Abstract
Adult neuronal stem cells (NSCs) hold great promise for brain repair because of their unique location within the central nervous system, their potential to proliferate and to differentiate into all major neural lineages, and their ability to functionally incorporate into existing neuronal circuitry after stroke. Nevertheless, the ability to exploit these cells for therapeutic purposes is hampered by the lack of knowledge about the signals that control the generation of a functional neuron from adult NSCs after stroke, particularly in the aged brain. Therefore, to further define the regulatory mechanisms that underlie neurogenesis after stroke, it is critically important to develop future NSC-based repair strategies. Notch signaling defines a fundamental pathway controlling cell fate acquisition. Studies have shown that Notch signaling pathways play critical roles during the maintenance, proliferation, and differentiation of NSCs in the developing brain. Recent evidence shows that Notch1 signaling is conserved in the regulation of adult neurogenesis. Here we summarize current knowledge about the role of Notch signaling in the regulation of neurogenesis in the normal and stroke brain.
Keywords: Neurogenesis, adult, Notch signaling, SVZ, stroke
Introduction
Stem cells are characterized by self-renewal and multi-lineage differentiation. Neural stem cells (NSCs) in the adult brain hold great promise for brain repair because of their unique location within the central nervous system (CNS), potential to proliferate and to differentiate into all major neural lineages, and ability to functionally incorporate into the existing neuronal circuitry. However, the capacity of such self-repair is limited. The successful development of NSC recruitment therapy will depend on our ability to manage the proliferation, migration, differentiation and functional integration of recruited cells. The first step towards this goal is to investigate the molecular mechanisms governing NSC behaviors. Therefore, to further define the regulatory mechanisms that underlie neurogenesis in the adult brain, it is critically important to develop future NSC-based repair strategies for treating neurological diseases such as stroke [1].
Growing evidence shows that biological behaviors, such as proliferation, migration and differentiation, of NSCs in the adult brain must be properly regulated by intrinsic signals and external factors [2-4]. Notch signaling defines a fundamental pathway controlling cell fate acquisition [5]. For example, Notch signaling pathways play critical roles during the maintenance, proliferation, and differentiation of NSCs in the developing brain. Recent studies show that Notch1 signaling is conserved in the regulation of adult neurogenesis. Here, we summarize the current knowledge about the role of Notch signaling in the regulation of neurogenesis in the adult brain under normal and pathological conditions such as stroke.
Notch family
The Notch gene was discovered in 1917 by Thomas Hunt Morgan, when it was first noticed in a strain of the fruit fly Drosophila melanogaster with notches apparent in their wing blades. However, its sequence was not determined until the 1980s. Studies show that the Notch signaling family is composed of a group of highly conserved proteins. So far, a number of Notch signaling family members, including Notch receptors, ligands and their corresponding intracellular signaling molecules have been identified.
Notch receptors in mammals
In mammals, the Notch genes encode large transmembrane proteins that act as receptors for the DSL (Delta, Serrate, Lag-2) family of ligands. There are four Notch membrane-bound type I receptors, referred to as Notch1, Notch2, Notch3, and Notch4 (Figure 1A). The Notch proteins are expressed on the cell surface as heterodimers composed of an extracellular region, a single transmembrane-pass, and a small intracellular region [5, 6]. The extracellular domain contains a variable number of epidermal growth factor (EGF)-like repeats, which are followed by three cysteine-rich LIN12/Notch repeats (LNR) that prevent signaling in the absence of the ligand [7, 8]. The Notch intracellular domain (NICD) contains a RAM23 domain [9], seven Ankyrin/CDC10 repeats involved in protein-protein interactions [10], and a PEST sequence [rich in proline (P), glutamic acid (E), serine (S) and threonine (T)] that negatively regulates protein stability [11]. In addition, Notch receptors 1-3 contain two nuclear localization signals (NLS) compared to one NLS in Notch4. The NSL is necessary to target the intracellular domain to the nucleus where the transcriptional activation domain (TAD) activates downstream events. Note that Notch3 and Notch4 contain no TAD domain.
Figure 1.

