Summary
Mammalian sleep is composed of two distinct states – rapid-eye-movement (REM) and non-REM (NREM) sleep – that alternate in cycles over a sleep bout. The duration of these cycles varies extensively across mammalian species. Because the end of a sleep cycle is often followed by brief arousals to waking, a shorter sleep cycle has been proposed to function as an anti-predator strategy. Similarly, higher predation risk could explain why many species exhibit a polyphasic sleep pattern (division of sleep into several bouts per day), as having multiple sleep bouts avoids long periods of unconsciousness, potentially reducing vulnerability.
Using phylogenetic comparative methods, we tested these predictions in mammals, and also investigated the relationships among sleep phasing, sleep-cycle length, sleep durations and body mass.
Neither sleep-cycle length nor phasing of sleep was significantly associated with three different measures of predation risk, undermining the idea that they represent anti-predator adaptations.
Polyphasic sleep was associated with small body size, shorter sleep cycles and longer sleep durations. The correlation with size may reflect energetic constraints: small animals need to feed more frequently, preventing them from consolidating sleep into a single bout. The reduced daily sleep quotas in monophasic species suggests that the consolidation of sleep into one bout per day may deliver the benefits of sleep more efficiently and, since early mammals were small-bodied and polyphasic, a more efficient monophasic sleep pattern could be a hitherto unrecognized advantage of larger size.
Keywords: mammalian sleep architecture, monophasic sleep, polyphasic sleep, phylogeny, sleep-cycle length
Introduction
Terrestrial mammals have two sleep states, rapid-eye-movement (REM) sleep and non-REM (NREM) sleep, which have different physiological characteristics and, possibly, distinct functions (Zepelin 1989). These two states alternate in a cycle during a sleep bout, which in adults always starts with an episode of NREM sleep (Zepelin 1989). Sleep-cycle length varies extensively across mammalian species (Zepelin 1989), from about 6 min (e.g. Chinchilla lanigera; Van Twyver 1969) to c. 90 min (e.g. Homo sapiens; Tobler 1995). Mammals also differ in how they accommodate sleep within their activity budgets (the ‘phasing’ of sleep; Stampi 1992); some species partition their sleep time into multiple bouts alternated with waking phases (polyphasic sleepers; e.g. cats; Ball 1992), while others concentrate the majority of their sleep into one bout per day (monophasic sleepers; e.g. chimpanzees; Ball 1992).
Whilst many studies have examined the correlates of species differences in the total daily amount of time spent sleeping and in REM and NREM sleep (Allison & Cicchetti 1976; Elgar, Pagel & Harvey 1988; Zepelin 1989; Lesku et al. 2006, Capellini, Barton, McNamara et al. 2008), less is known about interspecific variation in how sleep is organized through the daily cycle, specifically the duration of the REM–NREM sleep cycle and the number of sleep bouts per day (the ‘phasing of sleep’). One idea is that predation risk determines the length of sleep cycles (Tobler 1989; Voss 2004; Lima et al. 2005). At the end of a sleep cycle, episodes of REM sleep are followed by increased levels of consciousness and brief arousals to waking, allowing individuals to monitor their environment (Van Twyver & Garrett 1972; Voss 2004; Lima et al. 2005). In support of this idea, birds and laboratory rats wake from sleep more frequently and have shorter sleep cycles after encounters with predators, when sleeping in areas perceived to be less safe, or when sleeping in smaller groups (Broughton 1973; Lendrem 1983, 1984; Gauthier-Clerc, Tamisier & Cezilly 1998, 2000, 2002; Lesku et al. 2008). Thus, shorter sleep cycles may be adaptive for species with relatively high predation risk (Lima et al. 2005, page 728). We tested the following predictions of this hypothesis using proxy measures of predation risk developed in previous studies of the evolution of daily sleep quotas (Lesku et al. 2006; Capellini et al. 2008). (i) Sleep-cycle length is predicted to be shorter in animals that sleep in more exposed and vulnerable sleeping sites relative to those that sleep in enclosed and protected sites, and (ii) in ‘prey’ as compared to ‘predators’. (iii) Because predation risk is reduced in larger groups due to detection and dilution effects (Caro 2005), sleep-cycle lengths should be shorter in solitary species relative to species in which individuals sleep in groups. (iv) Finally, predation pressure is thought to be lower for larger animals because fewer predator species can successfully kill a large prey animal (Peters 1983; Owen-Smith 1988; Caro 2005). Across species, therefore, the predation risk hypothesis predicts shorter sleep cycles in species characterized by smaller body size.
