Abstract
Extraction of crude membrane fractions with alkaline solutions, such as 100–200 mM Na2CO3 (pH ~11), is often used to solubilize peripheral membrane proteins. Integral membrane proteins are largely retained in membrane pellets. We applied this method to the fractionation of membrane proteins of the plague bacterium Yersinia pestis. Extensive horizontal spot trains were observed in 2-DE gels. The pI values of the most basic spots part of such protein spot trains usually matched the computationally predicted pI values. Regular patterns of decreasing spot pI values and in silico analysis with the software ProMoST suggested `n-1' deamidations of asparagine (N) and/or glutamine (Q) side chains for `n' observed spots of a protein in a given spot train. MALDI-MS analysis confirmed the occurrence of deamidations, particularly in N side chains part of NG dipeptide motifs. In more than ten cases, tandem MS data for tryptic peptides provided strong evidence for deamidations, with y- and b-ion series increased by 1 Da following N-to-D substitutions. Horizontal spot trains in 2-DE gels were rare when alkaline extraction was omitted during membrane protein sample preparation. This study strongly supports the notion that exposure to alkaline pH solutions is a dominant cause of extensive N and Q side chain deamidations in proteins during sample preparation of membrane extracts. The modifications are of non-enzymatic nature and not physiologically relevant. Therefore, quantitative spot differences within spot trains in differential protein display experiments following the aforementioned sample preparation steps need to be interpreted cautiously.
Keywords: Alkaline membrane extraction, deamidation, membrane proteome, spot train, two-dimensional gel electrophoresis
INTRODUCTION
Proteomic analysis of bacterial pathogens is useful to determine the expression of virulence factors and to characterize their dynamic changes under varying environmental conditions [1]. Membrane-associated proteins often play important roles in virulence and are implicated in the modulation of cell surface characteristics of bacterial pathogens in their host environments. In the Gram− bacterium Yersinia pestis, such proteins are the plasminogen activator (Pla) [2], the capsular F1 antigen [3], components of the type III secretion system [4] and the outer membrane (OM) protein Ail [5]. Membrane proteins tend to be less abundant and are less soluble in aqueous buffers than cytoplasmic and periplasmic proteins. Membrane proteins are typically enriched in pellet fractions of cell lysates and selectively extracted from membrane phospholipids [6, 7]. To isolate proteins with different membrane association features, the crude membrane fraction is washed with high salt buffers (e.g. 2.5 M NaBr), followed by alkaline extraction (e.g. 180 mM Na2CO3, pH 11.3) and solubilization of the remaining membrane pellet with a denaturing buffer (e.g. 8 M urea, 2 M thiourea and 1% amidosulfobetaine-14). High salt buffers solubilize extrinsic and membrane-adsorbed proteins by weakening their electrostatic interactions [8]. Alkaline extraction generates open membrane sheets from sealed membrane vesicles, resulting in removal of the majority of membrane-attached proteins devoid of transmembrane domains (TMDs) [8–10]. Detergents have varying degrees of effectiveness to solubilize and separate TMD proteins from associated phospholipids.
Post-translational modifications (PTMs) are biologically important, often enzymatic events that alter the function or the susceptibility of a given protein to degradation. Even though PTMs are not as frequent and less complex in bacteria compared to eukaryotes, they are known to occur. This includes the removal of short N-terminal signal peptides sequences from proteins destined for export, the removal of longer peptides from the N- or C-termini of enzyme precursors, e.g. peptidoglycan hydrolases [11], the removal of C-terminal fragments from cell wall-anchored proteins in Gram+ bacteria [12], phosphorylation of T, S or Y residues [13], methylation [14] and even protein glycosylation [15]. Such PTMs can result in significant shifts of pI and Mr values giving rise to spot trains in 2-DE gels. Glycosylation [16], phosphorylation [17], carbamylation [18] and proteolytic truncation [19] are known causes for altered pI and Mr values of proteins. Phosphorylation results in acidic pI shifts by 1 to 2 pH units per phosphoryl group [20]. Most PTMs are relatively infrequent and occur in small ratios compared to the unmodified protein. Therefore, the display of spot trains in 2-DE gels suggests chemical protein modifications rather than true PTMs. Frequently reported chemical modifications are acetylation, deacetylation, deamidation, oxidation and dephosphorylation of amino acid side chains as well as tryptophan decomposition [21–23].
Spot trains of bacterial proteins with acidic pI shifts in 2-DE gels have been reported, e.g. for Escherichia coli [24]. Acidic pI shifts have been associated with side chain deamidation reactions in the amino acids asparagine (N) and glutamine (Q) for human blood plasma proteins [25]. Deamidation reactions of N-glycine (NG) sequences were linked to sample preparation issues [21] and reported for bacterial and eukaryotic proteins [24, 25]. A single deamidated N or Q residue should result in a net Mr gain of 1 Da for a protein (not detectable in 2-DE gels) and a pH-dependent net charge gain detectable as an acidic pI shift. The net Mr gain of 1 Da can be determined in MS and MS/MS spectra of peptides resulting from the digestion of deamidated proteins. Here, we investigated the molecular causes of the appearance of Y. pestis membrane proteins as horizontal spot trains with acidic pI shifts in 2-DE gels.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The Y. pestis strain KIM6+ used in this study is an avirulent derivative of the fully virulent KIM strain, which was cured of the pCD1 plasmid but retained the chromosomal pgm locus and the plasmids pMT1 and pPCP1 [26]. Aliquots of bacterial colony stocks grown on tryptose blood agar and stored at −80°C were used to grow 5–10 mL pre-cultures in PMH2 media containing 10 μM FeCl3 for 8–15 hours, followed by dilution into 0.5–1 L of the same media. Overnight cell cultures at 26°C were grown to an A600 of ca. 1.9–2.5. Cell pellets were harvested by centrifugation at 8,000 × g for 15 min at 4°C and washed with a ca. 30-fold volume of 33 mM K2HPO4 (pH 7.5) as previously described [27].