Schematic representation of the Notch receptor and ligand structure in mammals. A) There are four mammalian Notch receptors (Notch1-Notch4). Mature Notch proteins are comprised of the extracellular and transmembrane (intracellular) portions. Extracellular Notch proteins are characterized by numerous EGF-like repeats. Transmembrane Notch1 and Notch2 are almost identical in size and share many structural features, including the membrane-proximal RBP-J-associated molecule (RAM) domain, the Ankyrin (ANK) domain, two nuclear-localization sequences (NLSs); a carboxy-terminal transactivation domain (TAD), and a PEST (proline-, glutamate-, serine- and threonine-rich) domain. Transmembrane Notch3 and Notch4 are shorter and lack the TAD. The heterodimerization domain (HD) spans the region of interaction between the extracellular and transmembrane portions. B) Mammals have five mammalian Notch ligands (Jagged1, Jagged2, Delta-like 1 (DLL1), DLL3 and DLL4). Delta-likel, 3 and 4 are homologs of Drosophila Delta (dDelta), while Jagged1 and 2 are homologous to Drosophila Serrate. Notch ligands are transmembrane proteins of which the extracellular domain contains a characteristic number of EGF-like repeats and a cysteine rich N-terminal DSL domain. Jagged1 and Jagged2 contain an additional cysteine rich domain (CRD). SP: signal peptide.
Notch ligands in mammals
In flies, the two structurally related Notch ligands, Delta and Serrate, have both redundant and nonredundant functions [5, 12]. Notch ligands are conserved in vertebrates and include the Serrate orthologs Jagged1 and 2 and the Delta orthologs Delta 1, 3, and 4. In mammals, five structurally similar Notch ligands (Delta-like1, Delta-like3, Delta-Iike4, Jagged1, and Jagged2) have been identified (Figure 1B). Delta-likel, 3 and 4 are homologs of Drosophila Delta (dDelta), while Jagged1 and 2 are homologous to Drosophila Serrate (dSerrate). All Notch ligands are single pass transmembrane polypeptides, and their intracellular domains vary in length and do not display any significant sequence similarity except at the very end of the C-terminal. Notch ligands consist of multiple highly conserved EGF-like motifs and a conserved DSL domain [13]. In addition, Jagged1 and Jagged2 harbor an additional cysteine-rich domain. Because most ligands are also transmembrane proteins, the extracellular domain of Notch ligands interacts with Notch receptors expressed on neighboring cells that are in direct contact [13]. This way, groups of cells can organize themselves, such that, if one cell expresses a given trait, this may be switched off in neighboring cells by the inter-cellular Notch signal. Notably, there is little proof that Delta-like3 physically binds to the Notch receptors or that it truly functions as a Notch ligand [14].
Notch signaling
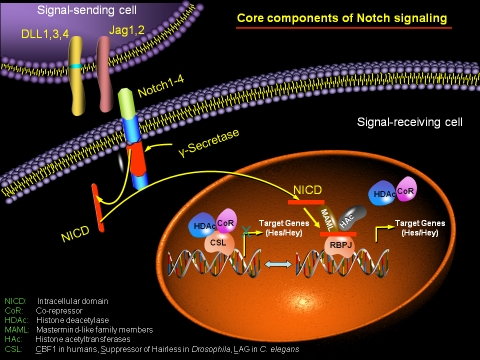
The Notch pathway is a complex signaling system, composed of a series of molecular events (Figure 2). Experiments in many different systems have provided a detailed model for Notch signaling that involves ligand-dependent cleavages of both the extracellular and intracellular domains of the receptor [15]. Notch signaling is initiated by direct cell-cell interactions that facilitate binding between the transmembrane Notch ligand Delta or Jagged on a signaling cell and the Notch receptor on a responding cell, leading to consecutive proteolytic cleavages of the receptor. The first cleavage occurs in the Golgi by the furin convertase enzyme (S1 cleavage), which results in the expression of a non-covalently linked Notch heterodimer receptor on the cell surface [16, 17]. In the absence of Notch ligand interaction, the cytoplasmic adaptor protein Numb interacts directly with the cytoplasmic domain of Notch and inhibits Notch activation. On the cell surface, proteolytic cleavage (S2 cleavage) by the TNF-α-Converting Enzyme (TACE) and ADAM17 metalloprotease occurs extracellularly (residue 1711). After cleavage, the extracellular portion of Notch continues to interact with the ligand and is then endocytosed by the ligand-expressing cell. After this first cleavage, Y-secretase, a complex composed of four different integral membrane proteins (presenilin, nicastrin (Nct), Aph-1, and Pen-2) cleaves the remaining part of the Notch protein just inside the inner leaflet of the cell membrane of the Notch-expressing cell (S3 cleavage). This occurs within the transmembrane domain (residue 1744) and leads to the release of the NICD into the cytoplasm [18, 19]. NICD subsequently translocates to the nucleus, where it binds via its RAM23 domain to the transcription factor CSL (CBF1 in humans, Suppressor of Hairless in Drosophila, LAG in C. elegans), also called RBPJK (Recombination