Abstract
Background
Circadian genes continue to gain attention as important transcriptional regulators with the potential to influence a variety of biological pathways, including many cancer-related processes. The core circadian gene cryptochrome 2 (CRY2) is essential for proper circadian timing, and is a key component of the negative arm of the circadian feedback loop. As such, aberrant expression of CRY2 may influence carcinogenic processes and thereby impact cancer susceptibility.
Methods
We silenced CRY2 in breast cancer cell lines (MCF-7) using small-interfering oligos (siRNA) and measured the impact of CRY2 knockdown on a number of cancer-relevant parameters. Cell cycle distribution, cell viability, and apoptotic response were measured in CRY2 knockdown (CRY2-) and normal (CRY2+) cell populations using flow cytometry in cells with and without exposure to a mutagen challenge. DNA damage accumulation was measured using the single cell gel electrophoresis (comet) assay, and damage was quantified using the Olive tail moment, which considers the amount and distance of DNA migration away from the nucleus, indicative of DNA strand breaks. Expression changes in cancer-relevant transcripts were measured by whole genome microarray. The Student's t-test was used for statistical comparisons, and P-values obtained from the microarray were adjusted for multiple comparisons using the false discovery rate correction, in order to obtain an adjusted Q-value for each observation.
Results
The comet assay results indicated that upon exposure to the same dose of chemical mutagen, CRY2- cells accumulate significantly more unrepaired DNA damage than CRY2+ cells (P = 0.040), suggesting that CRY2 may be important for DNA repair. In addition, a number of transcripts with relevance for DNA damage repair displayed altered expression following CRY2 silencing. These included BCCIP (Q = 0.002), BCL2 (Q = 0.049), CCND1 (Q = 0.009), CDKN1A (Q < 0.001), GADD45A (Q = 0.002), HERC5 (Q < 0.001), MCM5 (Q = 0.042), PPP1R15A (Q < 0.001), SUMO1 (Q < 0.001), and UBA1 (Q = 0.023). However, no significant influence of CRY2 knockdown on cell cycle distributions, cell cycle checkpoints in response to mutagen challenge, or apoptotic response was detected.
Conclusions
In total, these data suggest a limited, but potentially important role for CRY2 in the regulation of DNA damage repair and the maintenance of genomic stability. Future investigations may focus on identifying the mechanisms by which CRY2 may regulate the expression of transcripts with known relevance for carcinogenesis.
Background
Although our understanding of the molecular basis for the circadian rhythm is continually evolving, the current model involves a complex interplay between environmental and endogenous factors, which include a core set of circadian genes [1]. Transcriptional and post-transcriptional interactions among these gene products results in an autoregulatory feedback system, which allows for predictable cycling of the core circadian elements [2-4]. In addition, many of the circadian genes operate as transcriptional regulators for transcripts outside of the circadian system, and recent evidence indicates that as many as 10% of all mammalian genes may be regulated to some degree by the circadian oscillatory mechanism [5-7]. As a result, disturbance of the circadian system, either through environmental exposures, or through genetic alterations in the key circadian genes, may have important implications for a variety of biological pathways.
One such core circadian gene, CRY2, operates in the negative arm of the circadian feedback loop as a transcriptional repressor [8]. CRY2 has also been shown to be involved in cancer-relevant pathways including DNA damage checkpoint control [9] and regulation of genes important for cell cycle progression [10,11]. However, Cry1-/-, Cry2-/- transgenic mice do not display a cancer-prone phenotype in response to ionizing radiation exposure [10]. Here, we report findings from in vitro loss-of-function investigations into the phenotypic effects of CRY2 knockdown on cell cycle, apoptosis, and DNA damage response to mutagen challenge in a breast cancer cell line. We also investigate a whole genome expression array to interrogate the impact of CRY2 silencing on the expression of genes relevant to these pathways.
Methods
Cell culture and treatments
Human breast adenocarcinoma cells (MCF-7; American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen), 0.01 mg/ml bovine insulin, and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). siRNA oligos were designed and manufactured by Integrated DNA Technologies (IDT, Coralville, IA), targeting either CRY2 (Sense: 5'-UGCUUCAUUCGUUCAAUGUUAAGCCGG-3' Antisense: 5'-GGCUUAACAUUGAACGAAUGAAGCA-3') or a scrambled sequence negative control siRNA (Sense: 5'-CUUCCUCUCUUUCUCUCCCUUGUGA-3', Antisense: 5'-UCACAAGGGAGAGAAAGAGGGAAGGA-3'). Each oligo was complexed and reverse transfected using Lipofectamine RNAiMax transfection reagent (Invitrogen) at a final oligo concentration of 10 nM. Cells were either harvested 48 hours after transfection, to assay for knockdown efficiency by qPCR, or incubated with either PBS (neg. control) or 0.03% (v/v) methyl methanesulfonate (MMS, chemical mutagen) for use in subsequent assays.