Membrane protein extraction from Y. pestis cells
A lysozyme/EDTA spheroplasting method was used to generate spheroplasts [28]. Following cell lysis by sonication using 1 min on/off cycles (level 30, Branson sonifier), a mixed membrane pellet with inner and outer membranes (IM and OM, respectively) was separated from soluble cytoplasmic fractions by centrifugation at 108,000 × g for 60 min at 4°C. The crude membrane fraction was resuspended and homogenized in 10 mM Tris-HCl (pH 7.8), 5 mM EDTA, 0.2 mM DTT, 10 μg/ml Leupeptin, 5 μg/ml Pepstatin, 10 μg/ml TAME and 2 mM PMSF (ca. 10 mL/g pellet weight). Sodium bromide was added to a final concentration of 2.5 M, the membrane homogenate was stirred for 1h at 20°C and centrifuged at 51,000 × g for 60 min at 4°C. Proteins residing in the high salt-extracted membrane (hs-MBR) fraction were concentrated using Ultrafree-4 membrane filter devices (Mr cutoff of 10 kDa) to ca. 1–2 mg/mL and measured with the bicinchoninic acid reagent (Pierce Chemicals, Rockford, IL). The residual membrane pellet was re-homogenized in an ice-cold solution of 0.18 M Na2CO3 (pH 11.3), 50 mM DTT, 1 mM CaCl2, 1 mM MgCl2 and 1 mM MnCl. The membrane suspension was stirred for 1h at 4°C, followed by centrifugation at 108,000 × g for 60 min at 4°C. In a control experiment, the alkaline extraction step was substituted with a 2nd high salt extraction step with NaBr. Proteins solubilized with the Na2CO3 extraction step, called the high pH-extracted membrane (hpH-MBR) fraction, were concentrated to ca. 1–2 mg/mL in Ultrafree-4 devices as mentioned above. The membrane pellets were frozen at −80°C. Prior to protein analysis in 2-DE gels, these pellets – obtained from both the high pH and the control high salt extraction steps - were solubilized with 8 M urea, 2 M thiourea, 1% (w/v) amidosulfobetaine-14, 2 mM tributylphosphine and 0.5% Bio-Lyte pH 3–10 carrier ampholytes. Following incubation for 30 min at 20°C and centrifugation at 16,200 × g for 15 min, the supernatants (usb-MBR fractions) were analyzed, while the residual pellets were discarded.
Protein separation and differential protein spot display in 2-DE gels
Protein samples with an amount of ca. 100 μg were loaded onto 24 cm IPG strips (pH ranges of 4–7 and 3–10) and separated in 1st gel strips and 2nd dimension slab gels (25 × 19.5 × 0.15 cm). 2-DE gel separation conditions, the gel staining with Coomassie Brilliant Blue (CBB), gel imaging methods as well as spot matching and relative quantitation using the gel image analysis software Proteomweaver vs.4.0 (Bio-Rad, Hercules, CA) were previously described [29].
Mass spectrometry and bioinformatic protein analysis
Peptide digests were analyzed using MALDI-TOF/TOF mass spectrometry (4700 Proteomics Analyzer, Applied Biosystems, Framingham, MA) and LC-nESI-MS/MS mass spectrometry (LTQ, Thermo Fisher Scientific). Methods for spot cutting, protein digestion with trypsin and technical details for the MS analysis with the LTQ instrument were reported previously [29]. All MS spectra represented signal averaging of 1,000 laser shots and, for MS/MS spectra, 3,000 shots. In the MS/MS mode for MALDI analysis, the collision energy used to fragment peptides was 1 kV. A default calibration was applied using a six-component peptide standard spotted onto six edge or corner positions of the MALDI target plate. Data were searched against the latest release of the Y. pestis KIM strain subset of the NCBInr database, using the Mascot search engine vs.2.1 (Matrix Science, London, UK). Carbamidomethyl was invariably selected as a fixed modification and two missed tryptic cleavages were allowed, both for MALDI and LC-MS/MS. The MALDI mass range of data acquisition in the MS mode was m/z 800-4000. Search parameters included mass error tolerances of ±100 ppm for peptide ions and ±0.2 Da for fragment ions. Protein identifications were generally accepted as significant when a Mascot protein score >75 and one or more peptide e-values <0.1 were obtained. To search for protein deamidations, the web-based software Findmod (http://au.expasy.org/tools/findmod/) was used. Manual inspection of mass spectra was carried out using the Data Explorer® software vs.4.8 (Applied Biosystems). For bioinformatic predictions of protein export signal sequences, the algorithm SignalP (www.cbs.dtu.dk/services) was used [30]. The 2-DE gel analysis software ProMoST [31] was used to determine pI values and the pattern of spot trains in silico (http://prometheus.brc.mcw.edu/promost).