Signal-Binding Protein 1 for J-Kappa) in mice [20]. CSL is bifunctional. In the absence of NICD, CSL binds to at least four co-repressors, the silencing mediator of retinoid and thyroid hormone receptor (SMRT), histone deacetylase-1 (HDAC1), KyoT2, and Ski-interacting protein (SKIP), which suppresses transcription [21]. In contrast, the interaction of CSL with the RAM23 and ANK repeats of the NICD displace these repressors to generate a transcriptional activator complex, which in turn regulates expression of Notch target genes [5]. The nuclear protein Mastermind-like (MAML) also interacts with this complex to further increase transcription. However, the truncated version of MAML that maintains an association with the complex, behaves in a dominant-negative (DN-MAML) fashion and inhibits Notch activation. The most widely accepted Notch/CSL targets are members of the basic helix-loop-helix (bHLH) hairy/enhancer of the split (Hes) family and the related HRT/Herp (Hes-related repressor protein) transcription factor family [22-24]. Other proteins also participate in the intracellular portion of the Notch signaling cascade.
Figure 2.
Schematic representation of the Notch signaling in mammals. Notch signaling is triggered upon ligand-receptor interaction which induces two sequential proteolytic cleavages, the first in the extracellular domain mediated by metalloproteases of the ADAM family, and the second within the transmembrane domain mediated by a γ-secretase activity of presenilins (PS). This second cleavage allows the release of the NICD, which translocates to the nucleus and associates with the CSL family transcription factor complex, resulting in subsequent activation of the notch target genes Hey and Hes family members.
Notch signaling and cell fate
Notch signaling regulates a wide variety of mammalian cell fates and processes, and plays a fundamental role in development. Signals exchanged between neighboring cells through the Notch receptor can amplify and consolidate molecular changes that eventually dictate cell fates. Thus, Notch signals control how cells respond to intrinsic or extrinsic developmental cues that are necessary to unfold specific developmental programs. Notch signaling also has a role in the following processes: 1) stabilization of arterial endothelial fate and angiogenesis. The finding that Notch genes are robustly expressed in the vasculature suggests roles for Notch in guiding endothelial and associated mural cells through the myriad of cell-fate decisions needed to form the vasculature [25, 26]. In fact, mice with defects in genes encoding Notch, Notch ligands, and components of the Notch signaling cascade invariably display vascular defects. For example, the survival of Notch4-deficient mice shows that Notch4 is dispensable for vascular development [27], while expression of an activated form of Notch4 within the endothelium disrupts normal vascular development [28, 29]. In addition, mutations in Notch3 or Jagged1 lead to human cardiovascular diseases: cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and Alagille syndrome (paucity of intrahepatic bile ducts with cholestasis, cardiac disease, skeletal abnormalities, ocular abnormalities, and characteristic facies), respectively [30]. 2) Bone regeneration and osteoporosis. Notch activation reduces the surface expression of c-Fms, a receptor for macrophage colony-stimulating factor, in osteoclast precursor cells and enhances the expression of osteoprotegerin in stromal cells, which results in the down-regulation of osteoclastogenesis [31]. In addition, the hematopoietic stem cell compartment is expanded during bone development and participates in commitment to the osteoblastic lineage, suggesting a potential role for Notch in bone regeneration and osteoporosis [32]. 3) Roles in hematopoietic and immune systems. Notch signaling has a key role in the process of lymphocyte development. Notch signaling specifies T cell lineage fate, and controls several early steps of T cell development, as well as specific cell fate and differentiation decisions in other hematopoietic lineages [33]. Notch signaling is necessary for the self-renewal of hematopoietic stem cell/progenitor cell and the onset of definitive hematopoiesis in the embryo as well [34, 35]. 4) Notch signaling is a key determinant of muscle regenerative potential that declines with age [36]. It also has a role in the following processes: neuronal function and development, cardiac valve homeostasis, regulation of cell-fate decision in mammary glands. Faulty Notch signaling is implicated in many diseases including T-ALL (T-cell acute lymphoblastic leukemia), Multiple SclerosisJTetralogy of Fallot, Alagille syndrome and a myriad of other disease states.
