Abstract
Autophagy functions in antiviral immunity. However, it is not yet known whether endogenous autophagy genes protect against viral disease in vertebrates. Using three different approaches to inactivate the autophagy gene Atg5 in virally-infected neurons, we found that loss of Atg5 function increases mouse susceptibility to lethal Sindbis virus CNS infection. This phenotype is associated with delayed clearance of viral proteins, increased accumulation of the cellular p62 adaptor protein, and increased cell death in neurons, but not with altered levels of CNS viral replication. In vitro, p62 interacts with Sindbis virus capsid protein and genetic knockdown of p62 blocks the targeting of viral capsid to autophagosomes. Moreover, p62 or autophagy gene knockdown increases viral capsid accumulation and accelerates virus-induced cell death without affecting virus replication. These results suggest a novel function for autophagy in mammalian antiviral defense: a cell-autonomous mechanism in which p62 adaptor-mediated autophagic viral protein clearance promotes cell survival.
INTRODUCTION
Macroautophagy (herein referred to as autophagy) is an evolutionarily conserved pathway in which intracellular material, including pathogens, can be sequestered within double membrane-bound vesicles and targeted to autolysosomes for degradation. The identification of conserved autophagy (‘ATG’) genes has permitted genetic and functional dissection of this pathway, which has implicated autophagy in cell survival, development, aging, cancer, and neurodegenerative diseases (Levine and Kroemer, 2008). In addition, several studies have demonstrated a crucial role for ATG genes in host defense against diverse intracellular pathogens including bacteria, parasites, and viruses (Deretic and Levine, 2009).
The involvement of ATG genes in innate antiviral defense is phylogenetically conserved. In plants, the ATG genes BECLIN 1, ATG7, ATG3, and VPS34 are essential for controlling tobacco mosaic virus replication and limiting the spread of programmed cell death during the hypersensitive response (Liu et al., 2005). In Drosophila, ATG genes protect against vesicular stomatitis virus (VSV) infection (Shelly et al., 2009); the disruption of Atg5, Atg8 and Atg18 is associated with increased VSV replication both in cultured cells and in vivo, resulting in increased animal lethality. Autophagy may also function in vertebrate antiviral host defense. Overexpression of Beclin 1 (the mammalian Atg6 ortholog) in neurons protects neonatal mice against lethal Sindbis virus (SIN) infection (Liang et al., 1998). The herpes simplex virus type 1 (HSV-1) protein ICP34.5 inhibits autophagy by binding to Beclin 1 and an HSV-1 mutant virus lacking the Beclin 1 binding domain of ICP34.5 has decreased neurovirulence (Orvedahl et al., 2007). Additionally, in mouse models of peripheral HSV-1 infection, the Beclin 1-binding deficient mutant virus is cleared more rapidly and fails to counteract the adaptive immune response as efficiently as wild-type virus (Leib et al., 2009).
The ATG gene Atg5 encodes an essential component of the Atg5-Atg12-Atg16 conjugation system, is required for the formation of autophagosomes in mammalian cells (Mizushima et al., 2001), and has been shown to play diverse roles in immunity (Deretic and Levine, 2009). In addition to increased susceptibility to VSV infection, RNAi-mediated knockdown of Atg5 in Drosophila results in increased lethality following infection with the intracellular bacterium, Listeria monocytogenes (Yano et al., 2008). Targeted deletion of Atg5 in phagocytic cells renders mice more susceptible to infection with L. monocytogenes and with the parasite, Toxoplasma gondii (Zhao et al., 2008). Furthermore, Atg5 may either positively or negatively regulate type I interferon (IFN) production in a cell type-dependent manner during mammalian VSV infection (Deretic and Levine, 2009).
Despite these findings, the role of endogenous ATG genes in general, and Atg5 in particular, in the host antiviral response in vertebrates has not yet been examined in vivo. Therefore, we examined the role of Atg5 in protection against lethal SIN CNS infection in mice. SIN is a positive-sense enveloped RNA virus in the alphavirus genus and provides a unique mouse model system for studying virus-host interactions in neurons. We chose this system because: (1) neurons, as post-mitotic cells, may be more likely than dividing cells to be dependent on non-cytolytic mechanisms (such as autophagy) for antiviral defense (Orvedahl and Levine, 2008); and (2) SIN can be used simultaneously as a CNS pathogen and a vector for neuronal gene delivery in vivo, which is useful to probe the effects of host cell genetic manipulation on viral pathogenesis (Hardwick and Levine, 2000). Following intracerebral inoculation, SIN predominantly infects neurons (Jackson et al., 1988; Johnson, 1965) and exhibits age-dependent neurovirulence (Johnson et al., 1972). In neonatal mice, inflammatory infiltrates and classical signs of encephalitis are not observed (Johnson, 1965). Rather, lethal CNS infection is thought to be due to cell-autonomous virus-induced neuronal cell death, since using the SIN vector system, our laboratory and others have shown that virally-expressed inhibitors of apoptosis reduce the mortality of neonatal mice with CNS Sindbis virus infection (Griffin, 2005).
In this study, we used the SIN vector system to inactivate Atg5 in virally-infected neurons, and also used neuron-specific Atg5 knockout mice. Our results demonstrate that loss of neuronal Atg5 function results in increased susceptibility of neonatal mice to lethal SIN CNS infection. Atg5 disruption does not affect viral replication, but does result in increased neuronal death that is associated with the accumulation of SIN proteins and cellular p62, an adaptor protein that binds to the autophagosomal membrane protein LC3 and is degraded by the autophagy pathway (Bjorkoy et al., 2005). In vitro, we found that p62 interacts with SIN capsid and targets it to the autophagosome. The genetic knockdown of p62, or the essential autophagy protein, Atg7, results in increased SIN capsid accumulation and increased virus-induced cell death but not increased viral replication. Together, our in vivo and in vitro results suggest that autophagy protects against SIN pathogenesis by a cell-autonomous mechanism that facilitates viral protein clearance and prevents virus-induced cell death, without directly controlling viral replication.
RESULTS
Live, But Not UV-Inactivated, SIN Induces Autophagy in vitro
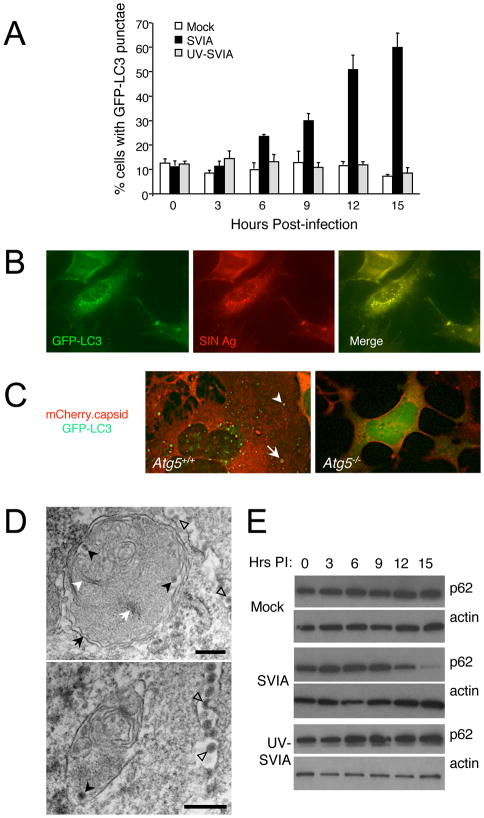
To determine whether SIN infection induces autophagy in vitro, we infected primary GFP-LC3 MEFs with SVIA or UV-inactivated SVIA (UV-SVIA). There was a significant increase in the percentage of cells with autophagosomes (GFF-LC3 dots) in MEFs infected with live SVIA as compared to mock-infected MEFs beginning at 9 h (Fig. 1A, p < 0.05); and more than 60% of SVIA-infected MEFs contained GFP-LC3 dots by 15 h post-infection (p < 0.001 vs. mock-infected controls). In contrast, there was no increase in the percentage of cells with autophagosomes in MEFs infected with UV-inactivated SVIA as compared to mock infection. These data indicate that SIN-induced autophagy requires viral replication, at least in MEFs. The lack of autophagy induction at 3 h post-infection (Fig. 1A), a time point when viral entry but not viral replication has occurred, is consistent with this conclusion.