RESULTS AND DISCUSSION
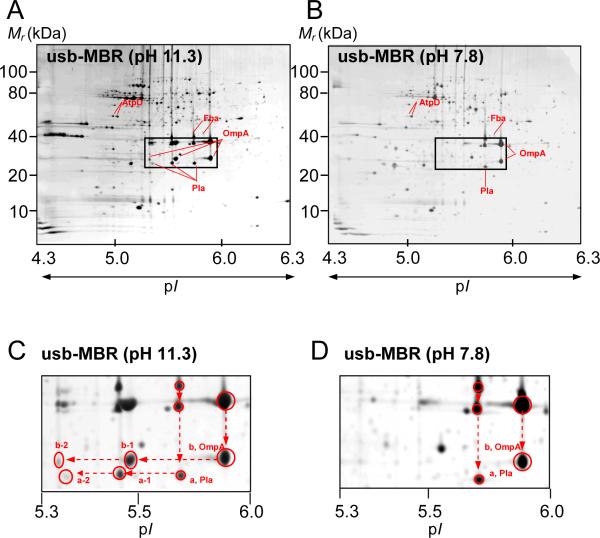
Proteins isolated in various subcellular fractions of the Y. pestis KIM6+ strain grown at 26°C were profiled using 2-DE gels, and spots were identified by tryptic peptide-based analysis of proteins with MS. This included the periplasm and cell culture supernatants [27], the cytoplasm and three fractions extracted from membrane pellets (unpublished data). Extensive protein spot trains were observed in 2-DE gels of those two membrane fractions whose proteins were exposed to a pH 11.3 during the extraction steps. Spot trains were far less prevalent in cytoplasmic, periplasmic and high salt-extracted membrane (hs-MBR) fractions which were not exposed to a highly alkaline pH. This study focused on the analysis of these spot trains and the inherent protein modifications from the high pH-extracted membrane (hpH-MBR) fraction and the urea/thiourea/amidosulfobetaine-14-solubilized membrane (usb-MBR) fraction. Proteins derived from the usb-MBR fraction were also exposed to the pH of 11.3, because the extraction step was performed subsequent to high pH membrane extraction with Na2CO3. After membrane extraction, proteins and peptides derived from them were not exposed to a pH higher than 8.8 -during sample preparation for 2-DE, the IPG and SDS-PAGE separation steps or protein digestion with trypsin. By omitting the high pH membrane extraction step with Na2CO3, a control usb-MBR fraction was isolated in which many of the same integral membrane proteins were identified as in the usb-MBR fraction. However, spot trains in the control usb-MBR fraction were far less prevalent than in the alkaline pH-extracted usb-MBR fraction. This is evident when comparing the spot patterns of the gel images in Figures 1A and B. While double spots with different pIs were observed in the control experiment (Figure 1B), more than 75% of the total spot volume for a double spot was usually associated with the more basic spot.
Figure 1.
Protein displays in 2-DE gels of Y. pestis KIM6+ membrane fractions isolated with (A,C) or without (B,D) extraction using an alkaline Na2CO3 solution. Circa 100 μg protein was loaded onto an IPG strip (24 cm, pH 4–7), separated in the first dimension, followed by SDS-PAGE (8–16%T) in the second dimension. Gels were stained with Commassie Brilliant Blue G-250. pI and molecular mass ranges for the profiled proteins are denoted. Magnified gel segments of the denoted rectangles in A and B are presented in C and D. The arrows indicate spots identified as OmpA and Pla. Vertical and horizontal spot trains for OmpA and Pla were reproduced in numerous 2-DE experiments.
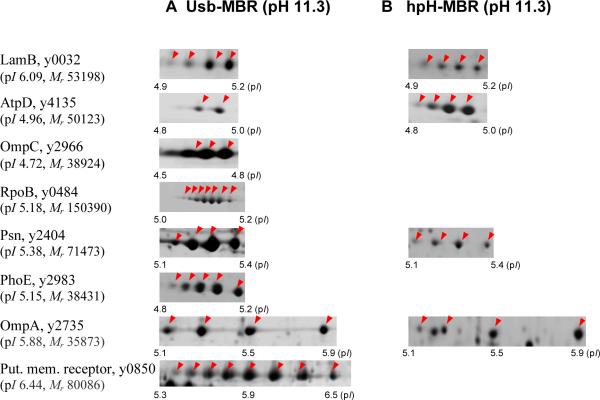
Protein identification of all the spots displayed in the gels of the hpH-MBR and usb-MBR fractions by MS revealed that many proteins partitioned into both fractions. This is illustrated with a few examples in Figure 2 (AtpD, OmpA, Psn and LamB). Proteins were displayed as spot trains with nearly identical experimental Mr and pI values. It suggests that differential extraction of protein isoforms in the two fractions is highly unlikely. This notion is further supported by the fact that the pI spot variants are nearly absent in the control usb-MBR fraction (Figure 1B). It can be concluded that the exposure to a high pH caused the emergence of protein variants with different pI values, displayed as horizontal spot trains. For example, OmpA and Pla did not display spot trains in the control usb-MBR fraction (Figure 1B), but in the usb-MBR fraction (Figure 1A). Acidic shifts for OmpA (b-1, b-2) and Pla (a-1, a-2) spots were observed with pI values at ~5.5 and ~5.9 (Figure 1A). These shifts could be due to the addition of one and two negative charges, respectively. Slight sample-to-sample variations of spot numbers and spot intensities in a given spot train were observed and seemed to be related to reproducibility differences in fractionation experiments rather than a lack of gel-to-gel reproducibility. For example, OmpA displayed a three-spot train in Figure 1A and a four-spot train in Figure 2. Differences in spot train appearance comparing distinct proteins were far greater. As shown in Figure 2, the outer membrane receptor y0850 featured eight spots, while AtpD featured only two spots. This suggested seven and one negative charge additions, respectively. Proteins with higher Mr values featured more extensive spot trains, suggesting a higher number of modifiable amino acid residues.