Extending the logic of these ideas, predation risk might also influence how sleep is distributed across the daily cycle (Tobler 1989; Ball 1992; Stampi 1992). In particular, polyphasic sleep might be a response to high predation risk, because dividing sleep time into more bouts would limit the vulnerability associated with prolonged periods of reduced consciousness. If this is true, then polyphasic sleep should be associated with smaller size, ‘prey’ rather than ‘predator’ status, and sleeping under more vulnerable conditions (in exposed sites and/or solitarily).
At present, the relationships among sleep phasing, sleep-cycle length and total sleep time are poorly understood. Ball (1992) found that species with polyphasic sleep spend more time asleep but have shorter sleep cycles when compared to species with monophasic sleep. However, this analysis was based on a relatively small sample size (22 species), did not examine the likely role of body size in sleep pattern differences between species, and did not account for phylogenetic history (see Methods section). We therefore examine the interrelationships among these traits using a larger data set and phylogenetic comparative methods.
Methods
Data Collection
We extracted data on sleep traits and ecological factors for mammals from the primary literature (data available at: http://www.bu.edu/phylogeny/index.html) following the protocol described in McNamara et al. (2008). We excluded monotremes and aquatic mammals from analyses as they exhibit unusual sleep architecture that prevents meaningful comparisons with terrestrial mammals (Zepelin 1989; Zepelin, Siegel & Tobler 2005). Following Stampi (1992), we classified species as monophasic if they concentrate at least 50% of their daily sleep into one bout alternated with a period of activity which may or may not be interrupted by short ‘naps’, and polyphasic if their sleep is divided into multiple bouts, each one accounting for less than 50% of total sleep time. Hence the distribution of sleep within the 24-h period was a discrete binary trait (0 for monophasic sleep, 1 for polyphasic sleep). The 50% criterion is arbitrary, but nevertheless provides a quantifiable measure of the extent of sleep concentration within single bouts. One person unaware of the research question and aims of the study coded the species as monophasic or polyphasic from the primary literature. Further data were extracted from the reviews by Ball (1992) and Tobler (1989). Our data set on phasing of sleep included 56 terrestrial mammals. Monotremes were however excluded when investigating the correlated evolution of sleep durations with sleep phasing and cycle length because it is uncertain whether REM and NREM sleep in these species is comparable to those of other mammals (Zepelin et al. 2005). Data on sleep traits are from laboratory studies, and it is unclear how well they reflect sleep pattern under natural conditions. However, there is some evidence that, despite differences in overall activity levels, sleep durations in laboratory shrews are similar to those observed in the field (Saarikko & Hanski 1990).
We have previously shown that laboratory conditions can impact estimates of sleep time (NREM and REM sleep duration), and this can in turn affect the outcome of comparative analyses (Capellini et al. 2008). Based on these findings, we restricted the analysis of sleep duration and sleep-cycle length to data collected with at least 12 h recording time and EEG recording equipment. Data on sleep-cycle length were available for 27 species, all of which had EEG estimates of sleep quotas. Among the 56 species with information on phasing of sleep, 45 species had EEG data on sleep time (Table S1 in Supplementary Material).
Data on body mass were extracted from the primary literature (all species; details in Capellini et al. 2008). Predation risk was assessed with three variables following a previous study (Capellini et al. 2008): social sleep behaviour (40 species), exposure of the sleeping site (56 species) and trophic level (35 species). The degree of social sleep behaviour was estimated with a three-point index. Sleep was considered ‘non-social’ when both males and females sleep alone, ‘partially social’ if females but not males sleep with conspecifics, and ‘social’ if both sexes regularly sleep with conspecifics. We did not consider sleeping with offspring as social sleep unless it persisted into adulthood. Sleep site exposure was coded as ‘low’ for fully enclosed sleeping sites (e.g. dens and tree holes), ‘intermediate’ for sites that provide partial shelter (e.g. vegetation on the ground or in trees), and ‘high’ for fully open sites that provide no protection (e.g. ground in open areas). Our index of exposure is based on fewer assumptions concerning vulnerability of sleeping sites, as compared to indices developed in previous studies (Allison & Cicchetti 1976; Lesku et al. 2006). For example, we did not consider sleeping in the tree canopy safer than sleeping below the tree canopy, because while the former sleep sites may better protect sleeping individuals against terrestrial predators (as argued by Lesku et al. 2006), they may also increase vulnerability to aerial predators. Data on social sleep behaviour and sleep site exposure were extracted from the literature using both primary and secondary sources (e.g. Nowak 1999, and the Mammalian Species monographs). Two independent observers coded the data on social sleep behaviour and three coded the data on sleep site exposure. Data were then checked and averaged if scores conflicted or indicated intraspecific variation. Finally, data on species' trophic level were taken from Lesku et al. (2006). Trophic level was a four-point diet-based index with the following ranks corresponding to increasing predation risk: (i) exclusively on vertebrate prey (carnivorous); (ii) large insects; (iii) small insects; (iv) exclusively vegetable matter (herbivorous). Trophic level was not significantly associated with sleep site exposure (P = 0·160) and social sleep behaviour (P = 0·332), indicating that these indices capture different aspects of predation risk.
