Abstract
We study the problem of classifying mild Alzheimer’s disease (AD) subjects from healthy individuals (controls) using multi-modal image data, to facilitate early identification of AD related pathologies. Several recent papers have demonstrated that such classification is possible with MR or PET images, using machine learning methods such as SVM and boosting. These algorithms learn the classifier using one type of image data. However, AD is not well characterized by one imaging modality alone, and analysis is typically performed using several image types – each measuring a different type of structural/functional characteristic. This paper explores the AD classification problem using multiple modalities simultaneously. The difficulty here is to assess the relevance of each modality (which cannot be assumed a priori), as well as to optimize the classifier. To tackle this problem, we utilize and adapt a recently developed idea called Multi-Kernel learning (MKL). Briefly, each imaging modality spawns one (or more kernels) and we simultaneously solve for the kernel weights and a maximum margin classifier. To make the model robust, we propose strategies to suppress the influence of a small subset of outliers on the classifier – this yields an alternative minimization based algorithm for robust MKL. We present promising multi-modal classification experiments on a large dataset of images from the ADNI project.
1 Introduction
Alzheimer’s Disease (AD) is a neurodegenerative disorder affecting over 5 million people in the United States, and is a leading cause of dementia worldwide. An emphasis in recent AD research, especially in the context of early diagnosis, has been placed on identifying markers of the disease (such as structural/functional changes in brain regions) using imaging data (e.g., MR, FDG-PET). Large scale studies such as the ADNI project [1] are collecting imaging data and associated clinical biomarkers in an effort to facilitate the development and evaluation of new approaches, and the identification of new imaging biomarkers. These advances are expected to yield important insights into the progression patterns of AD. One aspect of the ADNI project in particular is the acquisition and analysis of multi-modal imaging data: this includes Magnetic Resonance (MR), 18fluorodeoxyglucose-Positron Emission Tomography (FDG-PET), and Pittsburgh Compound B (PIB) PET image scans of the participants. The rationale is that because different modalities reveal different aspects of the underlying neuropathology, information from one modality adds to the diagnosis based on the other. For example, a patient may show only slight hippocampal atrophy in the MR images, but the FDG-PET image may reveal increased hypometabolism in medial-temporal and parietal regions (which is more suggestive of AD). Our objective here is to design machine learning algorithms which are (by design) cognizant of such multimodal imaging data, and “learn” the patterns differentiating controls from AD or MCI subjects using multiple modalities simultaneously: for predicting cognitive decline, or for identifying early symptoms of AD pathology.
The analysis of imaging data, in AD and aging research, has traditionally been approached by manual indication of brain regions suspected to be related to AD neurodegeneration, and performing statistical analysis to determine if group means (in those regions) are different [2]. Another approach is to automatically identify the discriminative regions using Voxel-based Morphometry (VBM) [3]. However, the diagnostic potential of group analysis is somewhat limited, usually by the degree of overlap in the group distributions. Therefore, a significant emphasis is being placed on determining and exploiting the predictive value of imaging-based biomarkers for diagnosis at the level of individual subjects. In this direction, a number of groups are exploring the applicability of machine learning ideas to this important problem. For instance, in [4], Support Vector Machines (SVM) were used to perform classification of structural MR scans after nominal feature selection. This procedure gave good classification accuracy on the Baltimore Longitudinal data set (BLSA). Recently [5] also used linear SVMs to classify AD cases from other types of dementia using whole brain MR images. High accuracy was obtained on confirmed AD patients and slightly less where post-mortem diagnosis was unavailable. Vemuri et al. showed promising evaluations on another data set, obtaining 88-90% accuracy [6], (also using linear SVMs). The authors of [7] proposed an augmented form of Linear Program Boosting (LP Boosting) which takes into account spatial characteristics of medical images, also reporting good accuracy on the ADNI data set. Observe that all of these methods have specifically focused on using a single imaging modality (e.g., MR or PET) for classification. One way to adapt such algorithms to make use of multiple imaging modalities (or additional clinical/cognitive data) is to “concatenate” the set of images for each subject into one feature vector. Not only does this increase the dimensionality of the distribution significantly, but it also requires finding a suitable “normalization” of each modality (to preserve its information content). Otherwise, features derived from one image type may easily overwhelm features from the other.
