Abstract
Rationale
The gene encoding the helix-loop-helix transcription factor, Id3, is located within atherosclerosis susceptibility loci of both mice and humans, yet its influence on atherosclerosis is not known.
Objective
The present study sought to determine if polymorphisms in the ID3 gene were associated with indices of atherosclerosis in humans and if loss of Id3 function modulated atherogenesis in mice.
Methods and Results
Six tagging SNPs (tagSNPs) in the human ID3 gene were assessed in participants of the Diabetes Heart Study. One tagSNP, rs11574, was independently associated with carotid intima-media thickness (IMT). The human ID3 variant at rs11574 results in an alanine to threonine substitution in the C-terminus. To determine the effect of this polymorphism on the basic function of Id3, site-directed mutagenesis of the human ID3 gene at rs11574 was performed. Results demonstrated a significant reduction in co-immunoprecipitation of the known E-protein partner, E12, with Id3 when it contains the sequence encoded by the risk allele (Id3105T). Further, Id3105T had an attenuated ability to modulate E12-mediated transcriptional activation compared to Id3 containing the ancestral allele (Id3105A). Microarray analysis of vascular smooth muscle cells from WT and Id3-/- mice revealed significant modulation of multiple gene pathways implicated in atherogenesis. Moreover, Id3-/- ApoE-/- mice developed significantly more atherosclerosis in response to 32 weeks of Chow or Western diet feeding than Id3+/+ApoE-/- mice.
Conclusions
Taken together, results provide novel evidence that Id3 is an atheroprotective factor and link a common SNP in the human ID3 gene to loss of Id3 function and increased IMT.
Keywords: atherosclerosis, diabetes mellitus, genetics
Introduction
Atherosclerosis is a chronic, inflammatory disease in which lipids, cells and fibrous elements accumulate in the intimal layer of large arteries1, 2. This process begins in adolescence and follows a variable clinical course that may ultimately result in myocardial infarction (MI) or stroke3, 4. Because of this, it is estimated that atherosclerosis is the underlying cause of 50% of all deaths in Westernized society1. Despite the magnitude of this problem, understanding mechanisms whereby specific genes or gene pathways modulate atherosclerosis in humans remains challenging. This is largely attributed to the fact that atherosclerosis, in its common form, is a multifactorial disorder. While recent studies have identified potential candidate genes in humans, few studies have been able to determine the impact of specific human gene variants on protein function and disease modulation.
Recent studies have identified susceptibility loci for atherosclerosis in both humans and mice5. Using genomewide linkage analysis in a Caucasian American population, Wang et al. reported a novel significant susceptibility locus for premature myocardial infarction at 1p34-36 (12-40 Mbp)6. Welch et al., used the LDLR-/- mouse in an interspecific genetic cross to identify a murine atherosclerosis susceptibility locus on mouse chromosome 4 (27-154 Mbp), which they named Athsq17. Intriguingly, comparison of human 1p34-36 with the Athsq1 locus in mice reveals that one major gene common to both loci is Inhibitor of Differentiation-3 (Id3).
Id3 is a member of the basic helix-loop-helix (bHLH) family of proteins. While Id3 contains the HLH domain required for protein:protein dimerization, it lacks the basic DNA-binding domain possessed by other members of the family, making it incapable of binding to DNA. Instead, Id3 binds to another subset of bHLH factors known as the E-proteins, including E12 and E47, thereby preventing their dimerization with tissue-specific bHLH factors and inhibiting subsequent DNA binding. These Id:E-protein dimers have been demonstrated to regulate many genes in a variety of cell types including B cells, T cells, adipocytes and smooth muscle cells8-10. Id3 itself has been implicated in the pathobiology of vascular disease and has been detected in vascular lesions of rodents, pigs and humans11-13. While it is undetectable in normal arteries, Id3 is expressed in response to wire endothelial denudation of the carotid artery in rodents12 and enhances vascular smooth muscle cell (VSMC) growth11, 14-16, implicating Id3 in promoting the neointimal response to injury. Animals fed a high fat diet leading to hyperlipemia express higher levels of Id3 in the atherosclerotic vessel wall compared with their normolipemic non-diseased controls13. Yet, the pathobiology and genetic determinants of injury-induced neotintimal formation and diet-induced atherosclerosis are quite distinct17, 18. Whether Id3 expressed in atherosclerotic lesions activates gene pathways to limit or promote atherosclerosis is unknown.