All continuous variables were log-transformed prior to statistical analysis with independent contrasts.
Phylogenetic Comparative Analysis
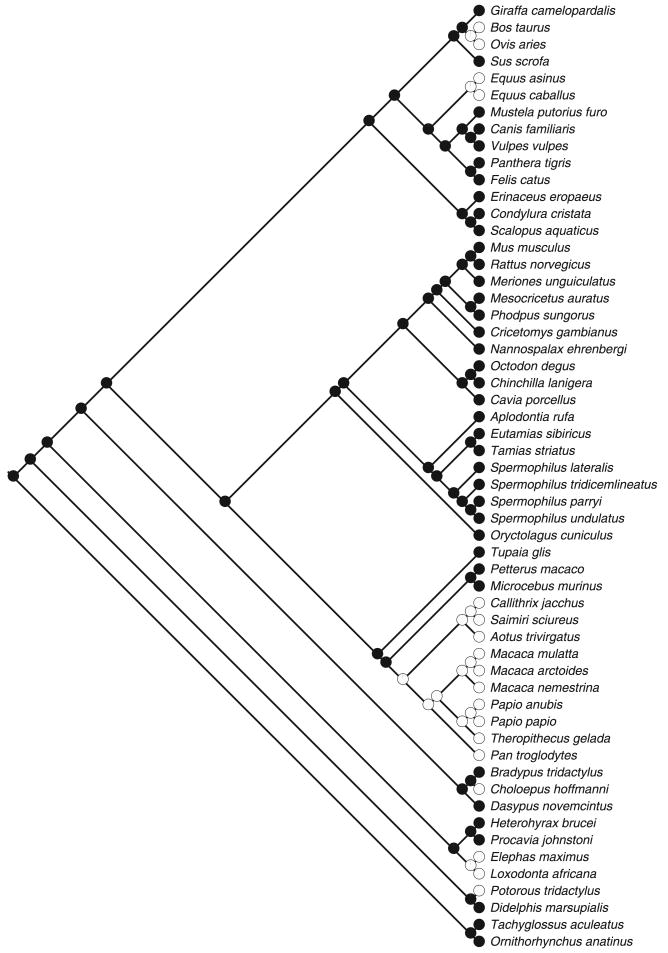
More closely related species tend to share traits through common descent (Felsenstein 1985; Harvey & Pagel 1991; Blomberg, Garland & Ives 2003) and this represents a problem that, if ignored, can inflate Type I error rates (Felsenstein 1985; Martins & Garland 1991; Harvey & Pagel 1991). We assembled a mammalian phylogeny for the species in our data base using published phylogenies (sources in Appendix S1). We reconstructed the ancestral character state of phasing of sleep using maximum likelihood (Pagel 1994, 1999; Schluter et al. 1997), as implemented in Discrete (Pagel 1994, 1999) and Mesquite (for graphical representation; Maddison & Maddison 2006). This approach estimates the probability of character state at the root under a stochastic model of evolution (Markov k-state 2 parameters). We allowed the rates of forward and backward transitions to be ‘unrestricted’, thus not constrained to be equal to each other or to a specified constant. Discrete also provides the likelihood for the ancestral character state at the root being either 0 (monophasic sleep) or 1 (polyphasic sleep); the reconstruction with the highest log-likelihood value is preferred and the difference between the two is considered significant if it is ≥2 log-units (Pagel 1999).
We accounted for shared evolutionary history of species by using phylogenetically independent contrasts, analysed with regression through the origin (Felsenstein 1985; Harvey & Pagel 1991; Garland, Harvey & Ives 1992; Nunn & Barton 2001). Contrasts of all variables were calculated using CRUNCH in CAIC, including phasing of sleep as a dummy variable (Purvis & Rambaut 1995; Midford, Garland & Maddison 2005).