Figure 2.
Spot trains for proteins extracted in two membrane fractions. In hpH-MBR and usb-MBR fractions, membrane-associated proteins were exposed to an alkaline pH solution. Gel separation conditions were identical to those described in the legend of Figure 1. For each of the gel segments, the arrows indicate spot positions with increasingly negative protein charges towards the acidic end of the gels. Protein names and gene loci are based on annotation data in the KIM genome. The calculation of Mr and pI values included absent signal peptides of exported proteins, if applicable.
We assessed whether the protein environment in the membrane mattered regarding spot train formation. Fba, a cytoplasmic enzyme and possibly a contaminant in the usb-MBR fraction, featured a double spot. AtpD, a peripheral IM subunit of the membrane-associated ATP synthase, featured a spot train. Both OmpA and Pla are integral β-barrel OM proteins and also featured spot trains. All proteins are visualized and denoted in the gels of Figure 1. Apparently, neither the membrane association type of a protein nor its localization (IM or OM) play a role in the emergence of spot trains after exposure to an alkaline pH. We focused on the spot train analysis of integral OM proteins and cannot confirm this notion with highly reproducible data and statistical significance. In summary, membrane protein extraction under alkaline pH conditions caused widespread protein modifications which were visualized as horizontal spot trains in 2-DE gels and, considering acidic spot shifts, related to quantifiable differences in protein acidification.
Horizontal spot trains for bacterial membrane proteins have been previously reported. Spot trains of membrane-associated proteins were profiled in 2-DE gels after extraction/wash steps with Na2CO3 at the pH of 11 in E. coli [32] and Pseudomonas aeruginosa [33]. Similar spot trains were also detected for proteins after extraction/wash steps with lauryl sarcosine at the pH of 7.5–7.8, in Helicobacter pylori [34] and Pasteurella multocida [35]. Two studies reported spot trains for membrane-associated proteins, following cell lysis at a pH of 7.5–8.0 without detergent or carbonate wash steps, in Borrelia burgdorferi [36] and E. coli [24]. Apparently, spot trains and the underlying chemical modifications in proteins can result from sample preparation processes at a neutral pH. In the E. coli proteome analysis [24], it was suggested that deamidation of N and Q side chains of proteins is a major cause of 2-DE spot trains. Deamidated human blood plasma proteins have also been linked to the formation of extensive horizontal 2-DE spot trains. Using MS analysis, Sarioglu et al. [25] determined a link between N side chain deamidation of proteins and the addition of negative charges which, in turn, results in spot trains with increasingly acidic pI values. Considering these reports, it was of particular interest to examine Y. pestis protein spot trains in the context of N and/or Q deamidations.
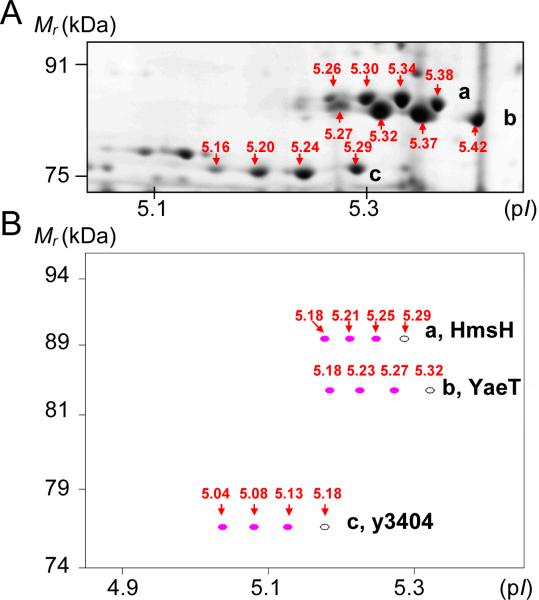
Using the ProMoST software, in silico predictions of N deamidation reactions supported the notion that such events had occurred in the examined Y. pestis proteome. As shown in Figure 3 for three integral OM proteins (HmsH, YaeT and y3404), differences in the experimental pI values and computed pI values of N-deamidated protein versions for a given spot train were in excellent agreement. In silico side chain phosphorylation of S, T or Y residues in the proteins resulted in spot trains stretched out across a wider pH range (not shown here). Gel staining with the dye ProQ Diamond did not provide any evidence for phosphorylation in membrane-associated proteins (data not shown). In summary, the simulation of N/Q deamidations using the ProMoST software was in good agreement with our experimental data. The number of spots (n) in a train appeared to correspond to the number of deamidations per protein molecule (n-1). The more acidic a given protein spot and the higher the Mr value of the protein, the smaller the pH distance to its nearest spot neighbor.
Figure 3.
Comparison of experimentally observed protein spot trains (A) and in silico predicted spot trains (ProMoST software) for equivalent proteins. Three in silico N deamidation events per protein were considered. The outer membrane proteins are HmsH (a), YaeT (b) and y3404 (c). Predicted spot patterns are similar to the experimental ones. The pI values denoted in (B) are calculated for proteins devoid of signal peptide sequences.