Results of the present study have identified an association between polymorphism at rs11574 in the ID3 gene and carotid intima-medial thickness (IMT) in participants from the Diabetes Heart Study (DHS). Mutation of the major allele of the human ID3 gene at rs11574 to the risk allele resulted in attenuated Id3 function. Moreover, deletion of the ID3 gene resulted in a significant increase in atherosclerosis formation in Western-fed ApoE-/- mice. Together, these results provide evidence that Id3 is an important atheroprotective factor in mice and in humans.
Materials and Methods
An expanded Materials and Methods section is available in the online data supplement at http://www.circres.ahajournals.org.
Human subjects
The Diabetes Heart Study (DHS) is an affected sib-pair study of subclinical atherosclerosis and its risk factors, consisting of families with two or more siblings with a diagnosis of T2D and lacking advanced renal insufficiency. Ascertainment and recruitment have been described previously19, 20. Details of the protocols for clinical data acquisition, IMT measurement, isolation of human genomic DNA and genotyping, and statistical analysis are provided in the online supplement.
Functional analysis of Id3105A vs. Id3105T
Plasmid constructs expressing products of the ancestral (Id3105A) and variant (Id3105T) alleles of ID3 were generated and analyzed for differences in expression, co-immunoprecipitation with E12, and dominant negative antagonism of E12 function as detailed in the online supplement.
Studies using Id3 null mice
Detailed methods for microarray analysis of VSMC from WT (C57BL/6) and Id3-/- mice and quantitation of atherosclerosis in Id3+/+ApoE-/- vs. Id3-/- ApoE-/- mice are provided in the online supplement.
Results
Analysis of ID3 SNPs in a Diabetic Human Population: Association with Subclinical Atherosclerosis
Two previous studies have identified regions of atherosclerosis susceptibility in mice and men which contain the ID3 gene, suggesting that Id3 may be a candidate gene for association with CVD6,7. We sought to determine whether polymorphisms in the human ID3 gene were associated with subclinical markers of atherosclerosis in humans by assessing tagSNPs in the participants of the DHS. The subjects for this analysis were all European American sib-pairs with T2D (n=780). Clinical characteristics of the sample are consistent with a population of subjects with T2D: older age (mean 62 yrs), increased BMI (32.4 kg/m2), elevated systolic blood pressure (139.8 mmHg). However, participants did not have overly aberrant lipid measures (LDL 104 mg/dL, HDL 42.7 mg/dL), likely due to the extensive use of lipid-lowering therapy in this group (44.7%)(Supplementary Table I). As a measure of subclinical atherosclerosis, intima-media wall thickness (IMT, mean 0.68 mm) was obtained on participants in this sample.