Contributions
The key contributions of this paper are (A) We propose an efficient multi-modal learning framework for AD classification based on multi-kernel learning (MKL). We cast the data from each imaging modality as one (or more) kernels, and solve for the support vectors (to maximize the margin) and the relative weights (importance) of each kernel; (B) To account for outliers (possibly misdiagnosed cases), the algorithm also incorporates a robustness parameter to identify such examples, and discount their effect on the classifier. This is tackled via alternative minimization; (C) We report the first set of multi-modal experimental results using robust-MKL learning on the ADNI dataset.
2 Preliminaries
We briefly review the underlying model for Support Vector Machines, before discussing the MKL setting and our construction. The SVM framework relies on the assumption that the concept (or classifier) we seek to learn (between two classes of examples) is well separated by a gap or margin in a certain feature space, and the algorithm can minimize the test error of a decision boundary by maximizing the width of the margin in the set of training examples provided. The decision boundary or separating hyper-plane, is parameterized by a weight vector w and offset (or bias) b. The classifier decides the possible class label for an unlabeled example x by calculating the inner product 〈w, x〉, and evaluating whether it is greater or less than b. SVMs operate on the principle that we must not only place examples on the correct side of the decision hyper-plane, but each example must also be far from the hyper-plane. In this case, the width of the margin is proportional to , see [8]. When choosing among two (or more) such decision boundaries (where both correctly classify all training data), the one with a smaller ℓ2-norm maximizes the margin and yields better accuracy. The SVM primal problem and corresponding dual problem are given as:
| (1) |
| (2) |
2.1 Multi-kernel learning
We first introduce Multi-kernel learning (MKL) [9], and then explain why it serves as a good basic formalization for our problem. In the next section, we will outline the main extensions. To motivate MKL, notice that in the dual of the SVM model (2), the dot product is replaced with a kernel function which expresses similarity among the the different data examples. This substitution offers a number of advantages, see [8]. Nonetheless, choosing the right kernel matrix for a given problem may not be straightforward, and typically requires adjustments. An attractive alternative is to represent each subset of features (e.g., each imaging modality) using its own kernel matrix and then seek an optimal combination of these kernels to form a single kernel matrix – one with the desirable properties of a good kernel (e.g., separable, maximizes the margin) for the complete data (i.e., all modalities). The process of choosing a set of coefficients, or sub-kernel weights, which are used to combine the candidate kernels into a single one, K̂ while simultaneously optimizing the expected test error is called Multi-Kernel Learning (MKL). The problem is formulated as follows.
| (3) |
Here, the coefficients βk are the sub-kernel weights. Notice that the squared ℓ1-norm penalty on the individual sub-kernel weights combined with the ℓ2-norm penalty on the weights in each individual view leads to sparsity among different kernels, but not among weights in each individual view [9]. In our AD classification problem, we use the set of images from each imaging modality to spawn a set of kernels. In other words, the distribution of the MR images of the set of subjects may give one (or a set of kernels). The same process is repeated for other types of images as well as any other form of demographic, clinical, or cognitive data available. The optimization problem then reduces to finding their weights (importance) while simultaneously maximizing the margin for the training data. The K̂ hence calculated is the combination of all available kernels.
3 Algorithm
3.1 Outlier Ablation
In addition to finding the optimal combination of kernels, we must also identify and suppress the influence of one or more mislabeled subjects (examples) on the classifier. This is important in the AD classification problem because of: (1) Co-morbidity: In some cases, AD is coincident with other neurodegenerative diseases such as Lewy bodies; (2) While the image data may suggest signs of pathology characteristic of AD, these usually precede cognitive decline. As a result, the subject may be cognitively normal (and labeled as control). To ensure that the algorithm is robust for this problem and other applications, we would like to identify such outliers within the model. In order to do this, one option within the SVM setting is to replace the regular loss function with the “robust” hinge loss function which differs only in that the “penalty” is capped at 1.