Figure 1. SIN Induces Autophagy in vitro.
(A) Quantitation of the percentage of GFP-LC3 MEFs with GFP-LC3 punctae (autophagosomes) after infection with indicated virus. Data shown represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were observed in 3 independent experiments.
(B) Representative fluorescent microscopic image demonstrating colocalization of SIN structural proteins (red) with GFP-LC3 (green) in GFP-LC3 MEFs at 12 h post-infection (p.i.).
(C) Representative fluorescent microscopic images showing colocalization in Atg5+/+ MEFs or lack of colocalization in Atg5−/− MEFs of SIN capsid (red) and GFP-LC3 (green) in cells infected with SIN-mCherry.capsid/GFP-LC3. Images shown represent a single time point at 12 h p.i. from live cell imaging. Arrowhead denotes colocalized puncta; arrow denotes capsid-positive GFP-LC3 ring structure. See movie S1 for dynamic representation of mCherry.capsid and GFP-LC3 localization between 16 and 17 h p.i. in Atg5+/+ MEFs.
(D) Representative EMs of wild-type MEFs at 12 h p.i. with SVIA. Left panel demonstrates a double-membraned autophagosome (black arrow) containing SIN nucleocapsids (black arrowheads), cellular membranes (white arrowhead), and aggregates (white arrow). Right panel demonstrates a single-membraned autolysosome with SIN nucleocapsid (black arrowhead). Open arrowheads denote virions budding from the plasma membrane. Scale bars, 200 nm.
(E) Measurement of autophagic protein degradation by p62 Western blot analysis in wild-type MEFs at serial time points after mock, SVIA, or UV-SVIA infection.
At time points when increased autophagosomes were observed in SVIA-infected MEFs, but not earlier, we observed intracytoplasmic expression of SIN structural proteins, which strongly colocalized with GFP-LC3 punctae (Fig. 1B). Similar results were also observed with an antibody specific for SIN capsid protein (data not shown). These structures likely represent classical autophagosomes rather than non-specific GFP-LC3 aggregates, since GFP-LC3 punctate structures that colocalize with SIN capsid were observed in wild-type but not in Atg5−/− MEFs (Kuma et al., 2004) infected with a recombinant SIN strain, SIN-mCherry.capsid/GFP-LC3, that expresses mCherry fused to the endogenous capsid protein and that expresses GFP-LC3 from a double-subgenomic promoter (Fig. 1C).
Since SIN replicates on the cytoplasmic surface of single-membraned vesicles (not double-membraned vesicles) (Gil-Fernandez et al., 1973), this colocalization likely reflects the targeting of newly synthesized SIN proteins or assembled SIN nucleocapsids to autophagosomes rather than an association of the SIN replication complex with LC3-positive membranes. Indeed, in live-cell time-lapse microscopy imaging of wild-type MEFs infected with SIN-mCherry.capsid/GFP-LC3, mCherry.capsid structures appear first and later are engulfed by GFP-LC3-positive membranes (Movie S1), suggesting these structures are actively targeted by autophagy. Furthermore, electron microscopic (EM) analyses of SIN-infected MEFs revealed autophagosomes and autolysosomes that contained Sindbis virions (Fig. 1D and data not shown). While autophagic structures contained Sindbis virions, there were also other cytoplasmic contents inside them, including cellular membranes and aggregates (Fig. 1D). Thus, the autophagic capture of SIN in MEFs is not entirely specific for viral contents.
Since some pathogens induce early stages of autophagy, but block later stages (i.e. autophagolysosomal fusion), we investigated whether SIN induces a complete autophagic response (i.e. autophagic flux) by measuring levels of the autophagic substrate, p62. We found that in live SVIA, but not UV-SVIA-infected MEFs, there was a gradual decline in levels of p62 protein with barely detectable levels observed by 15 h post infection (Fig. 1E). This decline was not due to altered levels of p62 mRNA expression (Fig. S1). We also observed a complete autophagic response in a cell type that is more relevant to the natural target of SIN in vivo. Mouse Neuro-2A cells, a mouse neuroblastoma cell line, infected with live SVIA, but not UV-SVIA, had increased autophagy induction and autophagic flux, as measured by LC3-II conversion and p62 degradation, respectively (Fig. S2). Together with the GFP-LC3 assays and EM observations, these data indicate that a complete autophagic response is induced in both MEFs and a neuronal cell line infected with replication-competent, but not UV-inactivated, replication-incompetent SIN.
SIN Replication in Atg5−/−-Deficient Cells
To evaluate the effect of autophagy on viral replication, we compared SIN growth curves in immortalized Atg5−/− and Atg5+/+ MEFs (Kuma et al., 2004). We found that SVIA replicated to significantly higher titers in a clone of immortalized Atg5−/− MEFs as compared to a clone of immortalized Atg5+/+ MEFs (Fig. S3A). However, in another cell line, Atg5−/− MEFs stably-transfected with a tetracycline-repressible Atg5-expressing plasmid, M5–7 cells (Hosokawa et al., 2006), we did not see any increase in SVIA replication with doxycycline-induced suppression of Atg5 expression (Fig. S3B). Also, we did not observe differences in SIN replication in Atg5−/− cells as compared to Atg5+/+ ES cells (Fig. S3C). Thus, although Atg5 may appear to restrict viral replication in certain immortalized MEFs, this effect is not conserved in different cells that are deficient in Atg5 expression. We therefore conclude that Atg5 may not play an essential role in controlling levels of SIN replication.
SIN Infection Induces Autophagy in vivo and Viral Antigen Co-localizes With Autophagosomes in Neurons
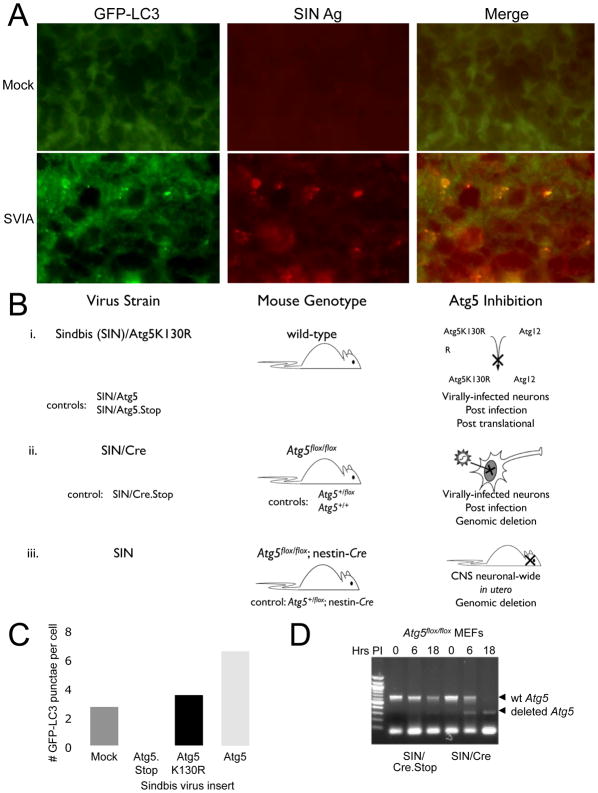
To determine whether SIN induces autophagy in virally-infected neurons in vivo, we infected GFP-LC3 transgenic mice with SVIA and examined GFP-LC3 subcellular localization and SIN antigen expression. In uninfected mouse brains, as described previously (Mizushima et al., 2004), we did not detect GFP-LC3 punctae in neurons (Fig. 2A, top). However, in the brains of SVIA-infected mice, we detected GFP-LC3 punctae specifically in neurons that also expressed SIN antigens (Fig. 2A, bottom) and SIN antigens colocalized with GFP-LC3 punctae. Thus, SIN infection induces autophagy in vivo in mouse brain and, similar to in vitro, SIN proteins colocalize with autophagosomes.