Adult neurogenesis in adult brain
Although Altman first observed the proliferative potential of the adult rodent brain in the 1960s, it has been thought for some time that the brains of adult mammals do not generate new neurons [37, 38]. After several years of debate, it is now accepted that the rostral subventricular zone (SVZ) surrounding the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyms (DG) are active proliferative regions that generate neurons [39], astrocytes [40] and oligodendrocytes [39, 41] continuously throughout life in mice [42], rats [43], non-human primates [44] and humans [45]. NSCs in the SVZ of the adult brain can be identified by 3H-thymidine and thymidine analog BrdU that is incorporated into the DNA of S-phase cells [37, 46], by expression of neural stem/progenitor marker such as nestin [47], and by their ability to form neurospheres that give rise to multiple cell types in vitro [48]. Four cell types have been identified in adult SVZ: (1) neuroblasts, or Type A cells; (2) astrocytes, or Type B1 and B2 cells; (3) undifferentiated, or Type C cells [49]; and (4) ependymal cells [40, 41]. The SGZ, a thin lamina between the hilar region and the granule cell layer of the hippocampal DG, retains the potential to form new neurons into adulthood [50, 51].
Stem cells in the SVZ generate immature neurons that aggregate to form an extensive network of neuroblast chains along the lateral wall of the lateral cerebral ventricle [52]. These chains of neuroblasts form a highly restricted migratory route, called the rostral migratory stream (RMS), which extends from the anterior SVZ into the olfactory bulb (OB). Unlike the radial glial-guided migration of young neurons during early brain development [53], neuroblasts undergo “chain migration” in the adult SVZ/RMS and migrate along one another, which involves interactions between the migrating cells and tube-like structures formed by specialized astrocytes [54]. Most of the neurons born in the adult SVZ migrate over a great distance to the OB through the RMS (2-6 Days). After immature neurons reach the OB, they begin to differentiate into two different types of local interneurons (15-30 Days). Over 95% differentiate into GABAnergic granule neurons whereas the remainder become periglomerular neurons expressing either GABA and/or dopamine as a neurotransmitter. Newborn granule cells and periglomerluar neurons become integrated into the OB circuitry and respond to olfactory stimuli (15-30 Days). Newborn granule cells can be classified into cells with dendrites that do not extend beyond the mitral cell layer and cells that possess non-spiny dendrites reaching into the external plexiform layer, whereas other cells in the SVZ may die shortly after their genesis [55-57]. However, the OB is not essential for proliferation and directed migration of SVZ precursors, since the proportion of dividing or dying cells in the RMS was not significantly affected after olfactory bulbectomy [58].
NSCs in the SGZ migrate into the granular cell layer (GCL) and undergo neuronal differentiation. However, the speed of maturation varies between neurons. The newborn cells eventually become physiologically indistinguishable from fully mature neurons (over 2 months old). In rodents, the rate of neurogenesis declines with age [51, 59, 60], but the number of granule cells in the DG increases into midlife and reaches a plateau thereafter [61]. Neurogenesis persists in the DG in elderly rodents [62] and humans [63] to maintain an equilibrium between the production of newborn cells and neuronal loss. Hippocampal neurogenesis in aged mice living in an enriched environment is higher by fivefold than in controls with significant improvements of learning parameters, exploratory behavior, and locomotor activity, suggesting that the old brain still has the ability to acutely react to functional challenges with a neurogenic response [64].