We checked that the main assumption of independent contrast analysis was met (i.e. no significant correlation between contrasts and their standard deviation) using different branch length options (Garland et al. 1992). We then set branch lengths to be equal because this option performed better in the assumption checks, with the sole exception of social sleep behaviour. When we found evidence that assumptions of regression analysis may not be met, we re-assessed the significance of our results using bootstrapped estimates of effects and 95% confidence intervals. These statistics do not make strict distributional assumptions about the data and reduce bias caused by outliers (Efron & Tibshirani 1993). Bootstrap analyses were implemented in Genstat v8 and we report statistics from bootstrap analyses only where they produced different results.
We controlled for multiple tests of each hypothesis by using the false discovery rate control (FDR). With FDR, the proportion of Type I errors is evaluated against all significant results in the analysis and an individual α threshold is adjusted for each result given the number of tests performed and their significance (Bejamini & Hochberg 1995; Verhoeven, Simonsen & McIntyre 2005). As a consequence, this method is more powerful than other methods, such as the Bonferroni correction (Verhoeven et al. 2005). Controlling for multiple testing produced no qualitative differences in the results.
Results
Sleep-Cycle Length
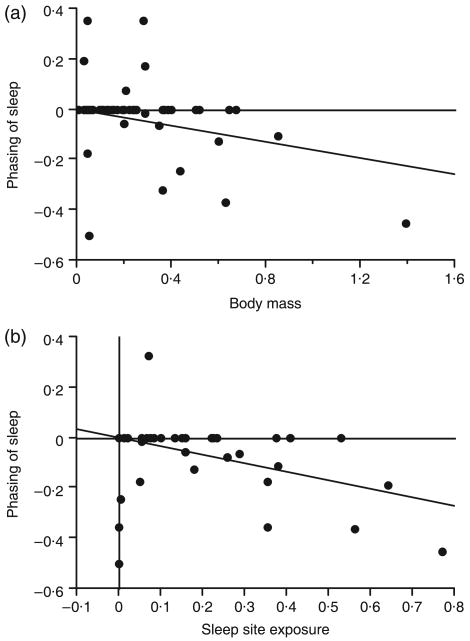
In agreement with previous studies (Elgar et al. 1988; Zepelin 1989), sleep-cycle length was positively associated with body mass (t25 = 4·54, R2 = 0·45, P = 0·0001; Fig. 1a). Contrary to predictions, sleep cycles were longer when sleeping sites were more exposed (t24 = 2·98, R2 = 0·27, P = 0·007; Fig. 1b), probably because body mass and exposure covaried (t52 = 4·98, R2 = 0·32, P < 0·0001). We thus calculated a relative measure of exposure, using residuals from the regression of contrasts in sleep site exposure index on contrasts in body mass. These residuals were not significantly related to sleep-cycle length (t24 = 0·79, R2 = 0·03, P = 0·437). Also, sleep-cycle length was not significantly correlated with social sleeping (t18 = 0·44, R2 = 0·01, P = 0·664) or trophic level (t19 = 0·13, R2 = 0·01, P = 0·718).
Fig. 1.
Phylogenetically independent contrasts of sleep-cycle length with (a) body mass and (b) sleep site exposure. The index of exposure quantifies vulnerability of sleeping sites from the least exposed site (lowest values) to the most exposed site (highest values). Sleep-cycle length and body mass were log-transformed.
Phasing of Sleep
Polyphasic sleep was reconstructed as the ancestral character state (ancestral state log-likelihood scores and probabilities: polyphasic sleep: log(L) = −43·02, probability = 99%; monophasic sleep: log(L) = −47·40, probability = 1%; difference in likelihood scores = 4·37) in our sample of mammals (Fig. 2). Polyphasic sleep was associated with smaller body mass (t53 = −3·19, R2 = 0·16, P = 0·002; Fig. 3a) and, contrary to predictions of the predation risk hypothesis, with sleeping in more protected sites (i.e. negatively correlated with sleep site exposure) both before (t52 = −4·61, R2 = 0·29, P < 0·001; Fig. 3b) and after controlling for body mass (see above; t52 = −2·94, R2 = 0·14, P = 0·005). We did not detect any significant effect of trophic level or social sleep behaviour on phasing of sleep (diet: t32 = −1·45, R2 = 0·06, P = 0·156; social sleep: t37 = −0·73, R2 = 0·01, P = 0·468).
Fig. 2.
