific biases, raw DE values for each tissue were replaced by Z-scores based on the expression distribution of each tissue. A Z-score of 1 represents one standard deviation above the mean. Finally, the total DE for each gene was calculated by taking the average of all absolute DE values for all 18 tissues.
Table I. Eighteen corresponding cancer and normal tissues in HPA.
Differential expression in cancer was measured by comparing the expression in the cancers with the corresponding normal tissues in the HPA database. Some of the tissues may have been renamed in the current on-line HPA database.
| Cancer type | Normal tissue counterpart |
|---|---|
| Breast cancer | Breast |
| Cervical cancer | Cervix uterine |
| Colorectal cancer | Colon and rectum |
| Endometrial cancer | Endometrium |
| Head and neck cancer | Oral mucosa and salivary gland |
| Liver cancer | Liver |
| Lung cancer | Lung and bronchus |
| Stomach cancer | Stomach |
| Malignant glioma | Hippocampus and cerebral cortex (non-neuronal cells) |
| Malignant lymphoma | Lymph node and spleen |
| Malignant melanoma | Skin |
| Ovarian cancer | Ovary |
| Pancreatic cancer | Pancreas |
| Prostate cancer | Prostate |
| Skin cancer | Skin |
| Testis cancer | Testis |
| Thyroid cancer | Thyroid gland |
| Urothelial cancer | Urinary bladder |
Analysis of Cancer-associated GO Terms
GO terms for all genes were retrieved from Ensembl via BIOMART (23), and the terms were expanded to include all higher level terms. All GO terms for the set of known cancer genes were tested for significant enrichment (fold change) with a hypergeometric test. The set of cancer-associated GO terms was then tested for significance (p < 0.05) for subsets of the candidates composed of all genes having a number of linked known cancer genes above or equal to a cutoff defining that subset. Relative fold changes for subsets were subsequently calculated for each GO term by taking the logarithm of the subset fold change divided by the fold change of the same term for the known cancer genes.
RESULTS
We have developed an analysis pipeline to identify and rank candidate cancer genes based on their connectivity to known cancer genes in the FunCoup network. By “known cancer gene,” we mean any gene with clear evidence for cancer involvement. To analyze the interconnectedness and clustering of the known cancer genes, we first explored their network topology. Then, using them as seeds, we extracted candidate novel cancer genes and refined this list by applying quality filters. Finally, to validate our approach, we used three types of independent validation tests: cross-validation, enrichment of cancer GO terms, and differential expression in cancer versus normal tissue.
New Compilation of Known Cancer Genes
Our approach starts with collecting known cancer genes. In a previous survey, the Cancer Gene Census, Futreal et al. (20) identified 364 cancer genes. By text mining Swiss-Prot for genes annotated to be involved in cancer, we identified 703 genes. Merging this list with the Cancer Gene Census resulted in 812 unique cancer genes (supplemental Table 1).
To analyze this set of genes in terms of network structure, we examined how they cluster into interconnected modules. This revealed one major component with 348 members, 12 small clusters with 2–9 members, and 429 singletons as shown in Fig. 1. Thus, 43% of the known cancer genes were interconnected in a single subnetwork that should represent processes central to cancer. To verify this, we analyzed enrichment of functional annotation terms in the Gene Ontology database (24) relative to all human genes. We observed strong enrichment (>5-fold enrichment, p < 10−10) for terms such as DNA repair and replication, cell cycle regulation, and apoptosis (supplemental Table 2). This is well in line with known cancer-associated processes.
Fig. 1.
Network layout of known cancer genes. The connections between genes represent links in FunCoup with confidence >0.75. The figure was made using Cytoscape (28). For a high resolution vector picture, see supplemental Fig. 1.