| (4) |
This means that once an example falls on the wrong side of the classifier there is no additional increase in penalty. To address the non-convexity of (4) the authors in [10] replaced the usual hinge loss function with the η–hinge loss function, which uses a discount variable ηi for each example. That is,
| (5) |
The result in [10] shows that η-hinge loss has the same optimum and value as robust-hinge loss. Our proposed model makes use of such a parameter to serve as both an outlier indicator and also to adjust the influence of this example on the classifier in the MKL setting. We present our optimization model next.
| (6) |
Here, wk is the set of weights for the kernel k, ξi is the slack for example i (similar to SVMs), and ηi,k is the discount on example i’s influence on training classification in view k (described in detail below).
Justification
Notice that η introduces a discount for every example’s influence on the classifier in each kernel. This is balanced by the positive reward for making η as large as possible. Therefore, an example which is badly characterized in some kernels can still be used effectively in other kernels where it is more accurately characterized. In this way, the proposed model performs automated outlier suppression in the MKL setting.
3.2 Alternative Minimization
We note that while (6) accurately expresses our problem, efficiently optimizing the objective function is rather difficult. To address this problem, we “relax” this formulation by treating the discount coefficients η fixed at each iteration. The value is iteratively updated according to the following expression.
| (7) |
Here, the denominator represents a normalization over all examples within a single kernel. This is necessary because different kernels have different variances, which must be accounted for (since we are combining kernels). Subsequent to setting the η variables, (6) can be solved to optimality, and η is again updated in the next iteration.
4 Experimental Results
In this section, we evaluate our multi-modal learning framework on image scans from the ADNI dataset. The Alzheimer’s disease neuroimaging initiative (ADNI) [1] is a landmark research study sponsored by the National Institutes of Health, to determine whether brain imaging can help predict onset and monitor progression of Alzheimer’s disease. The study is ongoing and will cover a total of 800 patients (200 healthy controls, 400 MCI individuals, and 200 mild AD patients). For our evaluations, we used MR and PET scans of 159 patients (77 AD, 82 controls) from this dataset. The data also provides a diagnosis for each subject based on clinical evaluations, this was used for training the classifier, and for calculating the accuracy of the system.
To evaluate our algorithm, we adopted a two fold approach. First, we measured the goodness of this approach w.r.t. to outlier detection, especially with respect to its effect on unseen test examples. In order to do this, we analyzed the variation in the kernel matrices as a response to outlier identification and suppression. Second, we evaluated the efficacy of the multi-kernel framework (with outlier detection) as a classification system, w.r.t. its accuracy using ROC curves. We discuss our experiments next.
4.1 Evaluation of Outlier Detection
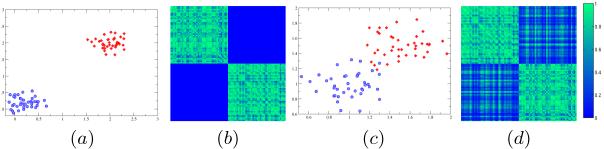
Here, we evaluate the usefulness of outlier detection in the classification model. Recall that an ideal input to any maximum margin classifier is a dataset where each class is separated from the other by a large margin. Since the MKL setup optimizes a collection of kernels, it is important to understand how a large margin in a data set translates to values in a kernel. To demonstrate this effect, we show two toy examples in Fig. 1. The first distribution is a setting where the classes are well separated (Fig. 1(a)): we see that the corresponding kernel matrix in Fig. 1(b) shows two distinct regions with high values (in green) for the two classes, where as the region pertaining to inter-class similarities shows no signal (in blue). In the second case (Fig. 1(c)), the classes are overlapping, which is also reflected in the kernel matrix being noisier as shown in Fig. 1(d).
Fig. 1.
Toy Example: (a) Well separated classes, (b) Kernel for well separated classes, (c) Overlapping Classes (d) Kernel for overlapping classes.