Evolutionary history of phasing of sleep (monophasic sleep in white, polyphasic sleep in black) reconstructed with maximum likelihood (see text). Areas of pies indicate the relative support for each of the two possible character states at each given node. Because all reconstructions at each node along the phylogeny strongly supported only one state (probabilities were 99% in favour of one state), circles appear to be filled. Support for polyphasic sleep as ancestral character state is 99% (actual calculations from Discrete; Pagel 1994, 1999; see text). Species with missing values for phasing of sleep are not shown. Phylogenetic tree assembled using published phylogenies (sources in Appendix S1).
Fig. 3.
Phylogenetically independent contrasts of phasing of sleep with (a) body mass and (b) sleep site exposure. Phasing of sleep was treated as a dummy variable and analysed with independent contrasts; lower values indicate monophasic sleep and higher values indicate polyphasic sleep (see text). The lowest values of sleep site exposure indicate most protected sites, the highest values the most exposed sites (see text).
Correlated Evolution of Sleep Traits
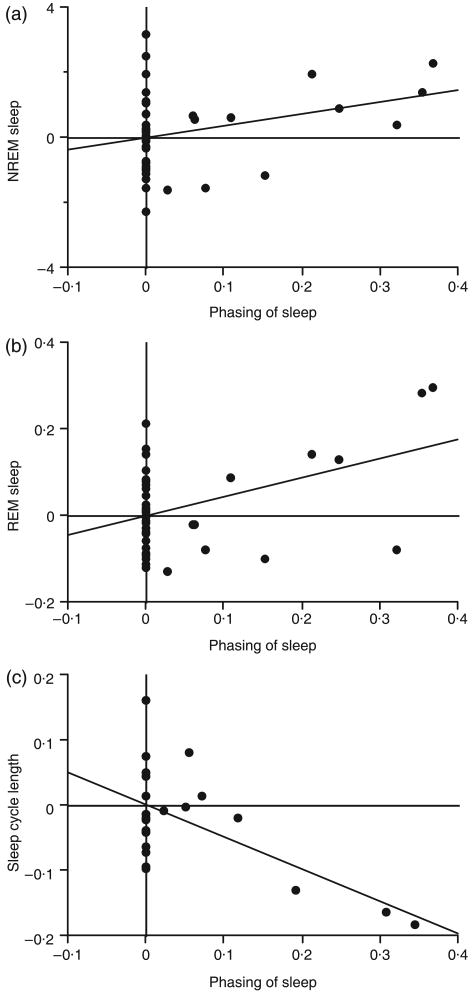
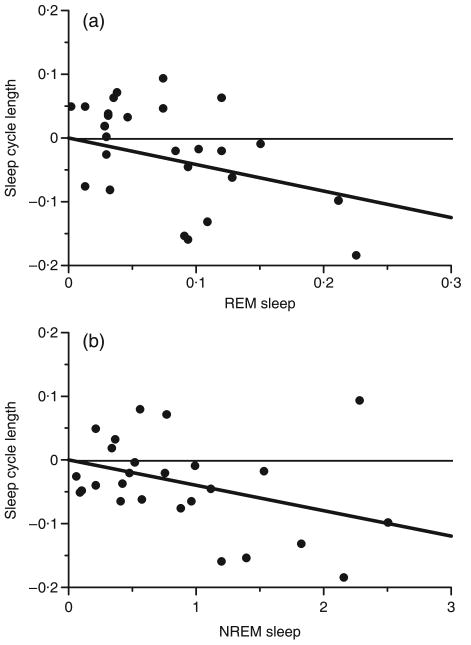
Polyphasic sleep was significantly associated with longer REM and NREM sleep quotas (NREM sleep: t43 = 2·35, R2 = 0·11, P = 0·024; REM sleep: t43 = 3·56, R2 = 0·23, P = 0·001; Fig. 4a,b). In addition, sleep-cycle length was shorter in species that sleep polyphasically relative to species that sleep monophasically (t22 = −4·07, R2 = 0·43, P = 0·001; Fig. 4c). Finally, sleep-cycle length was negatively correlated with both REM (t25 = −2·93, R2 = 0·26, P = 0·007; Fig. 5a) and NREM sleep quotas (t25 = −3·33, R2 = 0·31, P = 0·003; Fig. 5b).
Fig. 4.
Phylogenetically independent contrasts of phasing of sleep with NREM sleep (a), REM sleep (b) and sleep-cycle length (c). Phasing of sleep was analysed as a dummy variable; lower values indicate monophasic sleep and higher values polyphasic sleep (see text).
Fig. 5.
Phylogenetically independent contrasts of sleep-cycle length with contrasts of REM (a) and NREM sleep duration (b).