Figure 2. SIN-Induced Autophagy in Mouse Hippocampal Neurons and Scheme of Experimental Strategies to Inhibit Autophagy in Neurons in vivo.
(A) Colocalization of SIN structural proteins (red) with GFP-LC3 in hippocampal neurons of GFP-LC3 transgenic mice 24 h after mock infection (top) or infection with SVIA (bottom). Similar hippocampal regions are shown for mock and SVIA-infected brains. No GFP-LC3 punctae were observed in any regions of the mock-infected brains.
(B) Conceptual overview of strategies to inhibit or knock out Atg5 specifically in neurons in vivo, and the relevant control viruses and mouse strains. (i) SIN expressing a dominant negative mutant Atg5 (Atg5K130R). (ii) SIN expressing Cre recombinase in Atg5flox/flox mice. (iii) nestin-Cre mice crossed to Atg5flox/flox mice.
(C) Quantitation of the number of GFP-LC3 punctae (autophagosomes) per cell in GFP-LC3 MEFs at 12 h after infection with indicated virus below x axis. Data shown represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were obtained in 3 independent experiments.
(D) Detection of genomic Atg5 excision in primary MEFs obtained from Atg5flox/flox mice infected with SIN/Cre or SIN/Cre.Stop.
Strategies for Investigating the Effects of Neuronal Atg5 on the Pathogenesis of SIN CNS Infection
To investigate the role of neuronal autophagy in the pathogenesis of SIN CNS infection, we used three complimentary strategies to inactivate the ATG gene, Atg5, in virally-infected neurons in vivo (Fig. 2B). In all three models, we used a backbone strain of SIN, dsTE12Q, that is relatively avirulent in neonatal mice to facilitate the detection of increased neurovirulence in the setting of ATG gene inactivation. In the first model (Fig. 2Bi), we used the recombinant chimeric SIN system (Hardwick and Levine, 2000) to express a dominant-negative mutant of Atg5 (K130R, herein referred to as Atg5K130R). The K130R mutation inhibits a key step in the autophagic pathway by blocking covalent conjugation of Atg5 to Atg12 (Mizushima et al., 1998; Mizushima et al., 2001), resulting in impaired recruitment of LC3 to nascent autophagic isolation membranes and impaired autophagosome formation (Hamacher-Brady et al., 2006; Mizushima et al., 2001; Pyo et al., 2005). As controls, we constructed chimeric SIN constructs expressing wild-type Atg5 (SIN/Atg5) or a noncoding Atg5 sequence devoid of ATG codons (SIN/Atg5.Stop). We confirmed that SIN/Atg5K130R blocked virus-induced autophagy in vitro by counting the number of GFP-LC3 dots per cell in MEFs that were infected with SIN/Atg5K130R versus SIN/Atg5 or SIN/Atg5.Stop (p < 0.05, SIN/Atg5K130R vs. control viruses) (Fig. 2C). In the second model (Fig. 2Bii), we infected Atg5flox/flox, Atg5+/flox, or Atg5+/+ mice with recombinant chimeric SIN constructs that express either Cre recombinase (SIN/Cre) or a noncoding Cre sequence (SIN/Cre.Stop). After infection with SIN/Cre (but not with SIN/Cre.Stop), Atg5 is deleted only in infected neurons of Atg5flox/flox mice, whereas infected neurons from Atg5+/flox mice are rendered haploinsufficient, and there is no effect on Atg5 in wild-type (Atg5+/+) animals. We used PCR to confirm the excision of floxed Atg5 alleles in Atg5flox/flox MEFs after infection with SIN/Cre (Fig. 2D). In the third model (Fig. 2Biii), we assessed the effect of pre-existing deletion of Atg5 in both infected and uninfected neurons on SIN pathogenesis, using Atg5flox/flox; nestin-Cre mice, which undergo neuron-specific deletion of Atg5 in utero (Hara et al., 2006). These mice are normal at birth, and do not display signs of progressive neurodegenerative defects until after 3 weeks of age.
Inhibition of Neuronal Atg5 Results in Increased Mortality from CNS SIN Infection
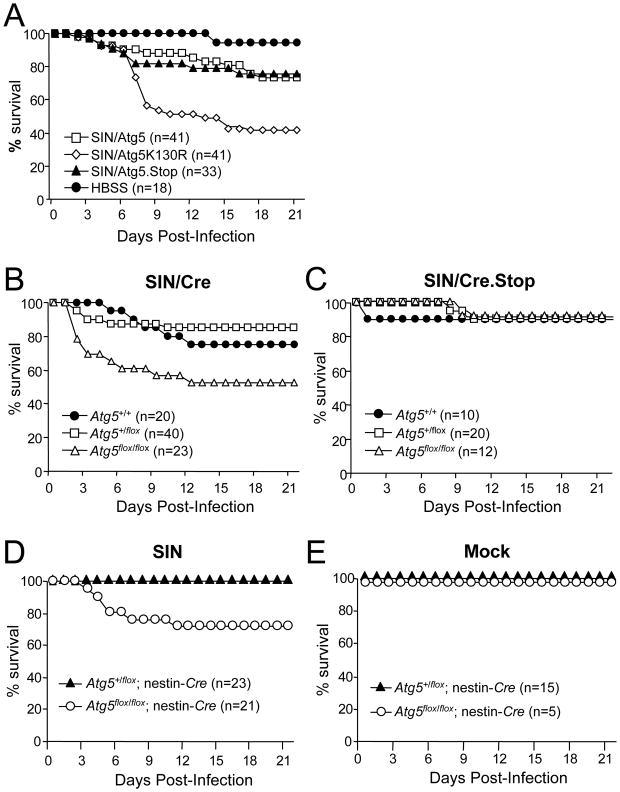
We assessed the effect of disruption of neuronal Atg5 function on mortality due to CNS SIN infection in neonatal mice. In each model, increased mortality was observed in the setting of neuronal Atg5 inactivation (Fig. 3). Wild-type mice infected with SIN/Atg5 and SIN/Atg5.Stop viruses had higher survival rates (73% and 76%, respectively) than those infected with SIN/Atg5K130R (41%; p < 0.001) (Fig. 3A). SIN/Cre-infected Atg5flox/flox mice were less likely to survive (52%; p < 0.001) than their Atg5+/+ (75%) or Atg5+/flox (85%) littermates (Fig. 3B). However, mice infected with the control virus, SIN/Cre.Stop, exhibited the same mortality (90%) across all genotypes (Fig. 3C). Finally, Atg5flox/flox; nestin-Cre mice were less likely to survive (71%; p < 0.001) than their control Atg5+/flox; nestin-Cre littermates (100%) (Fig. 3D). No mortality was seen in mock-infected Atg5flox/flox; nestin-Cre animals (Fig. 3E), indicating that this increased mortality was due to increased susceptibility to lethal SIN infection, rather than constitutive neuronal deletion of Atg5.
Figure 3. Increased SIN Neurovirulence in Mice with Inactivation of Neuronal Atg5.