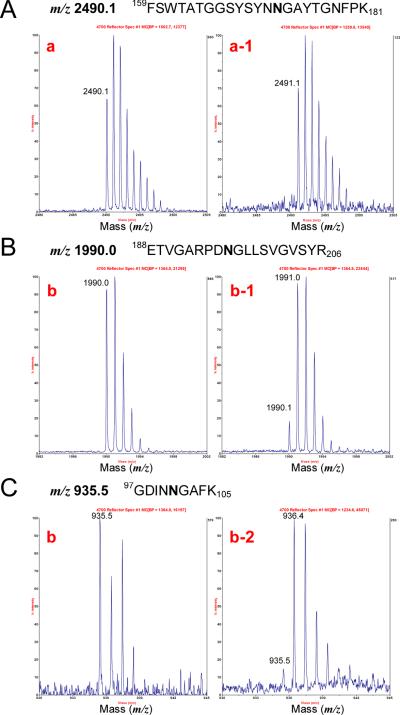
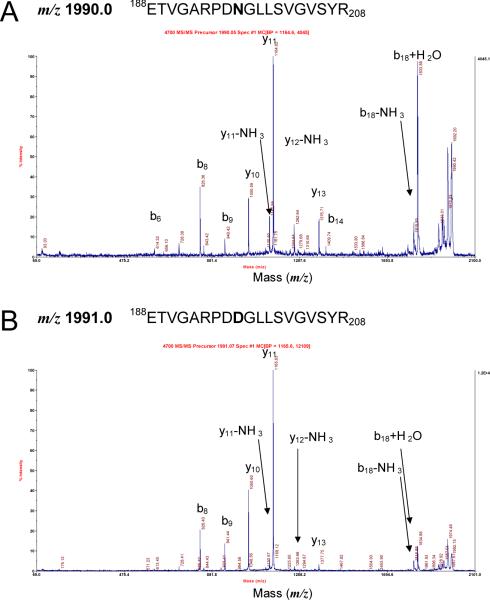
To determine whether spot trains (multiple charge variants) indeed constituted deamidated proteins, more than 100 peptide mass fingerprint (PMF) and MS/MS datasets were examined. N side chain deamidation theoretically results in a mass shift of +0.985 Da, and PMF data indeed revealed many such mass shifts in spots with acidic shifts on 2-DE gels. The detection of deamidations in high throughput MS analysis mode was difficult, however, because the mass change for a deamidated peptide is similar to that for a naturally occurring 13C isotope. Nonetheless, an irregular isotopic peak height pattern with an apparent mass shift of +1 Da was indicative of deamidation. This is illustrated for three tryptic peptides (one for Pla and two for OmpA) in the MALDI-MS spectra presented in Figure 4. A tryptic peptide of Pla (159FSWTATGGSYSYNN172GAYTGNFPK181) at m/z 2490.1 was present as a monoisotopic peak in the most basic spot (spot a), but absent in spot a-1 (see also Figure 1C). In contrast, the monoisotopic peak at m/z 2491.1, which was deamidated (N172→D), was detected in spot a-1. The peptides of OmpA (97GDINN101GAFK105 and 188ETVGARPDN196GLLSVGVSYR206) at m/z 935.5 and 1990.0, respectively, were present as monoisotopic peaks in the most basic spots (spots b), but weaker in spots b-1 and b-2 (Figure 1C). The corresponding deamidated peptide peaks (N101→D and N196→) at m/z 936.5 and 1991.0, respectively, were stronger in spots b-1 and b-2 (Figure 1C). Both of the deamidated peaks were observed in spot b-2. Lower peak heights of the unmodified peptide and the presence of more than one deamidated peptide in a given spot with one negative charge gain (m/z 936.5 and 1991.0, spot b-1) demonstrated that an individual protein spot was composed of a mixture of at least two distinct protein variants, each deamidated at a different N residue. Comparable results were reported in an analysis of human blood plasma proteins, but under experimental conditions without alkaline extraction [25].
Figure 4.
MS spectra for N-deamidated peptides derived from the Y. pestis outer membrane proteins Pla and OmpA. The numbers denoted in the spectra (a, a-1, b, b-1, b-2) correspond to protein spots displayed in the 2-DE gel image of Figure 1C.
Table 1 summarizes MS data for more than thirty peptides that featured isotope peak patterns with +1 Da mass shifts for monoisotopic peaks indicative of deamidation events. The list reveals that a G was frequently present C-terminal to the deamidated N residue. For example, all three above-mentioned OmpA and Pla peptides harbored NG motifs. The proteins PhoE and y0850, displayed as multi-spot trains in Figure 2, indeed harbored several NG motifs. MS data listed in Table 1 suggest that several proteins with acid-shifted spots on 2-DE gels featured one or more peptides with +1 Da m/z gains and NG motifs. Strong evidence for deamidations in NG motifs was obtained from the interpretation of MS/MS data. Fragment spectra for the unmodified OmpA peptide 188ETVGARPDN196GLLSVGVSYR206 at m/z 1990.03 and the deamidated peptide 188ETVGARPDD196GLLSVGVSYR206 at m/z 1991.02 are provided in Figure 5 and listed in Table 2. b- and y- ions increased by 1 Da starting with b9 and y11 ions, respectively, were observed and result in assignment of the deamidation to residue N196. Evidence from MS/MS spectra for deamidation in NG motifs was obtained for 11 peptides, derived from 10 different proteins (Table 1). As expected, the peptides were invariably associated with acidic spot shifts. MS and MS/MS data also suggested deamidations in Q side chains, e.g. for the proteins YaeT, OmpF and Psn (Table 1). We conclude that both Q and N side chains are susceptible to deamidation under alkaline conditions. MS data did not always allow conclusive evidence for the occurrence of N or Q deamidations, even if the associated spot trains were prominent and Mascot scores high. The addition of a negative charge occurring during deamidation from N to D and Q to E can result in depression of the ionization efficiency in the positive ion mode of the MS analysis, thus making the validation of deamidations more difficult. Tryptic peptides with NG motifs and less than seven or more than 35 amino acids were often not detected in MS spectra. In summary, MS data provided strong evidence for deamidation events in numerous proteins with acidic spot shifts. We noticed a prevalence of N deamidations in proteins harboring NG motifs, indicative of the fact that a small residue C-terminal to N facilitates deamidation. We have preliminary data supporting deamidations of Y. pestis proteins that were not exposed to alkaline solutions, in particular for proteins released from cells into extracellular media.