RNA isolation and quantitation
RNA samples were isolated from harvested cells using the RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions for mammalian cells, including on-column DNA digestion. Gene expression was quantified by two-step quantitative RT-PCR, beginning with first-strand cDNA synthesis using the AffinityScript cDNA kit (Stratagene, La Jolla, CA) with oligo-dT primers, followed by quantitative real-time PCR using the Power SYBR Green PCR master mix (Applied BioSystems, Foster City, CA). The primers used for CRY2 amplification were: (L: ACCGGGGACTCTGTCTACTG, R: GCCTGCACTGCTCATGCT). RNA quantity was normalized using HPRT1 content, and CRY2 silencing was quantified prior to each treatment according to the 2-ΔΔCt method. In each case, CRY2 was reduced to less than 20% of the levels seen in the mock siRNA-treated negative control (i.e. 5-fold downregulation).
Whole genome expression microarray and pathway-based expression analysis
A whole genome expression microarray (Agilent, Inc 44k chip, performed by MoGene, LC, St Louis, MO) was used to interrogate gene expression in cells with normal and reduced CRY2. The results of these experiments have been uploaded to the Gene Expression Omnibus (GEO) database, and can be accessed by referencing accession #GSE14617. In order to determine whether genes involved in cell cycle regulation and DNA damage response were influenced by CRY2 knockdown, we analyzed the microarray expression data for genes in SABioscience's "Human Cell Cycle" and "Human DNA Damage Signaling Pathway" arrays (catalog numbers PAHS-020 and PAHS-029, respectively). All fold changes are the result of two biological replicates of the microarray experiment, and all observations with low intensity (<50) in both CRY2 normal and CRY2 knockdown populations have been discarded. Significantly altered genes were confirmed by qPCR, and the primers used for these reactions can be found in Additional file 1.
Cell cycle, cell viability, and apoptosis assays
Cells with normal and reduced CRY2 levels were stained with propidium iodide (PI) and analyzed by flow cytometry using a fluorescence-activated cell sorter (FACS) flow cytometer (Becton Dickinson, San Jose, CA). Prior to analysis, cells were treated with either PBS or MMS for 1 hour, followed by duplicate washes and 24 hour incubation in normal growth medium. Cell populations from each treatment group were analyzed using the FlowJo flow cytometry analysis software (Tree Star, Inc., Ashland, OR), and cell phases were determined using the Watson pragmatic algorithm [12]. Cells from each of the four treatment groups (CRY2 +/-, MMS +/-) were also assayed for viability and apoptosis using the Vybrant Apoptosis Assay Kit #2 (Invitrogen). Briefly, cells were stained with PI and Annexin V according to the manufacturer's protocol, and scored as live, dead, apoptotic or ambiguous by flow cytometry. All results are based on the average of triplicate experiments. Raw flow cytometry images from each analysis are available in Additional file 2.
DNA Damage Assay
DNA damage accumulation was measured by alkaline single-cell gel electrophoresis (i.e. the comet assay). Cells with normal and reduced CRY2 levels were incubated with MMS for one hour, followed by two PBS washes and 3 hours of recovery time. Cells were then fixed onto slides with low-melting agarose, lysed, and treated with pH>14 solution at 4°C to denature the DNA. Slides were then subjected to electrophoresis to allow damaged DNA to migrate away from the nucleus, and then stained with ethidium bromide. In order to reduce the possibility of observer bias, the prepared slides were then given to a second person who scrambled them and assigned his own arbitrary numeric label to each slide before returning them for scoring. As such, the person responsible for generating the data was unaware of the treatment status for each slide. 50 cells from each treatment group were analyzed by fluorescence microscopy using the Komet 5 comet assay analysis software. DNA damage was quantified by the software using the mean Olive tail moment calculation for each cell, as previously described [13]. Once the scoring was complete, the data were sent back to the second person who replaced the numeric label with the treatment identity. All results are from duplicate experiments performed on 50 cells each.