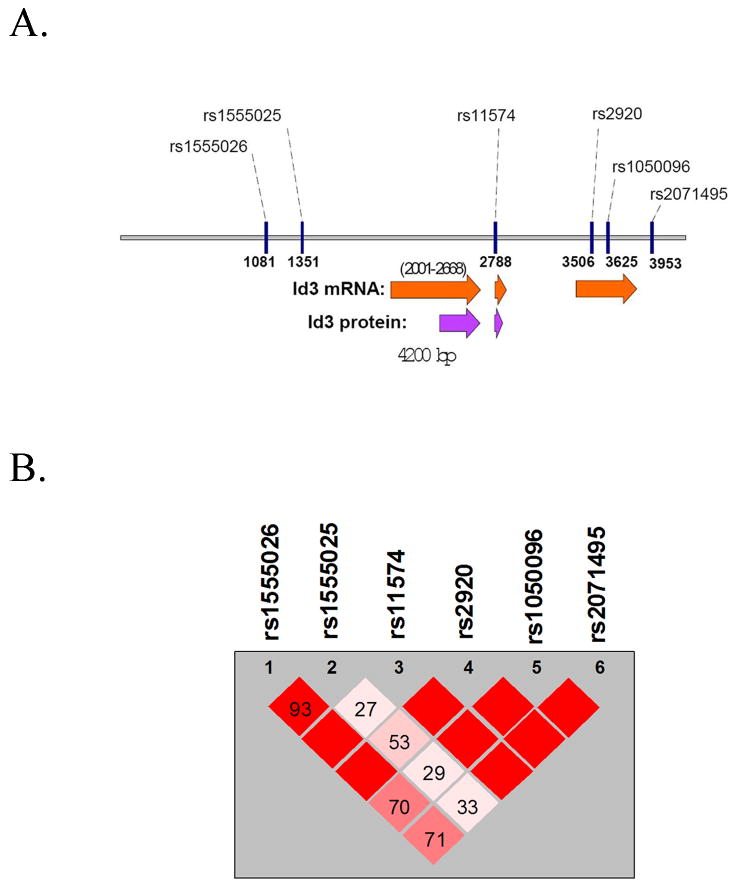
ID3 is a small gene, spanning only three exons, the first two of which are coding exons. Six SNPs were identified within the gene which captured all eight alleles and tagged haplotypes with a mean r2 of 0.967 (Figure 1A)21. All six tagSNPs were successfully genotyped in the samples from the DHS: rs1555026 and rs1555025 (5′ of the gene), rs11574 (exon 2), rs2920 and rs1050096 (exon 3) and rs2071495 (3′ to the gene). The total distance between rs155026 and rs2071495 is 2.9kb. The region consists of two blocks of linkage disequilibrium, the first two SNPs in one block and the last four SNPs in the other (Figure 1B).
Figure 1. Tagging SNPs within the human ID3 gene.
A, Schematic depicting the six tagging SNPs in the human ID3 gene. Approximate sites of RNA transcripts and protein products are noted beneath the gene sequence. B, LD plot depicting the two haplotype blocks encompassing the six ID3 tagging SNPS in the DHS analysis. Numbers reflect D' values.
To determine whether an association exists between any of the ID3 tagSNPs and subclinical atherosclerosis, the affected sib-pair families were subjected to a quantitative trait locus (QTL) association analysis. This analysis was performed in a model without covariates as well as in one that incorporated the effects of known risk factors (age, sex, BMI, systolic blood pressure, LDL and HDL). Of the six SNPs that were used for this analysis, one (rs11574) showed evidence of significant association with subclinical atherosclerosis as measured by IMT (Table 1, p=0.01). Incorporating the aforementioned covariates into the model, the association of rs11574 with IMT was shown to be independent of these risk factors (Table 1, p=0.005). When rs11574 was included in the model of IMT as a covariate, no other SNP was significantly associated with the residual phenotype (next most associated: rs2920, p=0.09). Pedigree-wide regression analyses demonstrated that there was significant heritability (h2) for IMT independent of the effects of known risk (h2 = 0.26 ± 0.12; p=0.02). Interestingly, there is a stepwise increase in mean IMT associated with the minor (variant) allele of rs11574. Subjects who are homozygous for the major (ancestral) allele (GG) have a mean IMT of 0.66mm, while mean IMT was 0.69mm for those who are heterozygous (GA) and 0.72mm for those homozygous for the minor allele (AA). Studentized range statistic for the 3 groups demonstrated these differences were significant (p<0.01)(Table 2).