Table 1.
Peptide masses indicative of N and Q deamidations of Yersinia pestis proteins.
| Protein name | # of NG motifs | # of observed spots | observed MS peaks indicative of deamidation (m/z) | deamidated peptides according to MS/MS spectra (m/z) |
|---|---|---|---|---|
| HmsH | 4 | 4 | 1101.5, 1571.8, 2061.9, 2076.9 | 1571.8, 2076.9 |
| YaeT | 3 | 4 or 5 | 1191.6, 1905.9†, 1954.9† | 1954.9† |
| y3404 | 4 | 4 | 1125.5, 1168.5, 1324.7, 1508.8 | 1125.5 |
| Pla | 1 | 2 or 3 | 2491.1 | 2491.1 |
| OmpA | 2 | 3 or 4 | 936.4, 1991.0 | 1991.0 |
| PhoE | 4 | 5 | 1119.5*, 1807.8, 2071.0* | 1807.8 |
| LamB | 3 | 3 or 4 | 1405.7 | 1405.7 |
| y0850 | 6 | 8 or 9 | 1486.8, 2677.4, 3163.5 | - |
| FadL | 1 | 2 | 1238.6, 1864.9 | 1864.9 |
| OmpF | 3 | 4 | 1492.7†, 2982.3* | 1492.7†, 2982.3 |
| OmpC | 4 | 3 – 5 | 912.4, 1123.5* | - |
| PldA | 1 | 2 | 1493.7 | 1493.7 |
| RpoB | 7 | 6 or 7 | 1339.6, 1608.8, 1623.8†, 1667.8, 1788.9, 1944.9†, 1957.9 | - |
| AtpD | 1 | 2 | - | - |
| Psn | 3 | 4 | 1396.7, 1732.9, 1906.9, 1983.0† | 1396.7, 1983.0† |
peptide contains two NG motifs;
deamidation was observed for peptides with the amino acid Q;
all N deamidations were related to NG motifs.
Figure 5.
Peptide fragment spectra for the unmodified tryptic peptide 188ETVGARPDN196GLLSVGVSYR206 and the corresponding deamidated peptide of OmpA. The MALDI MS/MS spectra of the peptides with m/z 1990.0 and 1991.0 correspond to the peptide peaks displayed in Figure 4B.
Table 2.
Daughter ions produced by MALDI-MS/MS for a deamidated peptide of OmpA at m/z 1991.0.
| Observed m/z | Calculated m/z | Δ obs-calc | Sequence | Ion type |
|---|---|---|---|---|
| 826.40 | 826.41 | −0.01 | ETVGARPD | b8 |
| 923.41 | 922.44 | 0.97 | ETVGARPDN-NH3 | b9- NH3 |
| 941.44 | 940.45 | 0.99 | ETVGARPDN | b9 |
| 1050.60 | 1050.60 | 0 | GLLSVGVSYR | y10 |
| 1148.61 | 1147.61 | 1.00 | NGLLSVGVSYR-NH3 | y11-NH3 |
| 1165.63 | 1164.64 | 0.99 | NGLLSVGVSYR | y11 |
| 1263.66 | 1262.64 | 1.02 | DNGLLSVGVSYR-NH3 | y12-NH3 |
| 1377.75 | 1376.72 | 1.03 | PDNGLLSVGVSYR | y13 |
| 1467.82 | 1466.76 | 1.06 | ETVGARPDNGLLSVG | b15 |
| 1604.93 | 1603.86 | 1.07 | ARPDNGLLSVGVSYR | y15 |
| 1653.90 | 1652.86 | 1.04 | ETVGARPDNGLLSVGVS | b17 |
| 1816.88 | 1815.92 | 0.96 | ETVGARPDNGLLSVGVSY | b18 |
It was previously reported that deamidation proceeds more quickly, if N is followed by a small, flexible residue and exposed to alkaline environments [38, 39]. NG and NS sequences are particularly susceptible to deamidation [40]. A 70–80% ratio of N deamidation in NG motifs was reported by Krokhin et al. after prolonged tryptic protein digestion at a pH of 8.2 and at 37 ° [21]. Here, we did not observe extensive N deamidation resulting from in-gel tryptic protein digestion reactions. In the analysis of horizontal spots trains of human blood plasma proteins, deamidation was reported to contribute significantly to spot trains with a growing number of negative charges [25]. Several reports indicated that N or Q deamidation can be relevant in protein aging and degradation [41, 42], protein turnover [43], protein regulation [44] and apoptosis [45]. Deamidation in the context of protein aging would constitute a natural, though non-enzymatic process. Enzymatic post-translational deamidations have been discussed in the literature [46, 47], but there is general agreement that most deamidation events are nonenzymatic and influenced by physiological conditions during cell growth [48] and sample preparation [49]. N and Q residues in peptides and proteins were reported to be susceptible to deamidation not only at alkaline pH values, but also under certain ionic strength, temperature, protein-denaturating and solvation conditions [49].