Screen for Candidate Cancer Genes
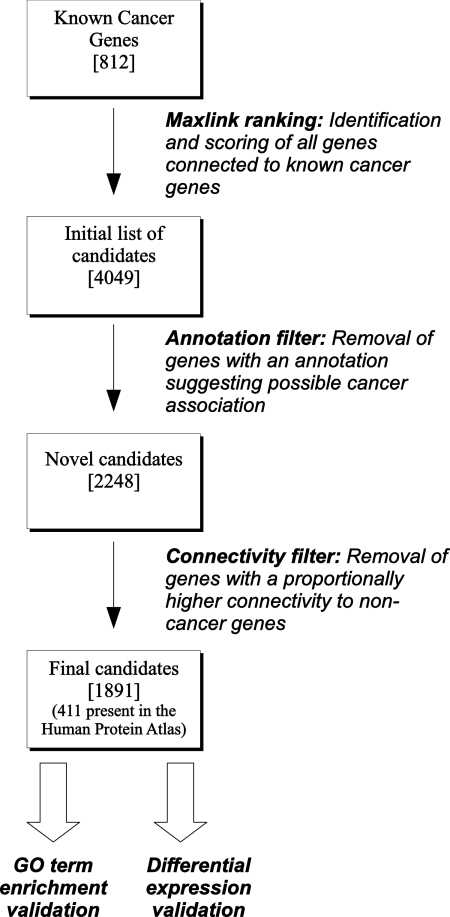
The FunCoup network was used to retrieve 4049 potential candidates connected to known cancer genes by high confidence links. Because our aim was to find genes previously not associated with cancer, the list was further refined by a number of filters. In the first step, 1511 genes that had any annotation suggesting a potential association with cancer were removed from the list. Because FunCoup was built using data sets of which some were linked to earlier versions of Ensembl, 254 genes were removed to ensure that the candidates are in sync with the current version. This constitutes a broad filter and would likely remove genes with only spurious cancer association. Hublike genes might be spuriously linked to many known cancer genes solely because they have many links and not because they are involved in cancer. Thus, 393 genes were removed in the second step because they had fewer links to known cancer genes than expected by chance given their connectivity in the entire network. Such genes may have been found simply because they are highly connected and not because of a preferential association to the known cancer genes. A schematic representation of the analysis work flow is shown in Fig. 2. After all filters, a final list of 1891 candidates remained with a maxlink score (links to known cancer genes) between 55 and 1 (see supplemental Table 3).
Fig. 2.
Schematic representation of analysis work flow. The number of genes remaining after each step is shown within brackets.
Validation by Cross-validation
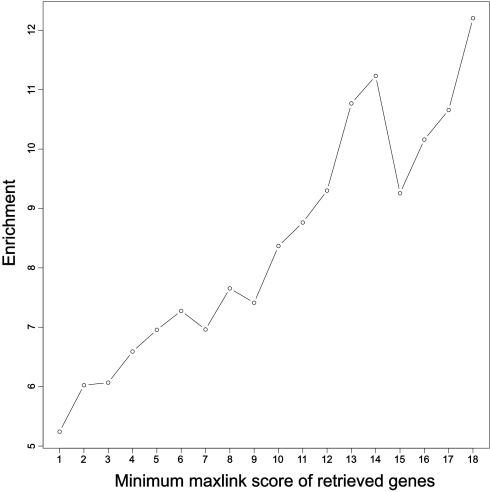
If our method works well, it should be able to detect the known cancer genes in a cross-validation test. We ran MaxLink five times, leaving out 20% of the known cancer genes each time. By doing so, we were able to identify 41.7% of the removed genes on average. However, only 47% of the known cancer genes had links to other input genes; thus, the obtained retrieval is close to the theoretical maximum restricted by the network. As we cannot assess false positives directly, we instead looked at enrichment of the removed known cancer genes among the retrieved genes, i.e. their frequency in the retrieved set relative to their frequency in the entire database. The average enrichment for all removed genes was more than 5-fold (p < 10−25). However, this increased to over 12-fold for the genes with highest maxlink score (see Fig. 3).
Fig. 3.
Enrichment of known cancer genes in cross-validation testing of MaxLink method. The enrichment is the frequency of cancer genes in the retrieved set relative to their frequency in the entire database. Overall, the enrichment of cancer genes with one or more links is just above 5. Restricting the retrieved set to genes with a higher maxlink score produces a proportionally increased enrichment. The enrichment levels at high maxlink scores are somewhat variable due to small amounts of data.
Validation by Differential Cancer Expression
The Human Protein Atlas (HPA) (25) contains protein expression in both normal tissues and cancers taken from a large number of tissues. Using these data, we calculated a normal versus cancer DE for the 411 candidate cancer genes with expression data in HPA using all 18 cancer types.