Table 1. Association of ID3 SNPs with IMT in the Diabetes Heart Study.
| Without Covariates | With Covariates | |||
|---|---|---|---|---|
| SNP | Effect | p-value | Effect | p-value |
| rs1555026 | 0.011 | 0.17 | 0.008 | 0.28 |
| rs1055025 | 0.002 | 0.59 | 0.000 | 0.79 |
| rs11574 | 0.011 | 0.01 | 0.011 | 0.005 |
| rs2920 | 0.003 | 0.47 | 0.003 | 0.55 |
| rs1050096 | 0.007 | 0.10 | 0.006 | 0.10 |
| rs2071495 | -0.004 | 0.49 | -0.004 | 0.34 |
Table 2. IMT increases stepwise with the minor allele at rs11574.
| Genotype | Resulting Amino Acid | Number of Patients | Mean IMT (mm) | Standard Deviation | Median IMT (mm) |
|---|---|---|---|---|---|
| GG | Ala/Ala | 419 | 0.66 | 0.13 | 0.64 |
| GA | Ala/Thr | 296 | 0.69 | 0.14 | 0.66 |
| AA | Thr/Thr | 49 | 0.72 | 0.12 | 0.71 |
| XX | --- | 16 | 0.64 | 0.10 | 0.61 |
p < 0.01 for comparison of GG vs. GA vs. AA
SNP rs11574 Does Not Affect the Expression of Id3
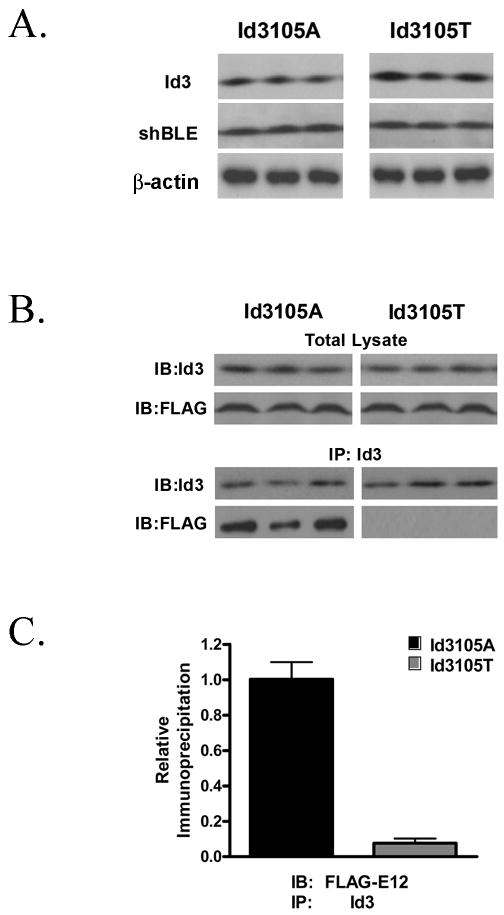
The rs11574 SNP is a nonsynonymous change, resulting in an alanine (ancestral) to threonine (variant) substitution at amino acid 105. In addition, this SNP is located in the second exon encoding the C-terminal portion of Id3, which has previously been shown to be essential for dominant negative function22. To determine functional significance of polymorphism at rs11574, the coding region of human ID3 (Id3105A) was cloned into a mammalian expression vector and site-directed mutagenesis was employed to generate a construct expressing the variant encoded by the minor allele (Id3105T). NIH3T3 fibroblasts were transfected with either expression vector and analyzed by Western blotting (Figure 2A). To control for potential differences in transfection efficiency, expression of Id3 was normalized to an internal control, the ShBle protein, which is expressed from the same plasmid. The results demonstrate no significant difference in the expression of Id3105A compared with Id3105T (Id3105A/ShBle ratio = 1.13 (95% CI 0.88 to 1.38), Id3105T/ShBle ratio = 1.19 (95% CI 0.91 to 1.47), p=0.62). β-actin levels were not significantly different between samples.
Figure 2. The variant ID3 allele at rs11574 demonstrates decreased binding to E12.