(A) Mortality of CD1 littermates infected with the indicated recombinant SIN strains or mock-infected with HBSS. Data shown represent combined mortality from 4 independent infections of 8–12 mice per group. Similar results were observed in each independent experiment.
(B–C) Mortality of Atg5flox/flox or littermate controls (Atg5+/flox and Atg5+/+) infected with recombinant SIN expressing Cre recombinase or SIN expressing non-coding Cre gene. Data shown in (B–C) represent combined mortality from infection of 22 and 8 separate litters, respectively.
(D–E) Mortality of littermates from Atg5+/flox; nestin-Cre transgenic mice crossed with Atg5flox/flox mice and infected with the dsTE12Q strain of SIN (D) or mock-infected (E). Data shown in (D–E) represent combined mortality from infection of 21 and 8 separate litters, respectively.
Neuronal Atg5 Inactivation Alters Viral Antigen Clearance and Neuronal Cell Death Without Affecting CNS Viral Replication or Type I IFN Production
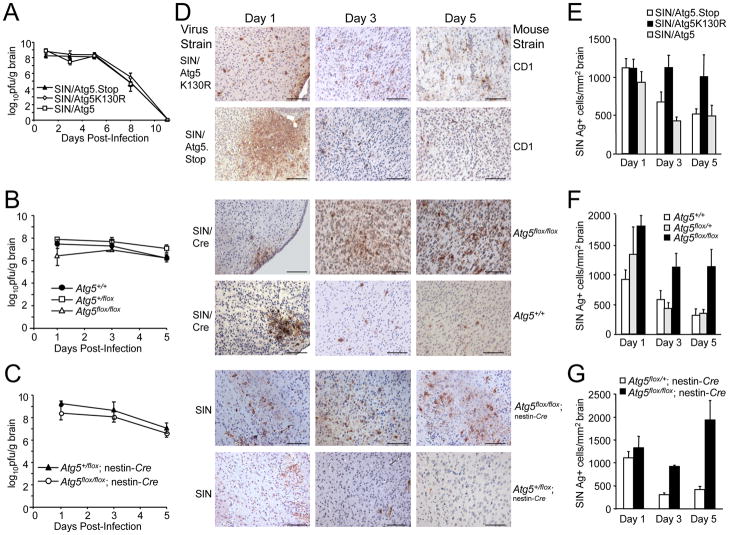
To gain insight into the mechanism of increased lethality in mice with neuronal Atg5 inactivation, we examined levels of CNS viral replication, type I IFN production, and brain histopathology at serial time points after infection. We observed no differences in any of the three model systems in the levels of infectious virus in the brains of control animals versus mice with disrupted neuronal Atg5 function (Fig. 4A-C). Thus, Atg5 does not seem to be required for the control of levels of infectious virus in vivo in virally-infected mouse brains. We also observed no differences in the levels of type I IFN in the brains of mice with intact neuronal Atg5 versus those with disrupted neuronal Atg5 function (Fig. S4). Also, as previously well-characterized in neonatal SIN infection (Johnson, 1965), perivascular and intraparenchymal inflammatory infiltrates were lacking in all experimental groups (data not shown). Thus, increased SIN-induced mortality in mice with neuronal Atg5 inactivation is unlikely due to alterations in viral replication, levels of innate immune signaling, or inflammation.
Figure 4. Atg5 Inhibition Delays Viral Antigen Clearance from Neurons Without Affecting SIN CNS Titers.
(A–C) SIN titers in mouse brains. Data shown represent geometric mean titers ± SEM for groups of 4–8 mice per time point.
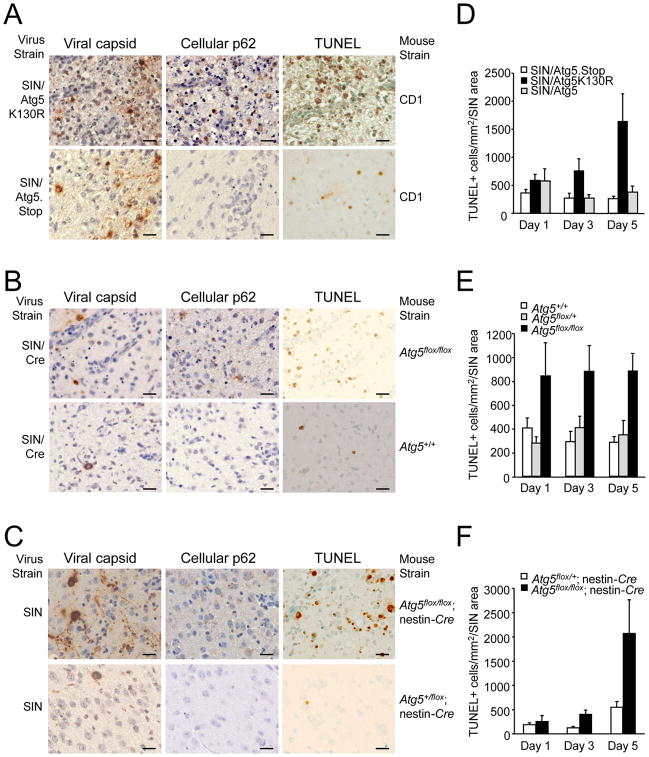
(D) SIN antigen staining of brains of mice of the indicated genotype (right labels) infected with the indicated virus (left labels). All micrographs in (D) are from the superior colliculus of the mouse brain, a region that is infected by SIN in all mice in this study. The images shown are representative of the data quantitated in (E–G) for the total mouse brain, with the exception that there is high degree of inter-mouse variability in the level of colliculus staining at day one. Scale bars, 100 μm.
(E–G) Quantitation of SIN antigen staining in the brains of mice treated as in (A–C), respectively. Data in (E–G) represent mean number of antigen-positive cells per unit area of mouse brain for 4–8 mice per experimental group.
Although no differences were observed in CNS viral titers, we observed marked differences in the numbers of viral antigen-positive cells in mice with intact versus disrupted neuronal Atg5 function (Fig. 4D–G). At day one after infection, no significant differences were observed in the number of viral antigen-positive cells between mice with intact Atg5 function versus disrupted neuronal Atg5 function. However, after day one, in all three models, the numbers of viral antigen-positive cells declined in mice with intact neuronal Atg5 function but remained persistently elevated in mice with disrupted neuronal Atg5 function. At days 3 and 5 after infection, a significant increase was observed in the numbers of viral antigen-positive neurons in mice with disrupted neuronal Atg5 compared to the control group(s) in each model (p < 0.05). These data indicate that, while intact neuronal Atg5 function is not essential for the control of replication-competent SIN, it is essential for the clearance of SIN antigen from neurons.
We further confirmed the delayed clearance of SIN antigen from neurons by performing immunohistochemical staining of SIN capsid protein (Fig. 5A–C left panels). Additionally, we found that in regions of SIN antigen and capsid staining, there was an accumulation of cellular p62 protein in mice with disrupted neuronal Atg5 function, but not in infected mice with intact Atg5 function (Fig. 5A–C, center panels). Higher power analysis of adjacent sections showed that identical cells displayed capsid and p62 immunoreactivity in each mouse model of neuronal Atg5 inactivation (Fig. S5). No increases in cellular ubiquitin immunostaining were observed among any of the groups of virus-infected mice (data not shown).
Figure 5. Increased SIN Capsid Staining, Cellular p62 Staining, and Cell Death in the Brains of Mice with Neuronal Atg5 Inactivation.
(A–C) Detection of SIN capsid (left column), cellular p62 (middle column), and cell death by TUNEL staining (right column) in mouse brain. Shown are representative photomicrographs for each experimental group of the superior colliculus at day 5 p.i. For (A–C) similar results were observed in 4–8 mice per group. Scale bars, 20 μm.