Statistical Analysis
All statistical analyses were performed using the SAS statistical software (SAS Institute, Cary, NC), unless otherwise noted. CRY2 knockdown was assessed using the 2-ΔΔCt method with RNA content normalized to the housekeeping gene HPRT1. Differences in cell cycle distribution were investigated by determining the percentage of cells in each phase for each treatment group. The CRY2 normal population was then compared to the CRY2 reduced population with and without mutagen challenge using the Student's t-test. Similar comparisons were made for cell viability and apoptotic response, comparing CRY2 normal and CRY2 reduced populations using the t-test. For the comet assay, comparisons were for the mean Olive tail moment in cells with reduced and normal CRY2, and again, the t-test was used. Due to the large number of observations present in the microarray, P-values were adjusted for multiple comparisons using the false discovery rate correction, as previously described [14], in order to obtain an adjusted Q-value for each observation.
Results
Silencing of CRY2 does not influence cell cycle distribution, cell cycle checkpoints, cell death, or apoptosis in response to mutagen challenge
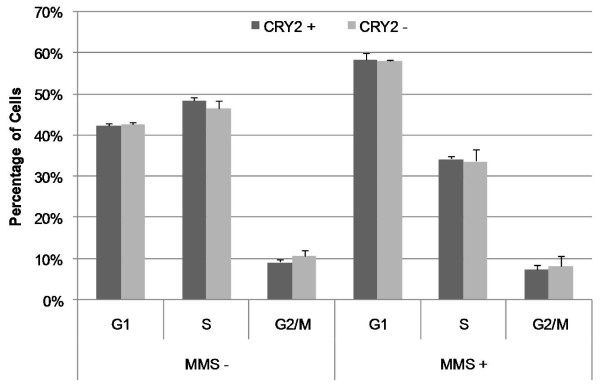
Cell cycle distributions in mock-treated or mutagen challenged cells were measured in populations with normal (CRY2+) and reduced CRY2 levels (CRY2-). In both treatment groups, similar distributions were observed for CRY2+ and CRY2- cell populations, indicating that reduction of CRY2 alone is not sufficient to significantly impact cell cycle regulation. The cell phase distributions in mock treated cells were: CRY2+: G1, 42.3%, S, 48.5%, G2/M, 9.2%; CRY2-: G1, 42.7%, S, 46.6%, G2/M, 10.8% (Figure 1). In addition, a similar magnitude of G1 delay was observed in both the CRY2+ and CRY2- populations following mutagen challenge, suggesting that CRY2 does not significantly influence DNA damage-induced cell cycle checkpoint control. Cell cycle distributions in mutagen treated cells were: CRY2+: G1, 58.3%, S, 34.3%, G2/M, 7.4%; CRY2-: G1, 58.1%, S, 33.7%, G2/M, 8.2%.
Figure 1.
Cell cycle distributions in CRY2- and CRY2+ cells in mock and mutagen treated cell populations. Both in the absence and presence of mutagen, cells with reduced CRY2 had a similar cell cycle distribution as cells with normal CRY2 levels, suggesting no direct impact of CRY2 on cell cycle progression or cell cycle checkpoints in response to DNA damage. All results are based on the average of triplicate experiments, and error bars are for SEM.
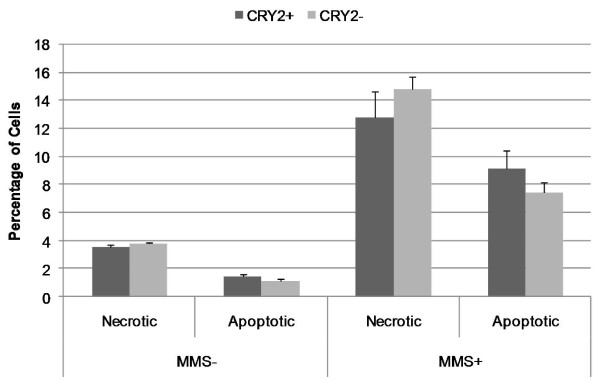
Flow cytometric analyses of necrosis and apoptosis in mock and mutagen treated cells also revealed no significant differences in CRY2+ and CRY2- cell populations. Both populations had similarly low percentages of necrotic and apoptotic cells in the mock treated populations: CRY2+: necrotic 3.5%, apoptotic, 1.5%; CRY2-: necrotic, 3.8%, apoptotic 1.2% (Figure 2). The percentage of necrotic and apoptotic cells in both these populations increased by similar margins following mutagen challenge: CRY2+: necrotic 12.8%, apoptotic, 9.1%; CRY2-: necrotic, 14.8%, apoptotic 7.5%. No comparisons, either in the percentage of necrotic or apoptotic cells, or in the magnitude of the response following mutagen challenge, were statistically significant.