A, NIH3T3 cells were transfected with either the pEF4-Id3105A or pEF4-Id3105T plasmids and analyzed by Western blotting 48 hours after transfection. To control for potential differences in transfection efficiency between Id3105A and Id3105T, expression of Id3 was normalized to expression levels of ShBle in the same samples. β-actin was used as a loading control. Results are representative of three independent experiments in triplicate. B, NIH3T3 fibroblasts were transfected with either Id3105A or Id3105T constructs and FLAG-E12. Forty-eight hours after transfection, total lysates were precipitated using G sepharose beads that had been pre-conjugated to an Id3 antibody. Immunoprecipitates and total lysates were separated by SDS-PAGE and immunoblotted with FLAG or Id3 antibodies. Results are representative of three independent experiments. C, Quantitation of co-IP experiments in B. p=0.0002.
SNP rs11574 Reduces the Binding of Id3 to E12
To determine whether differences exist in the ability of Id3105A and Id3105T to dimerize with typical Id protein partners, co-immunoprecipitation (co-IP) studies were employed. NIH3T3 fibroblasts were transfected with either Id3105A or Id3105T and FLAG-E12 and harvested after 48 hours. Lysates were then immunoprecipitated with sepharose G beads that were pre-conjugated with a monoclonal Id3 antibody generated within the lab (see methods). Id3105T demonstrated a robust 92% reduction in E12 co-IP versus Id3105A (Id3105A binding ratio = 2.58 ± 0.70, Id3105T binding ratio = 0.20 ± 0.20, p=0.0002) (Figure 2B). Quantitation of all of the co-IP experiments (n=3 in triplicate) is shown in Figure 2C.
Id3105T has a reduced ability to inhibit transcription compared with Id3105A
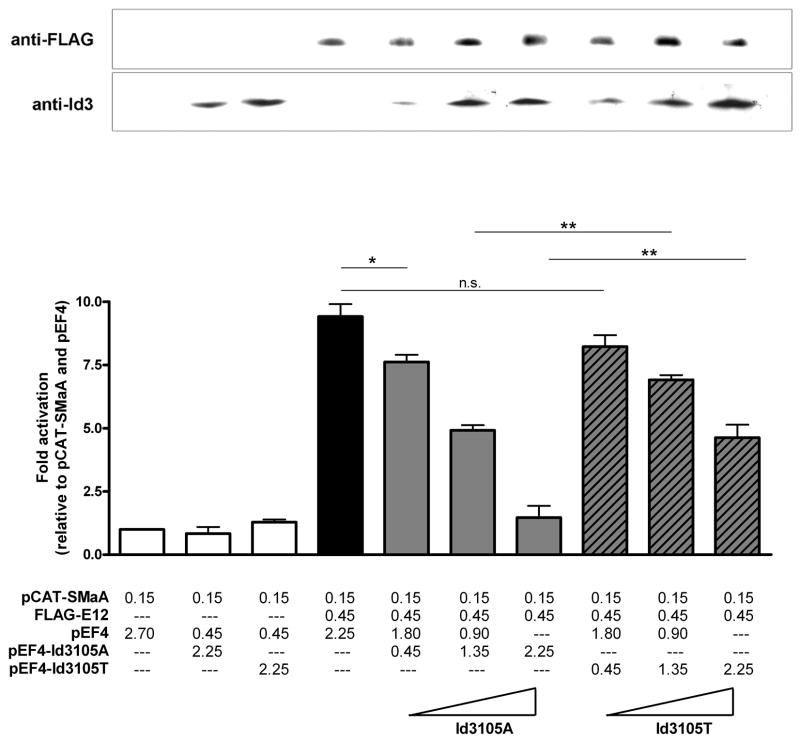
To determine if the reduced binding of Id3105T to its E12 partner may have functional consequences, promoter-reporter studies were undertaken. As a readout of transcriptional activity, the smooth muscle alpha actin (SMaA) promoter, which has previously been shown to be activated by E12, in an E-box dependent manner, was used9, 23. As anticipated, transfection of E12 resulted in activation of the SMaA promoter, an effect which was inhibited by Id3105A in a concentration-dependent manner. In contrast, the inhibitory ability of Id3105T was significantly attenuated, yielding only partial inhibition of E12-mediated SMaA promoter activity even at the highest concentration (Figure 3). Western blotting confirmed that the observed differences between Id3105A and Id3105T activity were not due to differences in expression.