Role of Notch signaling in neurogenesis in normal brain
Notch receptors, in combination with other cellular factors, affects the implementation of differentiation, proliferation, and apoptotic programs at all stages of development [5]. Notch signaling inhibits neuronal differentiation in vertebrates and invertebrates [5, 65] as well as suppresses oligodendrocyte development from precursors during gliogenesis [66]. In the vertebrate CNS, Notch receptors and their ligands are expressed in the proliferative zones of undifferentiated cells [67] and multiple Notch genes are expressed in all or most NSCs of the developing zebra fish neural tube [68]. Jagged homologues have been identified as ligands of Notch receptors [69]. The concurrent knockdown of multiple Jagged homologs results in a phenotype that serves as a model for Alagille's syndrome, which produces cholestatis, pulmonic stenosis and poor school performance, among other deficits [70]. Recently, Hitoshi et al. demonstrated that (a) NSCs are missing almost completely in the E10.5 Notch∼/∼ and E8.5 RBP-Jkappa∼/∼ transgenic brains; (b) the number of NSCs in the PSl—/∼ brains decreases across embryogenesis; (c) the dissociation of single primary PSl∼/∼ neurospheres from E14.5 brains produces fewer secondary neurospheres than do wild-type spheres. This decline of self-renewal ability of PSl∼/∼ NCSs is partially rescued by transducing a constitutively active form of the Notch1 gene, suggesting that diminished Notch signaling is responsible for the attenuated self-renewal of PSl∼/∼ transgenic mouse NSCs [71]. Notch1, therefore, is a crucial regulator of neural stem cell maintenance and self-renewal during the development stage. Consistent with this finding, deletion of the basic helix-loop-helix (bHLH) transcriptional repressor Hes1, a known mediator of Notch signaling, causes premature neuronal progenitor cell differentiation and a reduction in the self-renewal capacity of embryonic forebrain NSCs [72]. Overexpression of activated Notch1 in the embryonic cortex results in an increase of radial glial cells [73], which have been implicated in neurogenesis [74].
Recently, a bevy of publications has alluded to the necessity of Notch signaling in the regulation of neurogenesis in the adult brain. Evidence for its involvement includes the following:
(1). Notch signaling components are expressed in neuroproliferative regions of the postnatal brain. In situ mRNA hybridization revealed that Notch1 is associated with cells in the SVZ, DG and RMS [75], and that most Notch 1-positive cells in the adult SVZ express PSA-NCAM and GFAP [76] Notch1 is also found in PSA-NCAM-positive neuroblasts located within the RMS and, to a diminished extent, in those that have reached the OB. In addition, mRNA and protein for two of the Notch1 activators, Jagged1 and Delta1, are also expressed in the SVZ and the RMS in the adult brain [75, 76]. Downstream targets of the Notch1 signaling Hes1, Hes3 and Hes5, and the intrinsic Notch regulatory proteins Numb and Numblike are also associated with cells expressing Notch1 [75-77]. Notch1 was found to be mainly expressed in doublecortin (DCX)-positive cells corresponding to newborn neurons, whereas the Notch1 ligand, Jagged1, is predominantly expressed in GFAP-positive astrocytic cells in the SVZ of the normal adult brain [78]. These findings are confirmed by conditional depletion of DCX-positive cells in transgenic mice carrying herpes simplex virus thymidine kinase (HSV-TK) under the control of the DCX promoter [78].
(2). Notch1 expression in the SVZ is reduced with aging, in parallel with a reduction of neurogenesis [76].
(3). Disruption of Notch1 using antisense or a Y-secretase inhibitor demonstrated a requirement for Notch1 in the maintenance and proliferation of NSCs in the adult brain [79]. Ablation or overexpression of Notch1 in GFAP-expressing astroglial cells dramatically affects the proliferation, cell fate, and survival of progenitors as well as the maturation of newly generated neurons, displaying the central role of Notch1 in postnatal hippocampal plasticity [80]. In vitro studies show that blockage of the Notch pathway with DAPT not only significantly reduced the number of neurospheres, but also the diameter of spheres. In addition, attenuation of endogenous Notch with siRNA resulted in a significant reduction of the percentage of BrdU-positive cells compared with that in non-transfected ischemic neural progenitor cells [78].
(4). Transient administration of Notch ligands to the brain of adult rats increases the number of NSCs [81]. In addition, the astrogliogenic response of the SVZ to injury in a model of cortical stab wound is accompanied by activation of the Notch pathway [76]. These findings indicate that NSC expansion may be achieved by Notch ligands through a pathway that is fundamental to development and cancer [82].