Figure 2.
Cell death and apoptosis in CRY2+ and CRY2- cell populations. No differences were observed in CRY2 knockdown cells in terms of necrosis or apoptosis, with or without mutagen exposure. All results are based on the average of triplicate experiments, and error bars are for SEM.
CRY2 knockdown results in increased accumulation of mutagen-induced DNA damage
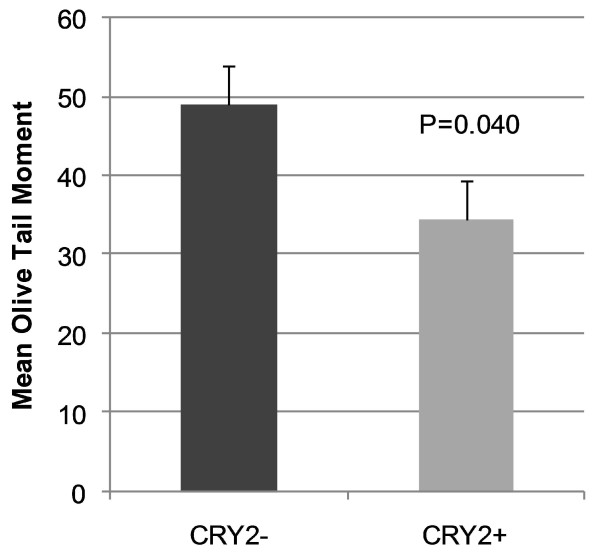
The single cell gel electrophoresis (comet) assay was used to evaluate the extent of DNA damage following mutagen challenge in CRY2+ and CRY-populations. DNA damage was quantified using the Olive tail moment measure, which incorporates the distance of DNA migration from the nucleus, as well as the percentage of total DNA which has migrated away from the nuclear core, indicating that it has been relaxed or broken. The mean Olive tail moment in CRY2- cells was significantly higher than that in CRY2+ cells (P = 0.04), indicating that the same levels of mutagen exposure result in greater DNA damage in cells with reduced CRY2 (Figure 3). This is especially notable given that CRY2- cells do not exhibit decreased survival or increased apoptosis, suggesting that they survive equally well despite the increased level of DNA damage; a phenotype which could lead to tumor promotion.
Figure 3.
Comet assay results for cells with reduced and normal CRY2. DNA damage was measured by the Olive tail moment, which considers the amount and distance of DNA migration away from the nucleus, indicative of DNA strand breaks. Higher values correspond to increased damage. Upon exposure to the same concentration of mutagen, CRY2- cells accumulated significantly more DNA damage compared to normal cells (P = 0.040). Comet assay results are from duplicate experiments performed on 50 cells each. All data are aggregated (i.e. 100 cells per treatment), and error bars are for SEM.
CRY2 knockdown results in expression changes in genes in the DNA damage response and cell cycle regulatory pathways
A whole genome expression microarray was performed using RNA isolated from CRY2+ and CRY2- cell populations. Using the genes in the SABioscience cell cycle and DNA damage response arrays, we identified 10 genes in these pathways which displayed significantly altered expression following CRY2 knockdown (Q-value < 0.05). Expression of each of these genes was also determined by qPCR for each cell treatment in order to confirm the array results. All genes were altered in the same direction when measured by array or qPCR, and all fold changes were of similar magnitude, with the exception of SUMO1, which was two-fold upregulated in the array, but only 1.32 upregulated when measured by qPCR (Table 1). Among the significantly altered gene set were cyclin dependent kinase inhibitor p21 (CDKN1A; fold change = 1.7, Q < 0.001) along with its interacting protein (BCCIP; fold change = 2.3, Q = 0.002). Perhaps most interesting, however, was the induction of cyclin D1 (CCND1; fold change = 1.5, Q = 0.009) in cells with reduced CRY2. CCND1 is a firmly established oncogene that is often overexpressed in primary breast cancers [15], and while aberrant overexpression of CCND1 is often due to gene amplification, up to 50% of breast cancers display increases in CCND1, many of which cannot be explained by copy number variations, indicating that alternative mechanisms such as transcriptional dysregulation must be involved [16].