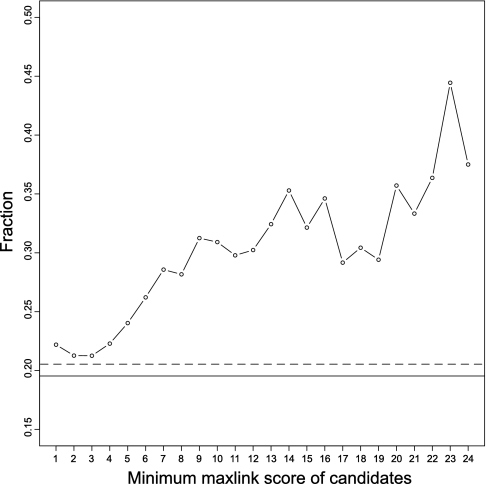
To examine the impact of a high maxlink score, we looked at the fraction of genes with DE above 1, i.e. when the differential expression exceeds one standard deviation on average for all cancer types. As seen in Fig. 4, this fraction increased for subsets of the candidates consisting of genes with a higher maxlink score and was considerably enriched compared with the known cancer genes and HPA as a whole. This indicates that candidates linked to a high number of known cancer genes are likely important for cancer.
Fig. 4.
Validation of method by differential cancer expression in HPA. The fraction of genes linked to a certain number of known cancer genes that is differentially expressed above one standard deviation is shown for subsets based on maxlink score. For comparison, the average DE values for all known cancer genes (dashed horizontal line) and all HPA genes (solid horizontal line) are shown.
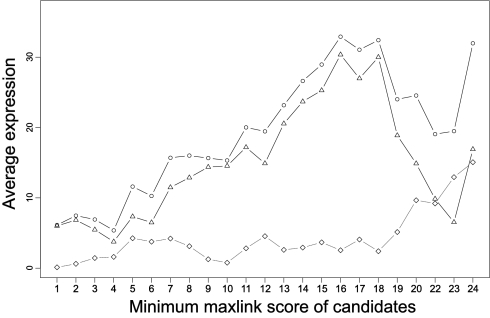
To investigate whether this trend is caused by a decrease of normal tissue expression or an increase in cancer expression, we plotted the absolute expression levels as a function of maxlink score (see Fig. 5). The overall trend is an increase of both normal and cancer tissue expression but with a relatively higher increase in cancers. Thus, the MaxLink approach can enrich genes differentially expressed in a wide range of cancers, and the maxlink score is a useful indicator of cancer relevance.
Fig. 5.
Relative protein expression levels of candidate cancer genes in cancer and normal tissues compared with HPA as a whole. The average expression in both cancer (circles) and normal (triangles) tissues was calculated for candidate subsets, binned by maxlink score, and normalized by subtracting the average expression of all genes in HPA for cancer and normal tissues, respectively. The relative expression levels are not strongly correlated with maxlink score, but the difference between cancer and normal expression (diamonds) is. The prevalence of high differential expression at high maxlink scores, as seen in Fig. 4, thus cannot simply be explained by increased cancer expression or decreased normal expression. The expression levels are discrete as used by HPA (29): 1 represents none, 10 represents low, 50 represents moderate, and 250 represents high expression. Expression in both normal and cancer tissues is generally increased for genes with higher maxlink score but with a relatively higher increase for cancer tissues.
We noted that the average DE of the known cancer genes was only slightly higher than that of HPA. This can be explained by the fact that HPA was started with a strong cancer focus and is highly enriched for cancer genes.
Validation by GO Terms
If our candidate cancer genes would show the same GO term enrichment as the known cancer genes, this would give further support to their relevance in cancer. To investigate this, we retrieved all GO terms for the known cancer genes and tested for a significant enrichment. Of a total of 4281 terms, enrichment greater or equal to 2-fold was significant at the 0.05 level for 1716 terms.