There is currently no evidence for the biological significance of deamidations in membrane-associated proteins of Y. pestis or other Gram− bacterial pathogens. Extracellular, outer membrane and periplasmic proteins are more exposed to environmental changes in pH, temperature, oxidative and osmotic stress [50]. Interestingly, the four N side chains which are part of the NG motifs in the E. coli OM porin OmpG for which a crystal structure exists [51] are exposed on the periplasmic side, the cell surface and to the lumen of this porin. N deamidation of β-barrel porins could influence diffusion rates of small ions. Yersinia pestis expresses at least three porins (OmpC, OmpF, and PhoE) all of which harbor NG motifs. All three proteins also formed marked spot trains under alkaline extraction conditions. Two β-barrel OM proteins (Pla and OmpA) not only displayed horizontal, but also vertical spot trains. In three repeated experiments, sequence coverage of the lower Mr spots was identical to that of the higher Mr spots, suggesting that incomplete unfolding accounted for the low Mr spots. Incomplete unfolding in SDS-PAGE gels was previously observed for the orthologous E. coli protein OmpA [37]. It is known that OmpA migrates as a compact folded species unless it is completely heat-denatured [52]. Thus, such vertical spot trains are unrelated to amino acid modifications.
CONCLUSION
We demonstrated that N or Q side chain deamidation in membrane proteins exposed to highly basic pH solutions accounts for the formation of extensive spot trains in 2-DE gels. The evidence for non-enzymatic deamidation and a preference for deamidation in NG motifs is strong. It is plausible that deamidation of Y. pestis proteins exposed to the extracellular environment occurs naturally. This may contribute to protein aging, protein turnover and altered functions. Nonetheless, differential 2-DE display data with abundance changes in distinct spots of a protein spot train with characteristic acidic pI shifts are likely not physiologically important, particularly when proteins were exposed to alkaline solutions.
ACKNOWLEDGEMENTS
This work was performed under the Pathogen Functional Genomics Resource Center contract (contract No. N01-AI15447), funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Dr. Keehwan Kwon for helpful discussions.
Abbreviations
- IM
(inner membrane)
- OM
(outer membrane)
- TMD
(transmembrane domain)
- hs-MBR
(high salt-extracted membrane)
- hpH-MBR
(high pH-extracted membrane)
- usb-MBR
(urea/thiourea/amidosulfobetaine-14-extracted membrane)
REFERENCES
- [1].Cash P. Adv Biochem Eng Biotechnol. 2003;83:93–115. doi: 10.1007/3-540-36459-5_4. [DOI] [PubMed] [Google Scholar]
- [2].Kukkonen M, Suomalainen M, Kyllonen P, Lahteenmaki K, et al. Mol Microbiol. 2004;51:215–225. doi: 10.1046/j.1365-2958.2003.03817.x. [DOI] [PubMed] [Google Scholar]
- [3].Garcia E, Nedialkov YA, Elliott J, Motin VL, Brubaker RR. J Bacteriol. 1999;181:3114–3122. doi: 10.1128/jb.181.10.3114-3122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cornelis GR. J Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bartra SS, Styer KL, O'Bryant DM, Milles ML, et al. Infect. Immun. 2008;76:612–622. doi: 10.1128/IAI.01125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Santoni V, Kieffer S, Desclaux D, Masson F, Rabilloud T. Electrophoresis. 2000;21:3329–3344. doi: 10.1002/1522-2683(20001001)21:16<3329::AID-ELPS3329>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [7].Fischer F, Wolters D, Rogner M, Poetsch A. Mol Cell Proteomics. 2006;5:444–453. doi: 10.1074/mcp.M500234-MCP200. [DOI] [PubMed] [Google Scholar]
- [8].Schluesener D, Fischer F, Kruip J, Rogner M, Poetsch A. Proteomics. 2005;5:1317–1330. doi: 10.1002/pmic.200400993. [DOI] [PubMed] [Google Scholar]
- [9].Howell KE, Palade GE. J. Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu CC, MacCoss MJ, Howell KE, Yates JR. Nat. Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- [11].Beukes M, Bierbaum G, Sahl HG, Hastings JW. Appl. Environ. Microbiol. 2000;66:23–28. doi: 10.1128/aem.66.1.23-28.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gaspar AH, Marraffini LA, Glass EM, Debord KL, et al. J. bacteriol. 2005;187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Nat. Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- [14].Paik WK, Kim S. Science. 1971;174:114–119. doi: 10.1126/science.174.4005.114. [DOI] [PubMed] [Google Scholar]
- [15].Benz I, Schmidt MA. Mol Microbiol. 2002;45:267–276. doi: 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- [16].Hsu YR, Hsu EWJ, Katta V, Brankow D, et al. Protein Expr. Purif. 1998;12:189–200. doi: 10.1006/prep.1997.0840. [DOI] [PubMed] [Google Scholar]