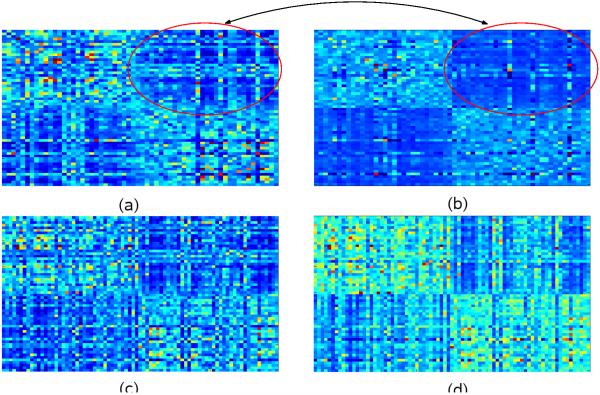
A similar effect can be observed in the kernels of our dataset as a response to outlier detection. Fig. 2 shows how outlier detection improves the signal quality in the kernel matrices. Fig. 2 (a) and (c) display the uncorrected train and test kernel matrices created simply by summing-up the set of individual kernel matrices. Fig. 2 (b) and (d) show the corresponding outlier-ablated train and test kernels. For visualization, the dataset is re-ordered with respect to groups before kernel creation, so that the kernel shows contiguous blocks (similar to Fig. 1). In 2 (a), we see vertical and horizontal lines of lighter color in the interclass region of the kernel, corresponding to outlier subjects who have a stronger resemblance to the opposite class. This effect is mitigated to a significant extent with outlier detection in Fig. 2 (b). Next, we analyze the effect of outlier ablation on unseen test items. For this, the test kernel is constructed with the training examples as rows and test examples as columns. In the kernel for the uncorrected case in Fig. 2 (c), the vertical lines correspond to unseen outlier subjects, whereas the horizontal lines are attenuated, indicating that in presence of training data, the non-outlier subjects have sharper contrast (causing an improved confidence in classification). Finally, the test kernel (after outlier detection) shown in Fig. 2 (d) shows a stronger within-class signal, and does not attempt to correctly classify the outliers, thereby discounting their effect on the decision boundary as desired (recall hinge loss from (4)).
Fig. 2.
(a) Sum of base kernel matrices on training examples. (b) Robust-MKL kernel matrix between training examples. Note that the two classes are clearly visible, and the vertical and horizontal lines corresponding to outliers are attenuated. (c) Sum of base kernel matrices on test examples. (d) Robust-MKL kernel matrix between test examples. Notice that while there are vertical lines corresponding to outlier test examples, the horizontal lines remain largely attenuated.
4.2 Efficacy of Multi-kernel framework
ROC curves and accuracy results
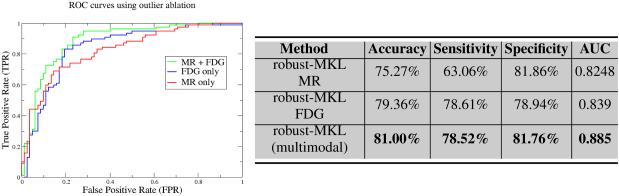
First, we evaluate the classification accuracy of our robust multi-kernel learning framework for single modality classification, using MR and FDG scans individually as well as both these modalities in a combined setting. We used a set of eight kernels each (linear and Gaussian with varying values of σ) for MR and FDG PET: 16 in all. Feature selection was performed using a simple voxel-wise t-test, and thresholding based on the p-values. We performed 10-fold cross-validation, and report the average of various error measures such as accuracy, sensitivity, and sensitivity (average over 25 runs). Our results are summarized in Figure 3. As expected, we can clearly see that the robust multi-modal framework with MR and FDG PET data out-performs the accuracy obtained using only one imaging modality (even when we use multiple kernels with each image type). The area under the curve (AUC) for the proposed algorithm is 0.885 suggesting that it is an effective method for AD classification.
Fig. 3.