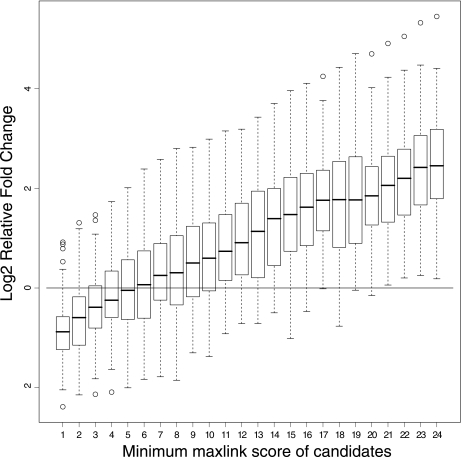
These cancer-associated GO terms were subsequently tested for enrichment in subsets of the candidate genes grouped by increasing numbers of links to known cancer genes. The average enrichment increased proportionally to the number of linked known cancer genes (Fig. 6), showing that genes more central in the cancer network are more functionally associated with cancer.
Fig. 6.
Validation of method by GO term enrichment. The relative fold change of cancer-related GO terms is shown for candidate cancer genes linked to a certain number of known cancer genes. The relative fold change of each cancer term is the base 2 logarithm of the fold change of the subset divided by the fold change in the known cancer genes. A relative fold change above 0 means that the cancer terms are more enriched in the candidate subset than in the set of known cancer genes. Candidates linked to more than five known cancer genes have a fold change of cancer terms on average greater than the known cancer genes.
Novel Candidate Cancer Genes
Our screen resulted in a list of 1891 novel candidate genes (supplemental Table 3). Given the above validations of the maxlink score as an indicator of cancer relevance, it makes most sense to focus on those candidates with the most linked known cancer genes. The list contains 185 candidates with 10 or more linked known cancer genes, and these should perhaps be seen as the most urgent targets for focused cancer studies.
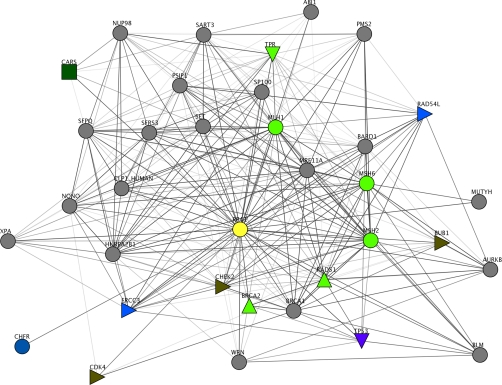
To illustrate how a candidate cancer gene may be analyzed further, we chose an example, RPA1 (UniProt accession number P27694), which is a DNA-binding subunit of replication protein A. It was functionally coupled to 34 known cancer genes (see Fig. 7).
Fig. 7.
FunCoup subnetwork of candidate cancer gene RPA1 surrounded by functionally coupled known cancer genes. RPA1 is the yellow circle, whereas the known cancer genes are colored/shaped according to KEGG (30) pathway membership. Note that KEGG contains a relatively small number of cancer pathways; hence, most genes are not assigned to any pathway (gray balls). All green genes are from cancer pathways, however. The figure was made using the jSquid applet (31).
Can we predict what cancer type RPA1 is most likely to cause or be associated with? According to HPA, RPA1 is expressed in all cancer types and has differential (|DE| > 1) expression compared with normal in seven tissues (colorectal, endometrial, head and neck, pancreatic, skin, testis, and urothelial cancer). Thus, even if RPA1 may play a more prominent role in some cancers, it is likely to be a universal cancer gene.
The network neighbors of RPA1 have diverse differential expression patterns, supporting the notion that it is not specific for a certain type of cancer. In Fig. 7, the KEGG pathway membership of the neighbors is indicated. Although this gives a very incomplete picture because of the low coverage of KEGG for cancer (for instance, breast cancer is absent), it does reveal several cancer types such as colorectal and pancreatic.
A literature search revealed that in mice RPA1 has been shown to cause defects in DNA double strand break repair, which can lead to leukemia (26). (This information is not present in UniProt.) In human, RPA1 is located in chromosomal region 17p13.3, which has been implicated in e.g. colorectal and breast cancer (26). These cancers had strong support by DE in HPA for both RPA1 and its neighbors as well as from annotation of many of the neighbors.