Role of Notch signaling in neurogenesis after stroke
The NSCs located in neurogenic regions increase in response to stroke and neurodegenerative diseases [43, 83, 84], and newborn cells can migrate into damaged brain regions [85], where they differentiate into mature neuronal cells and integrate into local neuronal circuits. We and others find that the activated form of Notch1 (NICD) and its downstream transcriptional targets, Hes1, are also expressed in SVZ cells after ischemia [77] [86-88]. Increased activation of Notch1 signaling increases SVZ cell proliferation, whereas inhibiting Notch1 signaling resulted in a reduction of proliferating cells in the SVZ. Levels of NICD, Hes1, and Shh are increased in the SVZ at 4 and 24 hr after focal cerebral ischemia. Interestingly, ischemia-induced cell proliferation in the SVZ is blocked by inhibition of the Notch1 signaling pathway [77]. An in vitro study shows that blockage of the Notch signaling by siRNA against Notch or a y-secretase inhibitor significantly reduces ischemia-mediated cell proliferation. During differentiation, Notch and Hes1 expression is down-regulated in neural progenitor cells after ischemia, which coincides with a significant increase in neuronal population, suggesting that the Notch signaling pathway mediates adult SVZ neural progenitor cell proliferation and differentiation after stroke [78]. These findings suggest that the Notch signaling pathway enhances the expansion of the neural progenitor pool and neuronal differentiation in adult neural progenitor cells after stroke.
Acknowledgments
This work was partially supported by Wenzhou Medical College 5010 grant and National Institute of Health (NIH) grant AG21980 and R01 NS057186 (K Jin).
References
- 1.Jagasia R, Song H, Gage FH, Lie DC. New regulators in adult neurogenesis and their potential role for repair. Trends Mol Med. 2006;12:400–405. doi: 10.1016/j.molmed.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- 3.Bordey A. Adult neurogenesis: basic concepts of signaling. Cell Cycle. 2006;5:722–728. doi: 10.4161/cc.5.7.2614. [DOI] [PubMed] [Google Scholar]
- 4.Hagg T. Molecular regulation of adult CNS neurogenesis: an integrated view. Trends Neurosci. 2005;28:589–595. doi: 10.1016/j.tins.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 6.Baker NE. Notch signaling in the nervous system. Pieces still missing from the puzzle. Bioessays. 2000;22:264–273. doi: 10.1002/(SICI)1521-1878(200003)22:3<264::AID-BIES8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 8.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 10.Blank V, Kourilsky P, Israel A. NF-kappa B and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- 11.Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- 12.Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- 13.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 14.Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand DII3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 16.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 18.Irvine KD. A notch sweeter. Cell. 2008;132:177–179. doi: 10.1016/j.cell.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 21.Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- 22.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 23.Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- 24.Nam Y, Aster JC, Blacklow SC. Notch signaling as a therapeutic target. Curr Opin Chem Biol. 2002;6:501–509. doi: 10.1016/s1367-5931(02)00346-0. [DOI] [PubMed] [Google Scholar]
- 25.Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M. Regulation of Notch1 and DII4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancini SJ, Mantei N, Dumortier A, Suter U, MacDonald HR, Radtke F. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 2005;105:2340–2342. doi: 10.1182/blood-2004-08-3207. [DOI] [PubMed] [Google Scholar]
- 27.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson TR, Yan Y, Wu X, Lam MT, Tang GL, Beverly LJ, Messina LM, Capobianco AJ, Werb Z, Wang R. Endothelial expression of constitutively active Notch4 elicits reversible arteriovenous malformations in adult mice. Proc Natl Acad Sci U S A. 2005;102:9884–9889. doi: 10.1073/pnas.0504391102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uyttendaele H, Ho J, Rossant J, Kitajewski J. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc Natl Acad Sci U S A. 2001;98:5643–5648. doi: 10.1073/pnas.091584598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 31.Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- 32.Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem. 2005;280:15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- 33.Sandy AR, Maillard I. Notch signaling in the hematopoietic system. Expert Opin Biol Ther. 2009 doi: 10.1517/14712590903260777. [DOI] [PubMed] [Google Scholar]