Table 1.
Cell cycle and DNA damage repair genes with altered expression following CRY2 silencing.
| Gene | RefSeq | Description | Array Fold Change | Q-Value | qPCR Fold Change |
|---|---|---|---|---|---|
| BCCIP | NM_078469 | BRCA2 and CDKN1A interacting protein | 2.33 | 0.0021 | 1.57 |
| BCL2 | NM_000633 | B-cell CLL/lymphoma 2 | -1.47 | 0.0487 | -1.50 |
| CCND1 | NM_053056 | Cyclin D1 | 1.51 | 0.0087 | 1.33 |
| CDKN1A | NM_000389 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 1.71 | 0.0001 | 1.98 |
| GADD45A | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 1.92 | 0.0015 | 2.16 |
| HERC5 | NM_016323 | Hect domain and RLD 5 | 3.93 | 0.0000 | 2.28 |
| MCM5 | NM_006739 | Minichromosome maintenance complex component 5 | -1.58 | 0.0415 | -1.46 |
| PPP1R15A | NM_014330 | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | 2.55 | 0.0004 | 2.45 |
| SUMO1 | NM_001005781 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 2.00 | 0.0002 | 1.32 |
| UBA1 | NM_003334 | Ubiquitin-like modifier activating enzyme 1 | 1.58 | 0.0232 | 1.43 |
Discussion
CRY2, in conjunction with CRY1 and the period genes, (PER1, PER2, and PER3) operates on the negative arm of the circadian system and is essential for maintaining proper circadian rhythm [17]. However, due to the complex nature of circadian gene interactions, which include pre- and post-transcriptional regulation, it remains difficult to determine the direct phenotypic impact of a single gene, especially in light of the potential for overlapping functions and compensatory mechanisms among the core circadian proteins. Nevertheless, the observation that cells with reduced CRY2 accumulate greater DNA damage is consist with the general understanding that circadian genes may directly influence organismal susceptibility to genotoxic stress [18].
The finding that CCND1 is induced following CRY2 knockdown, while not proof of direct inhibition of CCND1 by CRY2, does provide the intriguing possibility that the aberrant overexpression of CCND1 observed in several cancer types could be, in part, the result of circadian-mediated transcriptional dysregulation. Evidence for this association was provided by an earlier study which showed that enforced expression of PER2 resulted in a 56% reduction in CCND1 levels in vitro [19]. Interestingly, in addition to the positive regulator of cell cycle progression, CCND1, a crucial cyclin-dependent kinase inhibitor, CDKN1A (also known as P21), was also induced after CRY2 knockdown. This finding is consistent with a recent report demonstrating that clock-deficient mice have increased levels of p21, resulting in decreased cellular proliferation rates [20]. That both CCND1 and CDKN1A, which influence cell cycle progression in opposite directions, were each upregulated after CRY2 knockdown provides a potential explanation for the lack of observable phenotypic impact of CRY2 silencing on cell cycle distributions. Given the importance of these cell cycle regulators in a variety of cancer types, additional exploration into the nature of these associations is warranted.
Despite evidence that cryptochromes may be involved in cancer-associated processes [11], two in vivo studies did not find a cancer-prone phenotype in double mutant mice lacking both crytpochrome genes (i.e. Cry1-/-Cry2-/-). For example, Cry1-/-Cry2-/- mice did not have poorer survival rates than wild type (WT) mice following exposure to ionizing radiation [10]. In addition, fibroblasts derived from these mice did not have deficient DNA damage capacities compared to those derived from wild type mice, and cell cycle checkpoints were similarly unaffected. Another study of Cry1-/-Cry2-/- also showed that while these mice were significantly smaller than their WT counterparts, they did not have any obvious malignancies, and they remained reproductively fit [21]. Interestingly, in mice which are predisposed to cancer due to a mutation in p53, addition of the Cry mutation results in sensitization of p53 mutant cells to apoptosis and thus decreased cancer risk and increased survival [22]. In addition, there is a strong circadian rhythm in nucleotide excision repair activity in the mouse brain, caused at least partly by circadian regulation of xeroderma pigmentosum A (XPA) [23]. Cry negatively regulates this activity, and fibroblasts from Cry1-/-Cry2-/- mice exhibit 3-fold induction of XPA protein. It should be noted, however, that each of these studies employed double mutant mice, and are thus not necessarily reflective of the condition which may exist in the absence of Cry2 only. In fact, a behavioral and molecular study of the effect of cryptochromes on light entrainment and circadian regulation showed very different phenotypes for Cry1-/-Cry2-/- mice compared to mice lacking Cry1 only [24], and an earlier study of Cry-/- mice suggested that reductions in either cryptochrome alone have effects which are directly opposed to one another [8]. To the best of our knowledge, no study has yet explored the response of Cry2-/- only to mutagen challenge, or the effect of induced degradation of CRY2 on DNA repair. It should also be noted that our data are generated using cells with wild-type p53. Thus, future studies may wish to investigate the effect of CRY2 knockdown in mutant p53 cells, especially in light of the recent evidence suggesting differential effects of Cry mutations against the p53 mutant background, as outlined above.