Our analysis based on HPA expression, the gene subnetwork, and literature reinforces the connection between RPA1 and cancer, lending support for the cancer types associated with the RPA1 locus but suggesting that it may cause cancer in any tissue. Although it was one of the top ranking novel cancer gene candidates, there is no mention of any cancer association in UniProt or HPA. However, the presented evidences support that it plays a central role in tumorigenesis.
DISCUSSION
We have described a general network-based approach for identifying and ranking candidate novel genes relevant to a process or disease and have applied it to find novel cancer genes. The validations carried out show that the ranked list produced by our method is enriched for true cancer genes.
Cancer is in this study treated as one disease. This is obviously a simplification but is based on the notion that cancers originating in different tissues are often caused by perturbations in the same pathways, for instance DNA repair, cell cycle regulation, or apoptosis. It is supported by the fact that the network of known cancer genes only formed one large cluster (Fig. 1), which did not show very distinct subclusters. Also, Goh et al. (27) showed that different cancers are often caused by the same genes. The attractiveness of this approach is that genes found in most cancers have a greater potential for diagnostic and therapeutic value.
Because of the modularity of the MaxLink pipeline, other diseases or processes can easily be investigated. A more traditional approach to disease gene hunting is linkage analysis where the gene associated with a disease is known to be found in a genomic interval that can contain in the order of a hundred genes. MaxLink could also be used to prioritize genes for such projects as long as a fair number of genes are already known for the disease in question. The main advantage compared with other methods would be the richness of evidence integrated in the FunCoup links. For short lists, it may be necessary to lower the cutoff compared with this study to obtain a reasonable amount of links.
Many of our candidate genes were supported as cancer genes by the HPA database. However, even if a gene does not have differential cancer/normal expression in HPA, this does not disprove its potential implication in cancer. The protein level changes associated with the tumor progression may be too subtle to detect with the HPA technology. However, the predicted functional coupling to a cancer pathway is still valid, and the gene in question may well turn out to be a useful marker or therapeutic target.
The “total differential expression” measure used here was only intended to investigate the validity of the MaxLink approach and not as a definite indicator of cancer relevance. Because we average across all cancer types, a gene differentially expressed in only one or a few cancer types might receive an unfairly low total DE value.
The main result of this study is the ranked list of novel cancer gene candidates. The 185 candidates connected to 10 or more known cancer genes are prime targets for new experiments that will lead the way to better understanding cancer. Some of the candidates with a lower maxlink score may also develop into important cancer biomarkers or targets, but there is a rationale for focusing on the high scoring genes. A high maxlink score is an indication that the candidate acts as a hub and plays a central role in the process and is more likely to be of widespread importance in many different tumors. On the other hand, such functions are typically also important for healthy tissue homeostasis and may be unsuitable as targets for inhibition. An exception to this would be hubs that act in parallel in healthy tissue but only one is functional in a tumor. Such a situation would make a hub an excellent cancer-specific drug target.
Supplementary Material
Acknowledgments
We thank Vincent Devloo, Olof Karlberg, and Andrey Alexeyenko for assistance and helpful discussions. We also thank the Swedish Human Proteome Resource program for access to the protein atlas data.
 This article contains supplemental Tables 1–4 and Fig. 1.
This article contains supplemental Tables 1–4 and Fig. 1.
1 The abbreviations used are:
- GO
- Gene Ontology
- DE
- differential expression score
- HPA
- Human Protein Atlas
- KEGG
- Kyoto Encyclopedia of Genes and Genomes.