- [17].Immler D, Gremm D, Kirsch D, Spengler B, et al. Electrophoresis. 1998;19:1015–1023. doi: 10.1002/elps.1150190617. [DOI] [PubMed] [Google Scholar]
- [18].McCarthy J, Hopwood F, Oxley D, Laver M, et al. J. Proteome Res. 2003;2:239–242. doi: 10.1021/pr025564b. [DOI] [PubMed] [Google Scholar]
- [19].Phadke ND, Molloy MP, Steinhoff SA, Ulintz PJ, et al. Proteomics. 2001;1:705–720. doi: 10.1002/1615-9861(200104)1:5<705::AID-PROT705>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [20].Yamagata A, Kristensen DB, Takeda Y, Miyamoto Y, et al. Proteomics. 2002;2:1267–1276. doi: 10.1002/1615-9861(200209)2:9<1267::AID-PROT1267>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- [21].Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Analytical Chemistry. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- [22].Chu FS, Bergdoll MS. Biochem. Biophys. Acta. 1969;194:279–286. doi: 10.1016/0005-2795(69)90204-9. [DOI] [PubMed] [Google Scholar]
- [23].Khan MI, Surolia A. Eur. J. Biochem. 1982;126:495–501. doi: 10.1111/j.1432-1033.1982.tb06807.x. [DOI] [PubMed] [Google Scholar]
- [24].Lopez-Campistrous A, Semchuk P, Burke L, Palmer-Stone T, et al. Mol Cell Proteomics. 2005;4:1205–1209. doi: 10.1074/mcp.D500006-MCP200. [DOI] [PubMed] [Google Scholar]
- [25].Sarioglu H, Lottspeich F, Walk T, Jung G, Eckerskorn C. Electrophoresis. 2000;21:2209–2218. doi: 10.1002/1522-2683(20000601)21:11<2209::AID-ELPS2209>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- [26].Fetherston JD, Perry RD. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- [27].Pieper R, Huang ST, Clark DJ, Robinson JM, et al. Proteomics. 2008;8:1442–1458. doi: 10.1002/pmic.200700923. [DOI] [PubMed] [Google Scholar]
- [28].Lucier TS, Fetherston JD, Brubaker RR, Perry RD. Infect Immun. 1996;64:3023–3031. doi: 10.1128/iai.64.8.3023-3031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pieper R, Gatlin-Bunai CL, Mongodin EF, Parmar PP, et al. Proteomics. 2006;6:4246–4258. doi: 10.1002/pmic.200500764. [DOI] [PubMed] [Google Scholar]
- [30].Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- [31].Halligan BD, Ruotti V, Jin W, Laffoon S, et al. Nucleic Acids Research. 2004;32:W638–W644. doi: 10.1093/nar/gkh356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Molloy MP, Herbert BR, Slade MB, Rabilloud T, et al. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- [33].Nouwens AS, Cordwell SJ, Larsen MR, Molloy MP, et al. Electrophoresis. 2000;21:3797–3809. doi: 10.1002/1522-2683(200011)21:17<3797::AID-ELPS3797>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- [34].Baik SC, Kim KM, Song SM, Kim DS, et al. J Bacteriol. 2004;186:949–955. doi: 10.1128/JB.186.4.949-955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Boyce JD, Cullen PA, Nguyen V, Wilkie I, Adler B. Proteomics. 2006;6:870–880. doi: 10.1002/pmic.200401342. [DOI] [PubMed] [Google Scholar]
- [36].Nowalk AJ, Gilmore RD, Jr., Carroll JA. Infect Immun. 2006;74:3864–3873. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. J. Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Robinson NE, Robinson AB. Proc. Natl. Acad. Sci. USA. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Peters B, Trout BL. Biochemistry. 2006;45:5384–5392. doi: 10.1021/bi052438n. [DOI] [PubMed] [Google Scholar]
- [40].Steven DS. Rapid Commun. Mass Spectrom. 2007;21:830–836. doi: 10.1002/rcm.2901. [DOI] [PubMed] [Google Scholar]
- [41].Lerm M, Pop M, Fritz G, Aktories K, Schmidt G. Infect. Immun. 2002;70:4053–4058. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sharma S, Hammen PK, Anderson JW, Leung A, et al. J. Biol. Chem. 1993;268:17695–17704. [PubMed] [Google Scholar]
- [43].Gershon H, Gershon D. Proceedings of the National Academy of Sciences. 1973;70:909–913. doi: 10.1073/pnas.70.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Reissner KJ, Aswad DW. Cellular And Molecular Life Sciences. 2003;60:1281–1295. doi: 10.1007/s00018-003-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Deverman BE, Cook BL, Manson SR, Niederhoff RA, et al. Cell. 2002;111:51–62. doi: 10.1016/s0092-8674(02)00972-8. [DOI] [PubMed] [Google Scholar]
- [46].Yan C, Sloan DL. J. Biol. Chem. 1987;262:9082–9087. [PubMed] [Google Scholar]
- [47].Stewart AE, Arfin SM, Bradshaw RA. J. Biol. Chem. 1994;269:23509–23517. [PubMed] [Google Scholar]
- [48].Hochstrasser DF. Clin. Chem. Lab. Med. 1998;36:825–836. doi: 10.1515/CCLM.1998.146. [DOI] [PubMed] [Google Scholar]
- [49].Wright HT. Crit. Rev. Biochem. Mol. Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- [50].Beveridge TJ. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Koebnik R, Locher KP, Van Gelder P. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- [52].Freudl R, Schwarz H, Stierhof YD, Gamon K, et al. J. Biol. Chem. 1986;261:11355–11361. [PubMed] [Google Scholar]