ROC curves and Accuracy results for the single modal and multimodal classification using robust MKL
Interpretation of discriminative brain regions
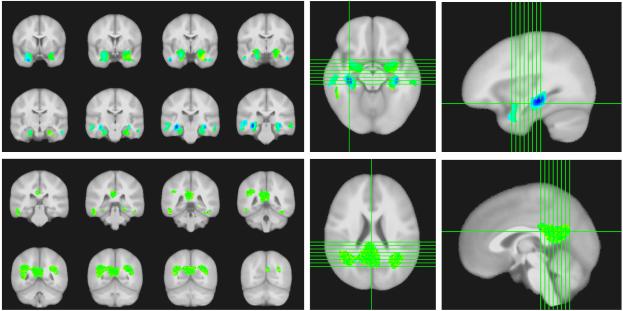
We evaluated the relative importance of various brain regions selected by the algorithm, and whether these regions are consistent with clinically accepted distributions of AD pathology. The classifier weights chosen by our algorithm are weights on different voxels, and therefore can be interpreted as distributions of weights on the brain regions. Fig. 4 shows the calculated weights for Gray Matter Probability (GMP) and FDG-PET images. For GMP, we see the hippocampus and hippocampal gyri are featured prominently, along with middle temporal regions. For FDG-PET, we see the posterior cingulate cortex and parietal lobules bilaterally are featured prominently. We find these results encouraging because the selected regions are all known to be affected in AD patients [11,12].
Fig. 4.
(Top) Classifier weights for gray-matter probability images shown overlaid on a template. (Bottom) Classifier weights for FDG-PET images shown overlaid on a template. The images (left) show the discriminative regions as a mosaic. The images (right) are provided for 3-D localization.
5 Conclusions
We have proposed a robust MKL framework for multi-modal AD classification. By framing each unique modality as one (or more) kernels, the scheme learns the kernel weights as well as a maximum margin classifier. Our framework also offers robustness to outliers, with the capability of automatically detecting them and partly discounting their influence on the decision boundary. Various sophisticated feature selection algorithms can be also be used as in [6] (discriminative image voxels) to further improve the accuracy of the model. Finally, rather than ad-hoc feature concatenation to make use of additional clinical and demographic data (if available), our algorithm allows an easy and intuitive incorporation – simply by constructing another kernel for such features.
Acknowledgments
This research was supported in part by the University of Wisconsin-Madison ICTR through an NIH Clinical and Translational Science Award (CTSA) 1UL1RR025011, a Merit Review Grant from the Department of Veterans Affairs, the Wisconsin Comprehensive Memory Program, and NIH grant AG021155. The authors also acknowledge the facilities and resources at the William S. Middleton Memorial Veterans Hospital. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI); CH was also supported by a UW-CIBM fellowship.
References
- 1.Jack CR, Bernstein MA, Fox NC, Thompson P, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008 doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apostolova LG, Thompson PM. Brain mapping as a tool to study neurodegeneration. Neurotherapeutics. 2007;4(3):387–400. doi: 10.1016/j.nurt.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner J, Friston KJ. Voxel-Based Morphometry - the methods. Neuroimage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 4.Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer’s disease: a high-dimensional pattern classification study. Neuroimage. 2008;41(2):277–285. doi: 10.1016/j.neuroimage.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klöppel S, Stonnington CM, Chu C, Draganski B, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vemuri P, Gunter JL, Senjem ML, Whitwell JL, et al. Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39(3):1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinrichs C, Singh V, Mukherjee L, Chung MK, Xu G, Johnson SC. Spatially Augmented LPBoosting with evaluations on the ADNI dataset. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.05.056. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schölkopf B, Smola AJ. Learning with kernels: Support Vector Machines, Regularization, Optimization, and Beyond. MIT Press; Cambridge, Mass: 2002. [Google Scholar]
- 9.Sonnenburg S, Rätsch G, Schäfer C, Schölkopf B. Large scale multiple kernel learning. Journ. of Machine Learning Research. 2006;7:1531–1565. [Google Scholar]
- 10.Xu L, Crammer K, Schuurmans D. Robust support vector machine training via convex outlier ablation; Proc. of AAAI; 2006. [Google Scholar]
- 11.Jack CR, Petersen RC, Xu Y, O’Brien PC, et al. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55(4):484–490. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minoshima S, Giordani B, Berent S, Frey KA, et al. Metabolic reduction in the posterior cingulate cortex in very early alzheimer’s disease. Ann Neurol. 1997:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]