Abstract
Finding relations among gene expressions involves the definition of the similarity between experimental data. A simplest similarity measure is the Correlation Coefficient. It is able to identify linear dependences only; moreover, is sensitive to experimental errors. An alternative measure, the Shannon Mutual Information (MI), is free from the above mentioned weaknesses. However, the calculation of MI for continuous variables from the finite number of experimental points, N, involves an ambiguity arising when one divides the range of values of the continuous variable into boxes. Then the distribution of experimental points among the boxes (and, therefore, MI) depends on the box size. An algorithm for the calculation of MI for continuous variables is proposed. We find the optimum box sizes for a given N from the condition of minimum entropy variation with respect to the change of the box sizes. We have applied this technique to the gene expression dataset from Stanford, containing microarray data at 18 time points from yeast Saccharomyces cerevisiae cultures (Spellman et al.,[3]). We calculated MI for all of the pairs of time points. The MI analysis allowed us to identify time patterns related to different biological processes in the cell.
Keywords: Gene expression, Mutual information
INTRODUCTION
An important problem in bioinformatics is that of finding relations among experimental data. A typical task is to find pairs of variables in an experimental dataset, which depend on each other, usually in the presence of a strong noise. A common way to characterize relations among experimental data is the correlation coefficient (CC). Unfortunately, it is designed to reveal linear dependence and fails to identify dependences which are significantly non-linear. Also, it is sensitive to outliers, i.e. points located strongly apart of the majority of the points. These exceptional points are often experimental errors and they can dramatically affect CC values. A well-known alternative to the CC, the Shannon mutual information (MI) [1], is free from these shortcomings. MI is widely used to describe dependences among discrete variables (see, e.g. [2]). However, the application of MI to continuous variables is less straightforward, due to some ambiguities which arise here. The problem is that the number of experimental points is always finite, N < ∞. Hence, one is forced to divide the range of values of the continuous variable into bins or boxes and to introduce the auxiliary discrete variable Nk equal to the number of points in every box. The resulting distribution depends on additional parameters: number of experimental points N and the box sizes. This dependence strongly affects MI.
Here, we seek a hopefully unique solution to the problem, of how, for a given value of N, to find the best values of the box sizes. Our motivation is to apply the potentially powerful tool of MI to the analysis of gene expression data. We confine our discussion by using the publicly available RNA expression dataset from Stanford, containing microarray data at 18 time points from yeast Saccharomyces cerevisiae cultures synchronized by α factor arrest (Spellman et al., [3]). This dataset has been thoroughly investigated and clusters of genes that represent similar behavior were found, using CC to measure similarity in gene expression patterns (Eisen et al., [4]). More general dependencies including time shifts and causal relationships (Local Invariants) for this dataset have been found (Fofanov et al., [5]). Clustering using the mutual information instead of the correlation coefficient to measure similarity among genes behavior has been also performed (Butte and Kohane, [6]). In section 2, we propose a method of defining the optimal box size for calculation of MI. In section 3, we apply this technique to the yeast dataset. We calculate the mutual information for expression values for all pairs of time points and discuss the time evolution of gene expression. The MI analysis allows us to identify three different time patterns: periodic behavior due to the cell division cycle, descending behavior due to the response to the initial synchronization (e.g. α factor arrest) and stationary behavior due to the group of genes not involved directly into the cell division cycle or response to initial synchronization.
MUTUAL INFORMATION AND BOX SIZES
We start with the one-dimensional case, recalling the definitions of Shannon entropy for the discrete and continuous random variable. Let K be a discrete random variable and k, its value. We have N experimental points and Nk of them correspond to the value k. The probability of k and entropy the probability distribution are defined as
| (1) |
For the case of a continuous random variable, X, which takes the values x, we introduce an auxiliary discrete variable K - the number of a box Δk of size Δ to which a given value of X belongs: K=k, if kΔ ≤ x < (k+1)Δ. Then, Nk is the number of experimental points in the box k. Probability distributions Pk and P(x) are connected as follows,
| (2) |
P(x̄) goes to P(x) in the limit Δ → 0. (Note, that first the limit N → ∞ and second, Δ → 0 have to be taken). We define the entropy of a continuous variable X as , i.e.,
| (3) |
In the case of two random variables x and y, with a joint distribution,
where Nkj is a number of experimental points in a box of size Δ = Δx×Δy,
joint entropy may be defined as
| (4) |
Mutual information is defined as follows,
where marginal entropies Sx and Sx have the form (3) (with ). Note, that we have from experiments samples of a random variable, and we need to work with finite values of N and Δ.
To chose the best box size for given N, we suggest that changing twice the sizes , of the rectangular box which contains the point (x,y) does not significantly affect the value of P(x,y). We calculate the corresponding variation of the entropy, δ(N, Δ) = S(N, Δ′)−S(N, Δ). Let us denote f(x) = x log x. It is clear that f(x+y) > f(x) + f(y), and moreover,
where and xi = c + δi. Using this equation to calculate the change of the entropy and reducing rectangular boxes from Δ= ΔxΔy to Δ/4= (Δx/2)(Δy/2) we have
| (5) |
where there are Nkj experimental points inside the box (kj) and they are distributed among sub-boxes as follows, Ni,kj = Nkj/4 +δi(k,j), i = 1,2,3,4.
It is natural to chose the box sizes which minimize the entropy variation δ(N, Δx, Δy). This leads to a simple and fast algorithm, which is in a very good correspondence with exactly solvable examples, such as normal or exponential distributions.