(D–F) Quantitation of number of TUNEL-positive cells per unit area of virus-infected region of brain at days 1, 3, and 5 p.i.. Data in (D–F) represent mean ± SEM for each brain from 4–8 mice per experimental group.
Brain regions that exhibited increased capsid and p62 staining also contained increased TUNEL-positive cells (Fig. 5A–C, right panels). We quantitated the number of TUNEL-positive cells per virus-infected region of the brain and found that in each model of neuronal Atg5 inactivation, significant increases in the numbers of TUNEL-positive cells were observed at day 3 post infection, and more strikingly, at day 5 post infection (Fig. 5D–F; p < 0.05 vs. controls). Higher power analysis of adjacent sections showed that identical cells displayed TUNEL and SIN antigen positivity (Fig. S6). Thus, in three different model systems in which neuronal Atg5 function is disrupted, SIN infection results in increased neuronal cell death.
p62 is Involved in SIN Capsid Targeting to Autophagosomes
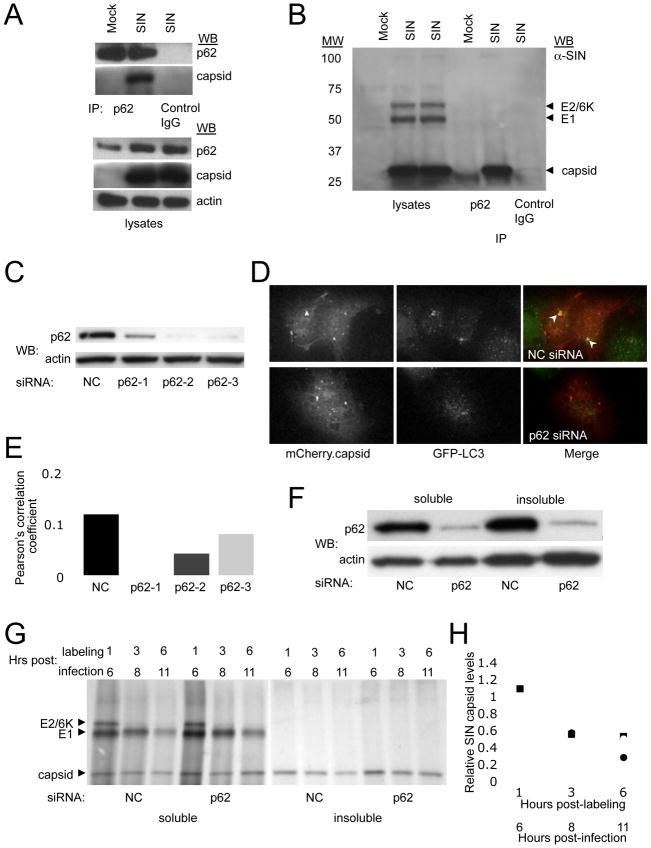
Our findings above indicate that: (1) Atg5 may perform a protective cellular housekeeping role during viral infection of post-mitotic cells such as neurons; and (2) p62 may serve as an adaptor protein, not only for the autophagic clearance of ubiquitinated cellular proteins, but also for the autophagic clearance of viral proteins. To investigate this latter possibility, we used an in vitro system that is amenable to biochemical and genetic knockdown studies. We found that in virally-infected HeLa cells, p62 coimmunoprecipitates with SIN capsid protein (Fig. 6A, B), but not with SIN E1 or E2 envelope glycoproteins (Fig. 6B). Furthermore, siRNA knockdown of p62 in HeLa cells (Fig. 6C) significantly decreases the colocalization of SIN capsid protein and GFP-LC3 punctae (p < 0.05) (Fig. 6D, E). Consistent with the degradative function of autophagy and our observations that SIN antigens persist in autophagy-deficient infected mouse brains in vivo, we found that siRNA knockdown of p62 delayed the degradation of SIN capsid (but not the E1 or E2 envelope glycoproteins) in HeLa/GFP-LC3 cells using a pulse-chase labeling radioimmunoprecipitation assay (Fig. 6F–H). This delayed clearance was most evident in the detergent-insoluble protein fraction, which was previously demonstrated to be cleared through p62-mediated autophagy (Komatsu et al., 2007). Thus, in vitro, p62 interacts with SIN capsid and is required for its targeting to autophagosomes. This interaction, if conserved in infected neurons in vivo, may potentially explain the accumulation of SIN capsid protein and cellular p62 in the brains of infected mice with disrupted neuronal autophagy.
Figure 6. p62 Interacts with SIN Capsid and Targets Capsid for Autophagy.
(A–B) Coimmunoprecipitation of SIN capsid with p62 in HeLa/GFP-LC3 cells mock-infected or infected with SVIA (labeled SIN) either by Western blotting with a polyclonal anti-SIN virus capsid antibody (A) or a polyclonal anti-SIN antibody that detects E2/6K, E1, and capsid protein (B).
(C–E) Detection of SIN capsid colocalization with GFP-LC3 after p62 knockdown. Western blot analysis of p62 expression in HeLa/GFP-LC3 cells treated with individual p62 siRNA oligos or non-silencing negative control oligos (NC) (C). Representative image of HeLa/GFP-LC3 cells treated with p62-2 siRNA (bottom) or NC (top), and infected with SIN-mCherry.capsid (D). Quantitation of mCherry.capsid colocalization with GFP-LC3 in cells treated with the indicated siRNA (E). Data shown represent the mean of at least 50 infected cells per condition ± SEM. Similar results were obtained in 3 independent experiments.
(F–H) Analysis of SIN protein degradation after p62 knockdown. Western blot analysis of p62 expression in the fractions used in (G) for radioimmunoprecipitation (F). Radioimmunoprecipitation with an anti-SIN antibody of soluble and insoluble fractions from HeLa/GPF-LC3 cells treated with NC or p62 siRNA, infected with SVIA, and pulse-chased for the indicated times (G). Quantitation of capsid levels in (G) relative to 1 h control levels for each fraction (H). Closed circles, NC soluble; closed squares, p62 soluble; open circles, NC insoluble; open squares, p62 insoluble. Similar results were observed in 3 independent experiments.
p62 and Atg7 are Important for the Survival of SIN-Infected Cells in vitro
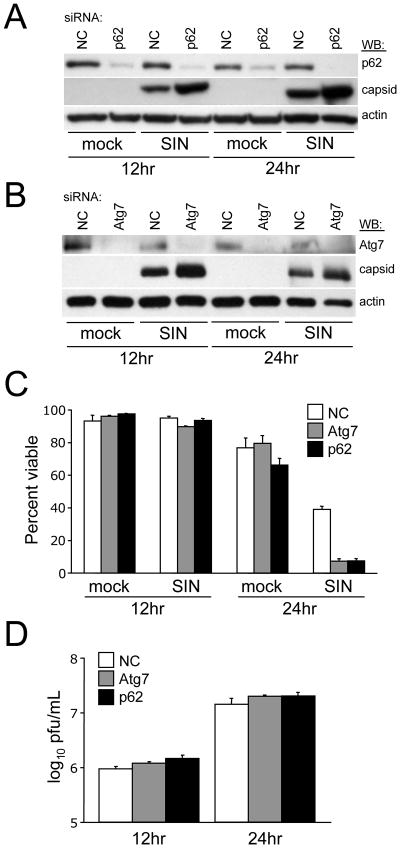
Our findings above indicate that p62 serves as an adaptor that targets SIN to the autophagosome. Next, we sought to evaluate the physiological consequences of p62 knockdown on SIN replication and SIN-induced cell death. Consistent with the delayed clearance of SIN capsid with p62 knockdown as measured by pulse-chase analysis (Fig. 6), we found that p62 siRNA results in an increase in steady-state levels of SIN capsid as compared to control siRNA at 12 and 24 h after infection (Fig. 7A). In these same conditions where p62 siRNA results in increased SIN capsid accumulation, we observed a significant increase in virus-induced cell death at 24 h after infection (p <0.001 vs. non-coding siRNA control) (Fig. 7C). However, we did not observe any alterations in levels of viral replication in p62 siRNA-treated cells at these time points (Fig. 7D) or in a 96 hour growth curve (data not shown). Thus, similar to neuronal Atg5 inactivation in vivo, we found that p62 inactivation in vitro results in SIN capsid accumulation, increased virus-induced cell death, and no effect on viral replication.