After bootstrapping, the association between phasing of sleep and REM sleep was not significant (bootstrapped coefficient = 0·454, SE = 0·263, P = 0·132), while the association between phasing of sleep and sleep-cycle length was marginally non-significant (bootstrapped coefficient = −0·816, SE = 0·355, P = 0·054).
Discussion
The phasing of sleep and sleep-cycle length are fundamental aspects of sleep architecture, as they reflect how the benefits of sleep are obtained throughout the 24-h cycle. Relative to studies of sleep durations, however, these traits have received much less attention in comparative analyses of sleep architecture. We tested the hypothesis that predation risk impacts the evolution of both sleep phasing and REM-NREM sleep-cycle length, predicting that species under higher predation pressure are polyphasic and have shorter sleep cycles. Our analyses with three independent measures of predation risk (exposure of the sleep site, social sleep behaviour and trophic level) failed to support the hypothesis that the phasing of sleep and sleep-cycle length represents anti-predator adaptations. We also found that polyphasic sleep was the ancestral character state in mammals and was associated with smaller body size, and that polyphasic sleepers and those with short sleep cycles had longer sleep durations. Collectively, our study suggests that energetic and foraging constraints associated with small size could explain some of the evolutionary patterns that we discovered. In what follows, we provide more details and interpretation of these main results.
First, the predation risk hypothesis predicts shorter sleep cycles in species under more intense predation risk, such as those that sleep in less protected sites and/or sleep solitarily, and in ‘prey’ relative to ‘predators’ (Lima et al. 2005). Contrary to predictions, sleep-cycle length increased in species that sleep in more exposed sleeping sites. Vulnerability of species that sleep in sites with different exposure depends on body size because sleep site exposure increased with body mass. While small-bodied animals probably invest in searching for protected sleeping sites to reduce their vulnerability while asleep, larger size is thought to also reduce predation pressure (Peters 1983; Owen-Smith 1988; Caro 2005). After controlling for body mass, sleep-cycle length and sleep site exposure were uncorrelated. In addition, sleep-cycle length was not significantly correlated with social sleep behaviour and trophic level. Taken together, these results do not support the predation risk hypothesis for the evolution of sleep-cycle length. We suggest that even the shortest cycles are probably too long to allow individuals to detect approaching predators successfully, given that the scanning rate of animals during waking periods can be as short as a few seconds (Caro 2005).
Second, we expected that predation risk would also impact sleep phasing. As predicted, smaller species are polyphasic but, contrary to the predation risk hypothesis, polyphasic species sleep in more protected sites. In addition, the phasing of sleep was not significantly correlated with social sleep behaviour and trophic level. Thus, our analyses failed to support the predation risk hypothesis for the evolution of the phasing of sleep. We suggest that polyphasic sleep, which is the ancestral character state in mammals, is associated with small body mass because small species are forced to forage more frequently than larger species due to their higher mass-specific metabolic rates and limited fat reserves (Lindstedt & Boyce 1984; Withers 1992; Blackburn & Hawkins 2004). Thus, sleep may be partitioned into multiple bouts per 24-h to allow animals to feed in between sleep bouts. In addition, digestion rate and gut capacity limit the rate of food ingestion in small mammals like shrews, forcing them to alternate short foraging bouts with short sleep (or rest) bouts to keep their digestive tract operating at constant high rate (Saarikko & Hanski 1990; Saarikko 1992).
Finally, we found that polyphasic sleep and sleep-cycle length are associated with longer sleep durations, and that polyphasic sleepers have shorter sleep cycles. The result that polyphasic sleepers exhibit longer sleep durations further argues against a major role of predation in driving the evolution of the phasing of sleep because total sleep time and sleep quotas are reduced in species that experience higher predation risk (Allison & Cicchetti 1976, Capellini et al. 2008).
An interesting corollary of the association of polyphasic sleep and sleep-cycle length with longer sleep time is that partitioning sleep into multiple bouts and more cycles will result in more frequent transitions from light sleep into deep sleep. Thus, to achieve the benefits of deeper stages of NREM sleep, polyphasic sleepers and those with shorter sleep cycles would require more total time in NREM sleep compared to monophasic sleepers and species with longer cycles. In other words, partitioning sleep into multiple bouts with shorter cycles may be less efficient than monophasic sleep because it requires more time spent in transitional sleep stages. Therefore, our results suggest that the evolution of monophasic sleep from the ancestral polyphasic sleep pattern may allow the benefits of sleep to be gained more efficiently (as proposed by Ball 1992). This interpretation is based on the assumption that transitional sleep stages cannot be skipped and/or compressed in time and that their primary function is to favour the transition from waking phase to deep sleep. These assumptions need to be tested in the laboratory.