REFERENCES
- 1.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 3.Bader G. D., Betel D., Hogue C. W. (2003) BIND: the biomolecular interaction network database. Nucleic Acids Res 31, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramani A. K., Bunescu R. C., Mooney R. J., Marcotte E. M. (2005) Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol 6, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peri S., Navarro J. D., Amanchy R., Kristiansen T. Z., Jonnalagadda C. K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T. K., Gronborg M., Ibarrola N., Deshpande N., Shanker K., Shivashankar H. N., Rashmi B. P., Ramya M. A., Zhao Z., Chandrika K. N., Padma N., Harsha H. C., Yatish A. J., Kavitha M. P., Menezes M., Choudhury D. R., Suresh S., Ghosh N., Saravana R., Chandran S., Krishna S., Joy M., Anand S. K., Madavan V., Joseph A., Wong G. W., Schiemann W. P., Constantinescu S. N., Huang L., Khosravi-Far R., Steen H., Tewari M., Ghaffari S., Blobe G. C., Dang C. V., Garcia J. G., Pevsner J., Jensen O. N., Roepstorff P., Deshpande K. S., Chinnaiyan A. M., Hamosh A., Chakravarti A., Pandey A. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13, 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehner B., Fraser A. G. (2004) A first-draft human protein-interaction map. Genome Biol 5, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K. R., Jurisica I. (2005) Online predicted human interactions database. Bioinformatics 21, 2076–2082 [DOI] [PubMed] [Google Scholar]
- 8.Persico M., Ceol A., Gavrila C., Hoffmann R., Florio A., Cesareni G. (2005) HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics 6, Suppl. 4, S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes D. R., Tomlins S. A., Varambally S., Mahavisno V., Barrette T., Kalyana-Sundaram S., Ghosh D., Pandey A., Chinnaiyan A. M. (2005) Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol 23, 951–959 [DOI] [PubMed] [Google Scholar]
- 10.Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexeyenko A., Sonnhammer E. L. (2009) Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res 19, 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emahazion T., Feuk L., Jobs M., Sawyer S. L., Fredman D., St Clair D., Prince J. A., Brookes A. J. (2001) SNP association studies in Alzheimer's disease highlight problems for complex disease analysis. Trends Genet 17, 407–413 [DOI] [PubMed] [Google Scholar]
- 13.Perez-Iratxeta C., Bork P., Andrade M. A. (2002) Association of genes to genetically inherited diseases using data mining. Nat. Genet 31, 316–319 [DOI] [PubMed] [Google Scholar]
- 14.Turner F. S., Clutterbuck D. R., Semple C. A. (2003) POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol 4, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George R. A., Liu J. Y., Feng L. L., Bryson-Richardson R. J., Fatkin D., Wouters M. A. (2006) Analysis of protein sequence and interaction data for candidate disease gene prediction. Nucleic Acids Res 34, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lage K., Karlberg E. O., Størling Z. M., Olason P. I., Pedersen A. G., Rigina O., Hinsby A. M., Tümer Z., Pociot F., Tommerup N., Moreau Y., Brunak S. (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol 25, 309–316 [DOI] [PubMed] [Google Scholar]
- 17.Oti M., van Reeuwijk J., Huynen M. A., Brunner H. G. (2008) Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics 9, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X., Jiang R., Zhang M. Q., Li S. (2008) Network-based global inference of human disease genes. Mol. Syst. Biol 4, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ideker T., Sharan R. (2008) Protein networks in disease. Genome Res 18, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futreal P. A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M. R. (2004) A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The UniProt Consortium (2008) The Universal Protein Resource (UniProt). Nucleic Acids Res 36, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bairoch A., Apweiler R., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., Yeh L. S. (2005) The universal protein resource (UniProt). Nucleic Acids Res 33, D154–D159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flicek P., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Eyre T., Fitzgerald S., Fernandez-Banet J., Gräf S., Haider S., Hammond M., Holland R., Howe K. L., Howe K., Johnson N., Jenkinson A., Kähäri A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A. J., Vogel J., White S., Wood M., Birney E., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Kasprzyk A., Proctor G., Smith J., Ureta-Vidal A., Searle S. (2008) Ensembl 2008. Nucleic Acids Res 36, D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7, 2019–2027 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Putnam C. D., Kane M. F., Zhang W., Edelmann L., Russell R., Carrión D. V., Chin L., Kucherlapati R., Kolodner R. D., Edelmann W. (2005) Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet 37, 750–755 [DOI] [PubMed] [Google Scholar]
- 27.Goh K. I., Cusick M. E., Valle D., Childs B., Vidal M., Barabási A. L. (2007) The human disease network. Proc. Natl. Acad. Sci. U.S.A 104, 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gry M. (2008) Ph.D. thesis, Royal Institute of Technology, Stockholm, Sweden [Google Scholar]
- 30.Kanehisa M., Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klammer M., Roopra S., Sonnhammer E. L. (2008) jSquid: a Java applet for graphical on-line network exploration. Bioinformatics 24, 1467–1468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.