Figure 7. p62 and Atg7 Promote the Survival of SIN-Infected cells.
(A–B) Western blot analysis of p62 and capsid expression (A) or Atg7 and capsid expression (B) in HeLa/GFP-LC3 cells treated with the indicated siRNA and infected for the time indicated.
(C) Cell death quantitation of cells treated as in (A) and (B) as measured by a trypan blue exclusion assay. Data shown represent mean ± SEM of at least 100 cells per sample for triplicate samples for each condition.
(D) Levels of infectious SIN in supernatants of cells in (C). Data represent geometric mean titers ± SEM for triplicate samples.
For (A–D), similar results were obtained in three independent experiments.
To further confirm that the p62 siRNA phenotype was a function of decreased autophagy, we performed similar experiments using siRNA against Atg7, another component of the protein conjugation system that is required for autophagosomal membrane expansion (Komatsu et al., 2005). Similar to siRNA against p62, siRNA targeted against Atg7 results in an increase in steady-state levels of SIN capsid protein (Fig. 7B), an increase in virus-induced cell death at 24 h post infection (p <0.001 vs. non-coding siRNA control) (Fig. 7C), and no changes in levels of infectious virus (Fig. 7D). Thus, genetic knockdown in vitro of either the p62-SIN capsid-interacting targeting factor or the Atg7 autophagy execution protein phenocopies in vivo disruption of neuronal Atg5 function with respect to increased virus-induced cell death in the absence of increased viral replication. Together, these data suggest that p62-mediated autophagic clearance of SIN capsid protein may be important for cell survival during viral infection.
DISCUSSION
Here we demonstrate that neuronal function of the ATG gene, Atg5 is essential to protect mice against fatal CNS alphavirus infection. While previous genetic knockout or knockdown studies have suggested an important role for ATG genes in the protection of mice, Drosophila, worms, and slime molds against bacterial or protozoal pathogens (Jia et al., 2009; Yano et al., 2008; Zhao et al., 2008), and in the protection of Drosophila (Shelly et al., 2009) and plants (Liu et al., 2005) against viral infection, our results provide the first evidence for a protective role of an endogenous ATG gene in antiviral host defense in mammals. Thus, ATG genes play a conserved role in antiviral immunity – ranging from plants, to Drosophila, to mice.
Our findings suggest a novel potential mechanism by which ATG genes protect host organisms against viral infection, which involves the clearance of viral proteins. In previous studies, the knockdown or knockout of ATG genes in plants and Drosophila has been shown to increase viral replication and increase animal mortality (in Drosophila) (Shelly et al., 2009) or pathology (in plants) (Liu et al., 2005). In addition, mice with Atg5-deficient plasmacytoid dendritic cells have impaired type I IFN production following infection with VSV (Lee et al., 2007). Surprisingly, although we observed increased SIN replication in vitro in one clone of immortalized Atg5−/− MEFs, we did not observe increased SIN replication in other Atg5-deficient cell lines or in the brains of mice with neuronal inactivation of Atg5 (nor, did we observe any defects in type I IFN production in vivo). We therefore conclude that the increased SIN-induced animal mortality is not due to a direct role of neuronal Atg5 in the control of viral replication or regulation of innate immune signaling. Rather, mice with disruption of neuronal Atg5 function had increased neuronal death associated with impaired clearance of SIN proteins. Although it is difficult to conclude the precise cause of either neuronal or organismal death in the context of in vivo mouse studies, one plausible explanation is that, in post-mitotic cells such as neurons, the failure to properly clear viral proteins by autophagy results in cellular toxicity and increased animal lethality.
Our in vitro data provide support for the hypothesis that autophagy-mediated clearance of SIN proteins is cytoprotective and identify a mechanism by which the SIN capsid protein is targeted for autophagic degradation. We found that the p62 cellular adaptor protein interacts with SIN capsid and is required for SIN capsid targeting to the autophagosome. Moreover, genetic knockdown of p62 or Atg7 increases capsid accumulation and cell death, without increasing levels of SIN infectious virus. Thus, p62-mediated autophagic targeting of SIN capsid may function to promote cell survival during SIN infection. While cell death inhibits viral replication in many settings, virus-induced cell death is an established important factor in the pathogenesis of neuronotropic viral infections (Levine, 2002). Accordingly, we propose that the cytoprotective function of autophagic-mediated degradation of SIN capsid may contribute to the protective role of endogenous ATG genes against lethal CNS SIN infection.
To our knowledge, our findings represent the first example of p62 targeting a specific microbial protein for autophagic degradation. Recent studies have indicated a role for p62 in targeting ubiquitin-coated Salmonella for autophagic degradation (Zheng et al., 2009). However, we are unaware of previous evidence of a biochemical interaction between p62 and a viral or other microbial protein. p62 is believed to function as an adaptor that binds ubiquitin and polyubiquitin through its C-terminal UBA domain and binds LC3 through a more N-terminal LC3 interaction region (LIR) domain (Pankiv et al. 2007). It is not yet known which domain of p62 is required for its interaction with SIN capsid, whether this interaction is direct or indirect, and whether p62 targets other structural proteins from other viruses to autophagosomes. Interestingly, our data suggest that, unlike p62-dependent autophagic targeting of cellular proteins or ubiquitin-coated bacteria, the interaction between p62 and SIN capsid and the targeting of SIN capsid may be ubiquitin-independent. In contrast to the neurodegenerative phenotype in older neuron-specific Atg5- or Atg7-deficient mice where there is accumulation of both ubiquitin aggregates and p62 aggregates (Hara et al., 2006; Komatsu et al. 2006), we only observed p62, and not ubiquitin, aggregates in SIN-infected mice with neuronal Atg5 inactivation. We were also unable to detect ubiquitination of SIN capsid in vitro in conditions where it interacts with p62. Thus, autophagy-dependent protein quality control in neurons may involve p62 in the clearance of both cellular and viral proteins, but other, as-of-yet undefined molecular tags besides ubiquitin may serve to link certain viral proteins to p62-dependent autophagy targeting pathways.
Unlike reports with Listeria or VSV infection in Drosophila, a pre-formed pathogen-associated molecular pattern in SIN does not appear to induce autophagy upon cellular entry, since UV-inactivated SIN does not induce autophagy in either MEFs or mouse neuronal cells. The requirement for viral replication in SIN-induced autophagy may suggest either (1) autophagy is induced through the delivery of viral nucleic acids to endosomal Toll-like-receptors (a process that in itself has been shown to require the autophagic machinery in VSV-infected pDCs (Lee et al. 2007); or (2) cytoplasmic sensors of viral RNA (i.e. retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs)) may play a heretofore-undefined role in autophagy stimulation. An important area of future research will be to elucidate the precise cytoplasmic signaling events that link detection of viral replication to autophagy activation during SIN and potentially other viral infections.