In conclusion, while studies on the plasticity of sleep have shown that birds and rats modify their sleep patterns in response to threat of predation, our comparative analyses suggest that predation risk is not responsible for the evolution of interspecific differences in sleep-cycle length or phasing of sleep in mammals. Polyphasic species are smaller and we argue that this is likely to reflect energetic constraints. In addition, shorter sleep cycles and polyphasic sleep are associated with longer sleep durations. We suggest that when sleep is partitioned into multiple cycles or more sleep bouts, more time in light sleep stages is needed overall. Monophasic sleep may therefore be a more efficient sleep pattern and an advantage of evolving a larger size.
Supplementary Material
Appendix S1. Data file and references for the phylogenetic tree
Table S1. Data set used for the analysis (Excel file)
Acknowledgments
Authors are grateful to Erica Harris, Nikita Patel, Sean O'Hara, Patrik Lindenfors, Joann Chang, Meike Mohneke and Timothy Morrison for help collecting the sleep and ecological traits, Mark Pagel and Andy Purvis for advice on the comparative analyses, Dr Charles Fox and one anonymous referee for their valuable comments on an earlier version of the manuscript. This research is funded by NIMH (Grant number 1R01MH070415-01A1) and the Max Planck Society (CN).
Footnotes
Supplementary material: The following supplementary material is available for this article:
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/full/10.1111/j.1365-2435.2008.01449.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Allison T, Cicchetti DV. Sleep in mammals: ecological and constitutional correlates. Science. 1976;194:732–734. doi: 10.1126/science.982039. [DOI] [PubMed] [Google Scholar]
- Ball NJ. The phasing of sleep in animals. In: Stampi C, editor. Why We Nap Evolution, Chronobiology and Functions of Polyphasic and Ultrashort Sleep. Birkhauser; Boston: 1992. pp. 31–49. [Google Scholar]
- Bejamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Blackburn TM, Hawkins BA. Bergmann's rule and the mammal fauna of northern North America. Ecography. 2004;27:715–724. [Google Scholar]
- Blomberg S, Garland T, Ives A. Testing for phylogenetic signal in comparative data: behavioural traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Broughton R. Confusional sleep disorders: interrlationship with memory consolidation and retrieval in sleep. In: Boag T, Campbell D, editors. A Triune Concept of the Brain and Behaviour. Toronto University Press; Toronto: 1973. pp. 115–127. [Google Scholar]
- Capellini I, Barton RA, Mcnamara P, Pretson BT, Nunn CL. Phylogenetic analysis of ecology and evolution of mammalian sleep. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00392.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro T. Antipredator Defences in Birds and Mammals. University of Chicago Press; Chicago: 2005. [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman and Hall; London: 1993. [Google Scholar]
- Elgar MA, Pagel MD, Harvey PH. Sleep in mammals. Animal Behaviour. 1988;36:1407–1419. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Garland T, Harvey PA, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Systematic Biology. 1992;41:18–32. [Google Scholar]
- Gauthier-Clerc M, Tamisier A, Cezilly F. Sleep-vigilance trade-off in green-winged teals (Anas crecca crecca) Canadian Journal of Zoology. 1998;76:2214–2218. [Google Scholar]
- Gauthier-Clerc M, Tamisier A, Cezilly F. Sleep-vigilance trade-off in gadwall during the winter period. Condor. 2000;102:307–313. [Google Scholar]
- Gauthier-Clerc M, Tamisier A, Cezilly F. Vigilance while sleeping in the breeding pochard Aythya ferina according to sex and age. Bird Study. 2002;49:300–303. [Google Scholar]
- Harvey PA, Pagel M. The Comparative Method in Evolutionary Biology. Oxford University Press; Oxford: 1991. [Google Scholar]
- Lendrem DW. Sleeping and vigilance in birds. I. Field observations of the mallard (Anas platyrhynchos) Animal Behaviour. 1983;31:532–538. [Google Scholar]
- Lendrem DW. Sleeping and vigilance in birds. II. An experimental study of the barbary dove (Streptopelia risoria) Animal Behaviour. 1984;32:243–248. [Google Scholar]
- Lesku JA, Bark RJ, Martinez-Gonzalez D, Rattenborg NC, Amlaner CJ, Lima SL. Predator-induced plasticity in sleep architecture in wild-caught Norway rats (Rattus norvegigus) Behavioural Brain Research. 2008;189:298–305. doi: 10.1016/j.bbr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Lesku JA, Roth TC, Amlaner CJ, Lima SL. A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology and ecology. American Naturalist. 2006;168:441–453. doi: 10.1086/506973. [DOI] [PubMed] [Google Scholar]
- Lima SL, Rattenborg NC, Lesku JA, Amlaner CJ. Sleeping under the risk of predation. Animal Behaviour. 2005;70:723–736. [Google Scholar]
- Lindstedt SL, Boyce MS. Seasonality, fasting endurance and body size in mammals. American Naturalist. 1984;125:873–878. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 1·11. 2006 http://mesquiteproject.org.