- 34.Thomas MD, Srivastava B, Allman D. Regulation of peripheral B cell maturation. Cell Immunol. 2006;239:92–102. doi: 10.1016/j.cellimm.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 2005;132:1117–1126. doi: 10.1242/dev.01660. [DOI] [PubMed] [Google Scholar]
- 36.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 37.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rat. J Comp Neurol. 1965;124:319–336. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 38.Altman J. Aremnew neurons formed in the brains of adult mammals? Science. 1962;135:1127–1128. doi: 10.1126/science.135.3509.1127. [DOI] [PubMed] [Google Scholar]
- 39.Chiasson BJ, Tropepe V, Morshead CM, van der Kooy D. Adult mammalian forebrain ependymal and subependymal cells demonstrate proliferative potential, but only subependymal cells have neural stem cell characteristics. J Neurosci. 1999;19:4462–4471. doi: 10.1523/JNEUROSCI.19-11-04462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 41.Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisen J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimura S, Takagi Y, Harada J, Teramoto T, Thomas SS, Waeber C, Bakowska JC, Breakefield XO, Moskowitz MA. FGF-2 regulation of neurogenesis in adult hippocampus after brain injury. Proc Natl Acad Sci U S A. 2001;98:5874–5879. doi: 10.1073/pnas.101034998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin K, Minami M, Lan JQ, Mao XO, Batteur S, Simon RP, Greenberg DA. Neurogenesis in dentate subgranular zone and rostral subventricular zone after focal cerebral ischemia in the rat. Proc Natl Acad Sci U S A. 2001;98:4710–4715. doi: 10.1073/pnas.081011098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDermott KW, Lantos PL. Distribution and fine structural analysis of undifferentiated cells in the primate subependymal layer. J Anat. 1991;178:45–63. [PMC free article] [PubMed] [Google Scholar]
- 45.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 46.del Rio JA, Soriano E. Immunocytochemical detection of 5'-bromodeoxyuridine incorporation in the central nervous system of the mouse. Brain Res Dev Brain Res. 1989;49:311–317. doi: 10.1016/0165-3806(89)90033-3. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 48.Craig CG, Tropepe V, Morshead CM, Reynolds BA, Weiss S, van der Kooy D. In vivo growth factor expansion of endogenous subependymal neural precursor cell populations in the adult mouse brain. J Neurosci. 1996;16:2649–2658. doi: 10.1523/JNEUROSCI.16-08-02649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gage FH, Kempermann G, Palmer TD, Peterson DA, Ray J. Multipotent progenitor cells in the adult dentate gyms. J Neurobiol. 1998;36:249–266. doi: 10.1002/(sici)1097-4695(199808)36:2<249::aid-neu11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Cameron HA, McKay RD. Restoring production of hippocampal neurons in old age. Nat Neurosci. 1999;2:894–897. doi: 10.1038/13197. [DOI] [PubMed] [Google Scholar]
- 52.Doetsch F, Alvarez-Buylla A. Network of tangential pathways for neuronal migration in adult mammalian brain. Proc Natl Acad Sci U S A. 1996;93:14895–14900. doi: 10.1073/pnas.93.25.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rakic P. Principles of neural cell migration. Experientia. 1990;46:882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- 54.Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- 55.Gould E, Beylin A, Tanapat P, Reeves A, Shors TJ. Learning enhances adult neurogenesis in the hippocampal formation. Nat Neurosci. 1999;2:260–265. doi: 10.1038/6365. [DOI] [PubMed] [Google Scholar]
- 56.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 57.Kornack DR, Rakic P. The generation, migration, and differentiation of olfactory neurons in the adult primate brain. Proc Natl Acad Sci U S A. 2001;98:4752–4757. doi: 10.1073/pnas.081074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirschenbaum B, Doetsch F, Lois C, Alvarez-Buylla A. Adult subventricular zone neuronal precursors continue to proliferate and migrate in the absence of the olfactory bulb. J Neurosci. 1999;19:2171–2180. doi: 10.1523/JNEUROSCI.19-06-02171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyms of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kempermann G, Kuhn HG, Gage FH. Experience-Induced Neurogenesis in the Senescent Dentate Gyrus. J. Neurosci. 1998;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kempermann G, Kuhn HG, Gage FH. Experience-induced neurogenesis in the senescent dentate gyrus. J Neurosci. 1998;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gould E, Reeves AJ, Fallah M, Tanapat P, Gross CG, Fuchs E. Hippocampal neurogenesis in adult Old World primates. Proc Natl Acad Sci USA. 1999;96:5263–5267. doi: 10.1073/pnas.96.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kukekov VG, Laywell ED, Suslov O, Davies K, Scheffler B, Thomas LB, O'Brien TF, Kusakabe M, Steindler DA. Multipotent stem/progenitor cells with similar properties arise from two neurogenic regions of adult human brain. Exp Neurol. 1999;156:333–344. doi: 10.1006/exnr.1999.7028. [DOI] [PubMed] [Google Scholar]