Our studies suggest that neuronal Atg5 protects against lethal SIN infection in a cell-autonomous manner. We used three complimentary models to inactivate neuronal Atg5; in the first two, Atg5 was specifically inactivated in virally-infected neurons exclusively after viral replication whereas in the third model, Atg5 was deleted in utero in neurons. The observation of a similar phenotype in all three models provides strong support that the observed increased neurovirulence, impaired viral protein clearance, and increased neuronal death is a direct consequence of neuronal Atg5 inactivation. These models have the advantage that they permit us to assess the direct effects of neuronal Atg5 inactivation on virus-host interactions in neurons, without confounding variables introduced by Atg5 deficiency in other cell types or on other stages of viral pathogenesis, such as peripheral replication and CNS invasion. However, the role of Atg5 deficiency in these other stages of pathogenesis that occur in the natural route of mosquito-borne arboviral infections is not yet known. Similarly, our experimental design does not permit us to definitively rule out a role for neuronal Atg5 deficiency in shaping innate immune responses other than type I IFN production or adaptive immune responses that may contribute to SIN pathogenesis.
Other groups have reported potential autophagy-independent functions of the ATG gene, Atg5, including a pro-apoptotic function by a calpain-mediated cleavage fragment (Yousefi et al., 2006) and autophagy-independent recruitment of Irga6 GTPase recruitment to T. gondii parasitophorous vacuoles (Zhao et al., 2008). Our findings are most consistent with an autophagy-dependent function of Atg5, as: (1) we observed increased, not decreased, cell death, in the setting of Atg5 disruption, suggesting that Atg5 is not acting as a cell death factor; (2) we confirmed that SIN nucleocapsids were captured inside classical double-membraned autophagosomes in wild-type MEFs and found that SIN capsid protein could not be targeted to autophagosomes in Atg5-deficient MEFs; and (3) we observed impaired clearance of viral proteins and an accumulation of p62 aggregates in virally-infected neurons lacking Atg5 function. While this phenotype is strongly consistent with a defect in the classical lysosomal degradative role of autophagy, we cannot exclude a contribution of autophagy-independent functions of Atg5 in protecting neurons against SIN infection. Further studies of SIN pathogenesis in mice with neuronal inactivation of other ATG genes will be important to confirm the role of the autophagy pathway in neuronal protection against SIN infection. However, our findings with Atg7 siRNA in HeLa cells provide in vitro evidence that other components of the autophagy pathway promote SIN capsid protein clearance and cell survival during virus infection.
In summary, our findings provide strong evidence for an important cell-autonomous role for neuronal Atg5 in in vivo protection against SIN infection. Based on our characterization of mouse brain titers and histopathology, this protection is associated with neuronal Atg5-dependent control of viral protein clearance and neuronal Atg5-dependent protection against cell death, but not neuronal Atg5-dependent control of virus replication. Moreover, our in vitro studies provide strong evidence that p62 functions to promote the survival of infected cells through autophagic targeting and clearance of viral proteins. Although further studies are required to definitively prove a direct cause and effect relationship between impaired viral protein (and/or cellular protein) p62-mediated clearance and increased cell death in SIN-infected mice with disrupted neuronal Atg5 function, our studies raise the intriguing hypothesis that autophagy-dependent protein quality control may be a previously unappreciated function in the vast repertoire of host antiviral immune responses.
EXPERIMENTAL PROCEDURES
Mammalian Cell Lines
Primary MEFs were established from GFP-LC3, Atg5+/flox, and Atg5flox/flox mice (Hara et al., 2006; Mizushima et al., 2004) at day e13.5 and cultured as described (Su et al., 2003). Atg5+/+ and Atg5−/− immortalized MEFs, Atg5−/− embryonic stem (ES) cells, and Atg5−/− MEFs stably transformed with tetracycline-repressible Atg5 (M5–7 cells) were described previously (Hosokawa et al., 2006; Kuma et al., 2004; Mizushima et al., 2001). MEFs and ES cells were cultured as described in the Supplemental Experimental Procedures. HeLa cells (provided by V. Stollar, (Li et al., 1997)) stably-expressing GFP-LC3 were generated and cultured as described in the Supplemental Experimental Procedures. Mouse Neuro-2A cells were obtained from ATCC and cultured according to the supplier’s instructions.
Mouse Strains
One to two day-old outbred CD1 mouse litters were obtained from Charles River. Atg5flox/+ mice (provided by N. Mizushima) were crossed with Atg5flox/+ mice to obtain Atg5+/+, Atg5flox/+, and Atg5flox/flox mice (Hara et al., 2006). Atg5flox/flox mice were crossed with B6.Cg-Tg(nestin-Cre)1Kln/J mice (Jackson Laboratories; stock#003771) to produce Atg5+/flox; nestin-Cre mice described previously (Hara et al., 2006). Atg5+/flox; nestin-Cre mice were crossed with Atg5flox/flox mice to obtain Atg5+/flox; nestin-Cre control and CNS-specific Atg5-deficient mice (Atg5flox/flox; nestin-Cre mice). Atg5flox/flox and Atg5flox/flox; nestin-Cre mice were genotyped as described (Hara et al., 2006).
Wild-type and Recombinant Chimeric SIN Strains
The SIN strain SVIA (ATCC) is derived from a low-passage isolate of the wild-type AR339 SIN strain (Taylor et al., 1955). To construct UV-inactivated SVIA, SVIA was treated for 5 minutes with a Stratalinker® UV Crosslinker 1800 (Stratagene), and the lack of infectious virus after UV irradiation was confirmed by plaque assay titration. The construction of the recombinant viruses, SIN/Atg5, SIN/Atg5K130R, SIN/Atg5.Stop, SIN/Cre, SIN/Cre.Stop, SIN-mCherry.capsid, and SIN-mCherry.capsid/GFP-LC3 was performed using the recombinant SIN vector dsTE12Q as a backbone (Liang et al., 1998) and is described in detail in the Supplemental Experimental Procedures. Infectious virus was produced from all SIN recombinant chimeric vectors as described (Hardwick and Levine, 2000), and recombinant viruses were titered by plaque assays on BHK-21 cells.
In Vitro Virus Infections
All infections (virus and mock) were performed in reduced serum (1% FBS)-containing media. For comparison of viral replication in Atg5-expressing and Atg5-deficient cells (MEFs, ES cells, M5–7 cells), infections were performed at a multiplicity of infection (MOI) of 0.1 plaque-forming unit (PFU) per cell. MEFs and Neuro-2A cells were infected at an MOI of 5 for EM, fluorescent microscopy and biochemical studies. HeLa/GFP-LC3 cells were infected at an MOI of 5 for coimmunoprecipitation and radioimmunoprecipitation experiments; at an MOI of 20 for colocalization studies; and at an MOI of 1 for viability assays and replication studies.
Animal Studies
All infections were performed by intracerebral (i.c.) inoculation of the designated number of PFUs of virus diluted in 30μl Hanks balanced salt solution (HBSS) into the right cerebral hemisphere. One to two day-old randomized CD1 litters were infected with 5000 PFUs i.c. of recombinant SIN constructs, SIN/Atg5, SIN/Atg5K130R, and SIN/Atg5.Stop. One day-old litters resulting from an Atg5+/flox x Atg5+/flox cross were infected with 1000 PFUs i.c. of SIN/Cre and SIN/Cre.Stop. One week-old Atg5flox/flox; nestin-Cre and Atg5+/flox; nestin-Cre litters were infected with 1000 PFUs i.c. of dsTE12Q. For mortality studies, mice were monitored daily for 21 days. For measurement of CNS viral titers, freeze-thawed 10% (weight/volume) homogenates of the right hemispheres were used for plaque assay titration. For histopathology studies, the left hemispheres were fixed in 4% paraformaldehyde, embedded in paraffin, and cut sagitally from the medial surface into 5 μM adjacent sections. For in vivo autophagy assessment, one day-old GFP-LC3 litters (Mizushima et al., 2004) were infected with 1000 PFUs i.c. of SVIA, then euthanized 24 h later by perfusion with 4% paraformaldehyde, and frozen brain sections were prepared. See Supplemental Experimental Procedures for additional details on tissue sample preparations. All animal procedures were performed in accordance with institutional guidelines and with approval from the Institutional Animal Care and Use Committee.