While changes were detected in DNA damage accumulation following CRY2 knockdown, no differences were observed in other cancer-related pathways. However, previous studies have shown that induction of the DNA damage response pathway is an important early event in determining whether precursor lesions will develop into malignancies [25], and those authors suggest that mutations which disrupt the DNA damage response may allow tumor progression. In another study, Gorgoulis et al. demonstrate that a DNA damage response is present in precancerous lesions, also suggesting that disruption of this pathway could be an important determinant of progression to carcinoma [26]. One interesting aspect of our phenotypic assays was the increase in DNA damage observed in CRY2- cell populations in the absence of decreased survival or increased apoptosis. If in fact reduced CRY2 results in increased DNA damage without triggering increases in cell death or apoptosis, this could potentially lead to cancer, as damaged cells could survive and be allowed to proliferate. As such, this phenotype warrants further investigation.
Conclusion
In summary, these data suggest a limited, but potentially important role for CRY2 in maintaining genomic stability. Future investigations may wish to focus on the transcriptional influence of CRY2 on oncogenic CCND1, and the relationship with CDKN1A, as these findings have the potential for broad impact on a number of cancer types. In addition, given the evidence that response to cancer therapy may be influenced by circadian cycling [18,27,28], and the fact that CRY2 may influence the accumulation of DNA damage, future investigations into the effects of CRY2 on response to treatment are also warranted.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AEH carried out the knockdown experiments, as well as the cell cycle and apoptosis studies and the analysis of the array data, and drafted the manuscript. YB, CY, and DL each participated in the comet assay. RS and TZ participated in the design of the study. YZ conceived the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Primer sequences used for qPCR confirmation of genes identified as significantly altered by microarray.
Flow cytometry images from the cell cycle (2A) and cell viability/apoptosis (2B) analyses.
Contributor Information
Aaron E Hoffman, Email: aaron.hoffman@yale.edu.
Tongzhang Zheng, Email: tongzhang.zheng@yale.edu.
Yue Ba, Email: bayue1963@gmail.com.
Richard G Stevens, Email: bugs@uchc.edu.
Chun-Hui Yi, Email: yich1@hotmail.com.
Derek Leaderer, Email: derekleaderer@yahoo.com.
Yong Zhu, Email: yong.zhu@yale.edu.
Acknowledgements
This work was supported by the US National Institutes of Health (grants CA122676, CA110937, and CA108369).
References
- Oster H. The genetic basis of circadian behavior. Genes Brain Behav. 2006;5(Suppl 2):73–79. doi: 10.1111/j.1601-183X.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. Embo J. 2001;20(24):7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–557. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282(5393):1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25(8):3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65(15):6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Watson JV, Chambers SH, Smith PJ. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 1987;8(1):1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the "comet" assay. Radiat Res. 1990;122(1):86–94. doi: 10.2307/3577587. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA, Sutherland RL. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8(8):2127–2133. [PubMed] [Google Scholar]
- Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23(18):4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Kondratov RV, Takahashi JS. Circadian clock genes as modulators of sensitivity to genotoxic stress. Cell Cycle. 2005;4(7):901–907. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4. doi: 10.1186/1740-3391-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283(8):4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. full_text. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106(8):2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci USA. 2009;106(8):2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Ørntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci USA. 2005;102(9):3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis CG, Grutsch JF, Wood P, You M, Rich I, Hrushesky WJ. Circadian timing in cancer treatment: the biological foundation for an integrative approach. Integr Cancer Ther. 2003;2(2):105–111. doi: 10.1177/1534735403002002002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for qPCR confirmation of genes identified as significantly altered by microarray.
Flow cytometry images from the cell cycle (2A) and cell viability/apoptosis (2B) analyses.