APPLICATION TO YEAST DATA
Now, we will apply this technique the analysis of time dependent gene expression data from yeast Saccharomyces cerevisiae cultures. Yeast time-series microarray experiments were performed by Cho (Cho et al., [7]) and Spellman (Spellman et al., [3]) using several methods of synchronization: α factor arrest, elutriation, cdc15 temperature sensitive mutant, and cdc28. There are many online sources from which these datasets can be downloaded. A common dataset contains 2467 genes. We briefly recall the experimental procedures from [3] for the α factor arrest. The α factor was added to asynchronous yeast culture for 120 min. The α factor was then removed, and the arrested cells were placed in fresh medium. Samples were taken every 7 min for 119 min. The synchronization experiments produced cell cycle synchrony through two cycles, as verified by bud count, FACS analysis and/or nuclear staining. Evidence for cell cycle count was not published for elutriation and α factor arrest, but has been performed and supports the cell cycle count (personal communication).
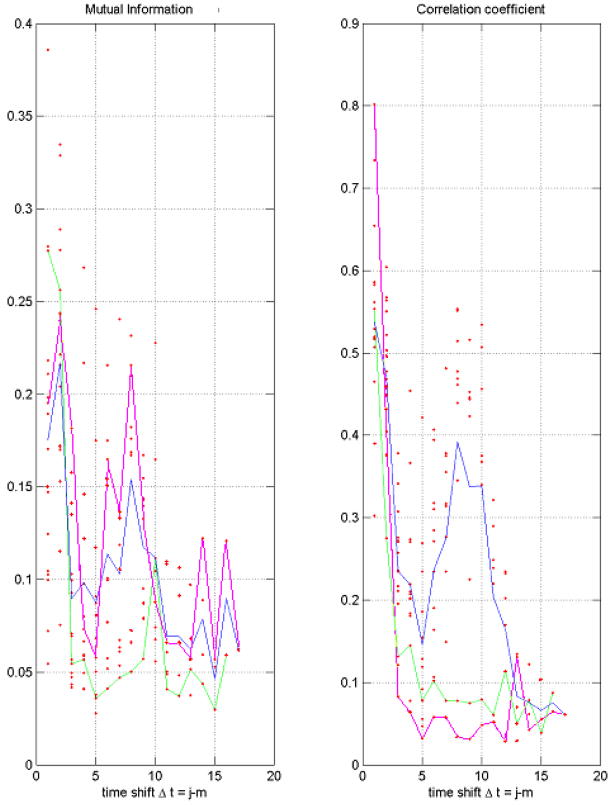
Here we restrict ourselves with the results for α factor arrest dataset. We calculated the mutual information for all of the pairs time points (i.e., for all the pairs of microarray chips). Consider the left graph in Fig 1. Ideally, one would expect high dependence (correlation) between one time point and its immediate neighbor, when “Δt = 1”. Dependence between a time point and another point two time-intervals down, Δt = 2, would expectedly be lower. Plotting MI versus Δt would produce a typical relaxation curve. However, since the α-experiments span approximately two cell cycles, and we expect the period of genes involved in cell cycle regulation to coincide with the length of the cell cycle, we see a second, but lower peak (hump) around Δt = 9. This time interval (9×7min = 63min) indicates the period of one cell cycle, which is approximately half of the duration of the experiment (119min). This reaffirms the authors’ claim that α factor experiment spanned two cell cycles. The correlation coefficient for the same data is shown on the right graph of Fig. 1.
FIGURE 1.
A pairwise Mutual Information (left) and Correlation Coefficient (right) as functions of the time shift, Δt, for the yeast dataset with α factor arrest. Five strongest outliers were removed. Every red point represents the pair of time points. The pink line marks the similarity in gene expression at the first time point with the rest of the points; the green line – the second time point with the rest of the points; the blue line – average over all the pairs of time points with a given value of Δt.
Both, MI and CC graphs reveal the periodic behavior due to the cell cycle. It is interesting to note, that the correlation graphs of time point t=1 and time point t=2 drop dramatically versus Δt, and show virtually no correlation after Δt =3, whereas the rest of time points show a periodic trend. On the other hand, the MI graph for the point t=1 with the rest of time points, t=2÷18 does reveal the periodic behavior whereas the MI for the second time point t=2 with the rest of time points, t=3÷18 shows no recurrence at the cycle period.
The analysis of scatter plots of one time point against another shows that nonlinear dependences cannot play a significant role for this data. Thus, the difference between the MI and CC graphs is caused by outliers. Note, that there are two types of outliers here. Several genes (numbered as 1770, 183, 462, 1771, and 2305) reveal very high expression level (>5), many genes show the moderately high expression (2÷4), still the most of the genes are expressed at lower level, (<2). The exclusion of the five most highly expressed genes (Fig. 1) does not change qualitatively the figures. However, the numerous outliers from the second group destroy the recurrence of correlation between the first time point and the rest of the points at the cycle period time. This does not affect the MI, which is determined by the total number of genes expressed at a lower level.
Acknowledgments
The authors acknowledge the support of the Texas Learning and Computation Center. BMP acknowledges support from NIH. SCh is grateful to the University of Houston for hospitality.
References
- 1.Shannon CE. The Bell System Technical Journal. 1948;27:379–423. 623–656. ibid. [Google Scholar]
- 2.Herzel H, Grosse I. Phys Rev E. 1997;55:800–809. [Google Scholar]
- 3.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Molecular Biology of the Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95(25):14863–148683. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fofanov YV, Montgomery Pettitt B. Journal of Biomedical Informatics. 2002;35:343–351. doi: 10.1016/s1532-0464(03)00035-2. (2003) [DOI] [PubMed] [Google Scholar]
- 6.Butte AJ, Kohane IS. Proc Pacific Symposium on Biocomputing. 2000;5:415–426. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 7.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Convay A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. Molecular Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]