Immunohistochemical and Immunofluorescent Studies
TUNEL staining of brain sections was performed according to manufacturers’ instructions (Apoptag® peroxidase In Situ Apoptosis Detection Kit; Chemicon International), using Sigma FAST™ 3, 3′-diaminobenzidine (DAB) tablets as the peroxidase substrate. Immunohistochemical staining of paraffin-embedded brain sections was performed using a rabbit polyclonal anti-SIN antibody (provided by D. Griffin) (1:500 dilution) (Jackson et al., 1988), a rabbit polyclonal anti-SIN capsid antibody (provided by M. MacDonald) (1:2000) (Rice and Strauss, 1982), and a polyclonal guinea pig anti-cellular p62 antibody (Progen, 1:1000 dilution). Primary antibodies were detected with the ABC Elite kit (Vector Laboratories) according to the manufacturer’s instructions. The number of SIN antigen-positive cells per mouse brain sagittal section and the number of TUNEL-positive cells per virus-infected area per mouse brain sagittal section was quantified as described in the Supplemental Experimental Procedures. Immunofluorescence staining of SVIA-infected GFP-LC3 MEFs and SVIA-infected GFP-LC3 neonatal mouse brains was performed using a rabbit polyclonal anti-SIN antibody as described in the Supplemental Experimental Procedures.
siRNA Treatment
p62, Atg7, and non-silencing negative control siRNA’s were purchased from Dharmacon. All siRNA experiments were performed using reverse transfection at a final concentration of 53 nM siRNA, Lipofectamine 2000 (Invitrogen) at a dilution of 1:1500, and otherwise according to manufacturer’s instructions. At 48 hours after siRNA transfection, protein knockdown was assessed by Western blot analysis and viral infections were performed.
Fluorescence, Light, and Electron Microscopy
Please refer to the Supplemental Experimental Procedures for details on the methods of sample preparation, microscopic imaging, and quantitative image analyses.
Coimmunoprecipitation and Radioimmunoprecipitation
For coimmunoprecipitation studies, mock- and virus-infected cell lysates were precleared with normal rabbit IgG and then immunoprecipitated using anti-SIN capsid or anti-SIN polyclonal antibodies. For radioimmunoprecipitation studies, infected lysates were fractioned into Triton X-100 soluble or insoluble fractions, precleared with normal rabbit IgG, and immunoprecipitated with anti-SIN polyclonal antibody. Western blot analyses were performed as described below. Autoradiographs were quantitated using ImageJ software (NIH). See Supplemental Experimental Procedures for additional details on coimmunoprecipitation and radioimmunoprecipitation methods.
Western Blot Analyses
Western blot analyses were performed using the methods described in the Supplemental Experimental Procedures with the following primary antibodies: anti-p62 (1:500 dilution) (Progen for MEFs and Neuro-2A cells, BD Biosciences for HeLa/GFP-LC3 cells), anti-Atg7 (1:500 dilution) (Sigma), anti-SIN virus capsid (1:10,000 dilution) (provided by M. McDonald), anti-Atg5 (1:2000) (Mizushima et al., 2001), and anti-actin (1:2000 dilution) (Santa Cruz).
Cell Viability Assays
Cell viability of HeLa/GFP-LC3 cells at serial time points after infection with dsTE12Q was determined by a Trypan Blue exclusion assay. Triplicate samples with a minimum of 100 cells per sample were counted for each experimental group.
Genomic PCR for Atg5
Genomic PCR for the Atg5flox/flox allele was performed as described (Hara et al., 2006). For in vitro analysis of SIN/Cre-mediated excision, primary Atg5flox/flox MEFs were infected with SIN/Cre or SIN/Cre.Stop at an MOI of 30, and genomic DNA from cell lysates were prepared using the Genomic DNA Buffer Set (Qiagen). Samples were then analyzed as described for mouse genotyping (Hara et al., 2006).
Autophagy Assays
Autophagosome accumulation was measured in GFP-LC3 MEFs by counting the percentage of cells with one or more green punctae and the number of punctae per positive cell by an observer blinded to experimental condition. Autophagic flux was measured by p62 Western blot analysis in MEFs and Neuro-2A cells. The presence of autophagic structures in MEFs was also confirmed by EM analyses.
Statistical Analyses
Log-rank tests were used to analyze all mortality studies, and student t-tests were performed for all other experiments using Prism software. A p value of < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Diane Griffin, David Leib, Margaret MacDonald, Noboru Mizushima, Victor Stollar, and Tamotsu Yoshimori for providing critical reagents. We thank Abhijit Bugde and Kate Luby-Phelps for assistance with live cell imaging, and Laurie Mueller for assistance with electron microscopy. This work was supported by the Ellison Medical Foundation Senior Scholars Award in Infectious Diseases (B.L), NIH RO1 AI151367 (B.L), and NIH T32 AI007520 (A.O.)
Footnotes
The Supplemental Data include Supplemental Experimental Procedures, a Supplemental Movie (S1) and Supplemental Figures (S1–6).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Fernandez C, Ronda-Lain C, Rubio-Huertos M. Electron microscopic study of Sindbis virus morphogenesis. Arch Gesamte Virusforsch. 1973;40:1–9. doi: 10.1007/BF01242630. [DOI] [PubMed] [Google Scholar]
- Griffin DE. Neuronal cell death in alphavirus encephalomyelitis. In Current Topics in Microbiology and Immunology. 2005;289:57–77. doi: 10.1007/3-540-27320-4_3. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hardwick JM, Levine B. Sindbis virus vector system for functional analysis of apoptosis regulators. Methods Enzymol. 2000;322:492–508. doi: 10.1016/s0076-6879(00)22045-4. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–9. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jackson AC, Moench TR, Trapp BD, Griffin DE. Basis of neurovirulence in Sindbis virus encephalomyelitis of mice. Lab Invest. 1988;58:503–9. [PubMed] [Google Scholar]
- Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–9. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RT. Virus invasion of the central nervous system: a study of Sindbis virus infection in the mouse using fluorescent antibody. Am J Pathol. 1965;46:929–43. [PMC free article] [PubMed] [Google Scholar]
- Johnson RT, McFarland HF, Levy SE. Age-dependent resistance to viral encephalitis: studies of infections due to Sindbis virus in mice. J Infect Dis. 1972;125:257–62. doi: 10.1093/infdis/125.3.257. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–71. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Wang HL, Stollar V. Complementation of and interference with Sindbis virus replication by full-length and deleted forms of the nonstructural protein, nsP1, expressed in stable transfectants of HeLa cells. Virology. 1997;227:361–9. doi: 10.1006/viro.1996.8342. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–77. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Levine B. Autophagy and viral neurovirulence. Cell Microbiol. 2008;10:1747–1756. doi: 10.1111/j.1462-5822.2008.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–9. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Rice CM, Strauss JH. Association of Sindbis virion glycoproteins and their precursors. J Mol Biol. 1982;154:325–48. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–98. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:7824–9. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Hurlbut HS, Work TH, Kingston JR, Frothingham TE. Sindbis virus: a newly recognized arthropod-transmitted virus. Am J Trop Med Hyg. 1955;4:844–62. doi: 10.4269/ajtmh.1955.4.844. [DOI] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–16. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–16. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.