- 64.Kempermann G, Gast D, FH G. Neuroplasticity in old age: Sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann Neurol. 2002;22 doi: 10.1002/ana.10262. [DOI] [PubMed] [Google Scholar]
- 65.Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 66.Tseng CK, Tsang NM, Kao SC, Chen SY, Chen YP. A quick method to extract DNA from paraffin-embedded tissues. Changgeng Yi Xue Za Zhi. 1998;21:63–66. [PubMed] [Google Scholar]
- 67.Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 68.Appel B, Givan LA, Eisen JS. Delta-Notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 70.Lorent K, Yeo S-Y, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 71.Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura Y, Sakakibara S-i, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH Gene Hes1 as a Repressor of the Neuronal Commitment of CNS Stem Cells. J. Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murineforebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 74.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 75.Stump G, Durrer A, Klein AL, Lutolf S, Suter U, Taylor V. Notch1 and its ligands Delta-like and Jagged are expressed and active in distinct cell populations in the postnatal mouse brain. Mech Dev. 2002;114:153–159. doi: 10.1016/s0925-4773(02)00043-6. [DOI] [PubMed] [Google Scholar]
- 76.Givogri MI, de Planell M, Galbiati F, Superchi D, Gritti A, Vescovi A, de Vellis J, Bongarzone ER. Notch signaling in astrocytes and neuroblasts of the adult subventricular zone in health and after cortical injury. Dev Neurosci. 2006;28:81–91. doi: 10.1159/000090755. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, Mao X, Xie L, Greenberg DA, Jin K. Involvement of Notch1 signaling in neurogenesis in the subventricular zone of normal and ischemic rat brain in vivo. J Cereb Blood Flow Metab. 2009 doi: 10.1038/jcbfm.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Chopp M, Zhang RL, Zhang L, Letourneau Y, Feng YF, Jiang A, Morris DC, Zhang ZG. The Notch pathway mediates expansion of a progenitor pool and neuronal differentiation in adult neural progenitor cells after stroke. Neuroscience. 2009;158:1356–1363. doi: 10.1016/j.neuroscience.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chojnacki A, Shimazaki T, Gregg C, Weinmaster G, Weiss S. Glycoprotein 130 Signaling Regulates Notch1 Expression and Activation in the Self-Renewal of Mammalian Forebrain Neural Stem Cells. J. Neurosci. 2003;23:1730–1741. doi: 10.1523/JNEUROSCI.23-05-01730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 82.Alexson TO, Hitoshi S, Coles BL, Bernstein A, van der Kooy D. Notch signaling is required to maintain all neural stem cell populations-irrespective of spatial or temporal niche. Dev Neurosci. 2006;28:34–48. doi: 10.1159/000090751. [DOI] [PubMed] [Google Scholar]
- 83.Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- 84.Jin K, Wang X, Xie L, Mao XO, Zhu W, Wang Y, Shen J, Mao Y, Banwait S, Greenberg DA. Evidence for stroke-induced neurogenesis in the human brain. PNAS. 2006;103:13198–13202. doi: 10.1073/pnas.0603512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jin K, Sun Y, Xie L, Peel A, Mao XO, Batteur S, Greenberg DA. Directed migration of neuronal precursors into the ischemic cerebral cortex and striatum. Mol Cell Neurosci. 2003;24:171–189. doi: 10.1016/s1044-7431(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 86.Kawai T, Takagi N, Nakahara M, Takeo S. Changes in the expression of Hes5 and Mash1 mRNA in the adult rat dentate gyms after transient forebrain ischemia. Neurosci Lett. 2005;380:17–20. doi: 10.1016/j.neulet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Guo YJ, Zhang ZJ, Wang SH, Sui YX, Sun Y. Notch1 signaling, hippocampal neurogenesis and behavioral responses to chronic unpredicted mild stress in adult ischemic rats. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:688–694. doi: 10.1016/j.pnpbp.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 88.Chen J, Zacharek A, Li A, Cui X, Roberts C, Lu M, Chopp M. Atorvastatin promotes presenilin-1 expression and Notch1 activity and increases neural progenitor cell proliferation after stroke. Stroke. 2008;39:220–226. doi: 10.1161/STROKEAHA.107.490946. [DOI] [PMC free article] [PubMed] [Google Scholar]