- Martins E, Garland T. Phylogenetic analyses of the correlated evolution of continuous characters: a simulation study. Evolution. 1991;45:534–557. doi: 10.1111/j.1558-5646.1991.tb04328.x. [DOI] [PubMed] [Google Scholar]
- McNamara P, Capellini I, Harris E, Nunn CL, Barton RA, Preston BT. The Phylogeny of Sleep Database: a new resource for sleep scientists. The Open Sleep Journal. 2008;1:11–14. doi: 10.2174/1874620900801010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midford PE, Garland T, Maddison PW. PDAP Package of Mesquite. Version 1·07 2005 [Google Scholar]
- Nowak RM. Walker's Mammals of the World. The John Hopkins University Press; Baltimore and London: 1999. [Google Scholar]
- Nunn CL, Barton RA. Comparative methods for studying primate adaptation and allometry. Evolutional Anthropology. 2001;10:81–98. [Google Scholar]
- Owen-Smith RN. Megaherbivores: The Influence of very Large Body Size on Ecology. Cambridge University Press; Cambridge: 1988. [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B. 1994;255:37–45. [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Systematic Biology. 1999;48:612–622. [Google Scholar]
- Peters RH. The Ecological Implication of Body Size. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrasts (CAIC): an Apple Macintosh application for analysing comparative data. Computer Applications in the Biosciences. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Saarikko J. Risk of predation and foraging activity in shrews. Annales Zoologici Fennici. 1992;29:291–299. [Google Scholar]
- Saarikko J, Hanski I. Timing of rest and sleep in foraging shrews. Animal Behaviour. 1990;26:861–869. [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Stampi C. Evolution, chronobiology and functions of polyphasic and ultrashort sleep: main issues. In: Stampi C, editor. Why We Nap Evolution, Chronobiology and Functions of Polyphasic and Ultrashort Sleep. Birkhauser; Boston: 1992. pp. 1–20. [Google Scholar]
- Tobler I. Napping and polyphasic sleep in mammals. In: Dinges DF, Broughton RJ, editors. Sleep and Alertness: Chronobiological, Behavioral and Medical Aspects of Napping. Raven Press; New York: 1989. pp. 9–30. [Google Scholar]
- Tobler I. Is sleep fundamentally different between mammalian species? Behavioural Brain Research. 1995;69:35–41. doi: 10.1016/0166-4328(95)00025-o. [DOI] [PubMed] [Google Scholar]
- Van Twyver H. Sleep patterns in five rodent species. Physiology and Behavior. 1969;4:901–905. [Google Scholar]
- Van Twyver H, Garrett W. Arousal threshold in the rat determined by ‘meaningful’ stimuli. Behavioral Biology. 1972;7:205–215. doi: 10.1016/s0091-6773(72)80200-1. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Simonsen KL, Mcintyre LM. Implementing false discovery rate control: increasing your power. Oikos. 2005;108:643–647. [Google Scholar]
- Voss U. Fuctions of sleep architecture and the concept of protective fields. Reviews in the Neurosciences. 2004;15:33–46. doi: 10.1515/revneuro.2004.15.1.33. [DOI] [PubMed] [Google Scholar]
- Withers PC. Comparative Animal Physiology. Saunders College Publishing; Orlando, Florida: 1992. [Google Scholar]
- Zepelin H. Mammalian sleep. In: Kryger MH, Roth T, Dement WC, editors. Principles and Practices of Sleep Medicine. Saunders; Philadelphia: 1989. pp. 30–49. [Google Scholar]
- Zepelin H, Siegel JM, Tobler I. Mammalian sleep. In: Kryger MH, Roth T, Dement WC, editors. Principles and Practices of Sleep Medicine. Saunders; New York: 2005. pp. 91–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Data file and references for the phylogenetic tree
Table S1. Data set used for the analysis (Excel file)