Figure 3. Id3105T has a decreased ability to inhibit transcription compared with Id3105A.
NIH3T3 cells were transfected in triplicate with a smooth muscle alpha actin promoter-reporter (pCAT-SMaA) as well as with Id3 and FLAG-E12 expression vectors as indicated (values in μg). Thirty-six hours after transfection, cells were harvested and assayed for CAT activity. CAT activity was normalized to protein concentration and is presented as fold activation relative to the first group (promoter plus vector only). Samples were immunoblotted with anti-FLAG and anti-Id3 antibodies in parallel to confirm the expected expression patterns. Representative Western blots are shown above their corresponding samples. *:p=0.034. **:p=0.002.
Loss of Id3 alters expression of genes in multiple pathways linked to atherogenesis
The attenuated ability of Id3105T to antagonize E12-mediated SMaA promoter activation provided proof of concept that, in addition to attenuating dimerization with E12, Id3 polymorphism at rs11574 had functional consequences on gene expression. Yet, atherosclerosis is a complex process involving many pathways and cell types, and it is not likely that transcriptional regulation of a single gene would result in the magnitude of difference in IMT seen in the DHS population. To determine if loss of Id3 has a significant impact on other genes in major atherogenic pathways, we performed microarray analysis of primary aortic VSMCs from WT (C57/BL6) or Id3-/- mice. Gene expression was assessed using the Affymetrix MG430A GeneChip representing ∼ 22,600 transcripts in duplicate samples in each group to confirm reproducibility. After quality of the array data was confirmed, the data set was analyzed using Ingenuity Pathway Analysis software and pathways known to be involved in atherogenesis were assessed. Results demonstrate that VSMCs from Id3-/- mice have significant modulation of genes involved in adhesion, immune trafficking, apoptosis, cellular proliferation, and cell movement (Table 3). Genes from these pathways with the greatest modulation and p<0.05 (WT vs. Id3-/-) are listed in Supplementary Tables II and III.
Table 3. Loss of Id3 alters expression of genes in multiple pathways linked to atherogenesis.
| Atherogenic Process | p-value |
|---|---|
| Cell to cell signaling and interaction (adhesion of cells) | 3.9 × 10-6 |
| Immune cell trafficking (cell movement of leukocytes) | 1.8 × 10-6 |
| Cell death (apoptosis of eukaryotic cells) | 8.5 × 10-11 |
| Cellular growth and proliferation (proliferation of eukaryotic cells) | 6.3 × 10-7 |
| Cellular movement (cell movement of eukaryotic cells) | 7.4 × 10-9 |
ApoE-/- Mice Null for Id3 Develop Significantly More Atherosclerosis than Control Mice
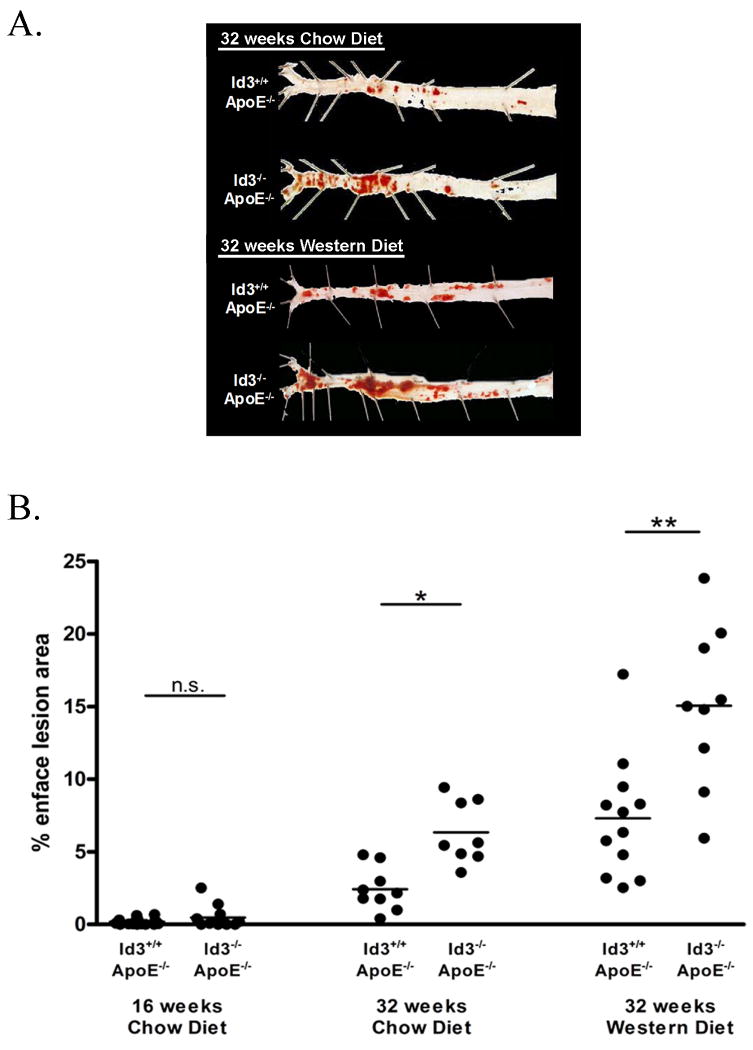
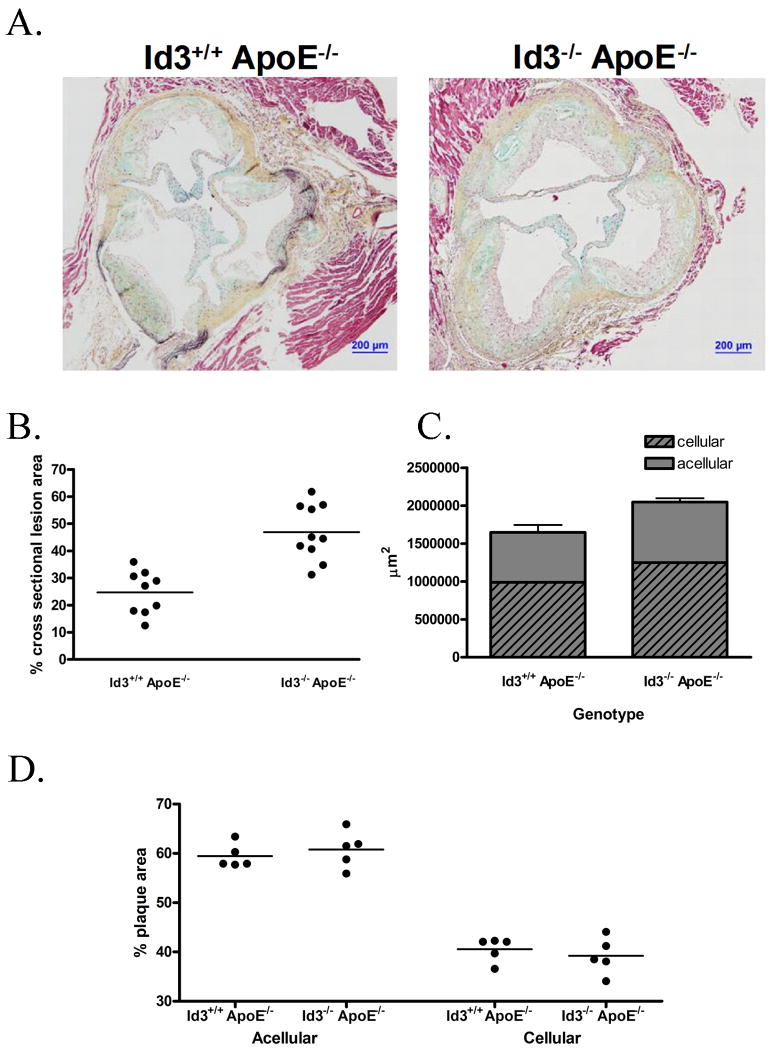
To evaluate the impact of loss of Id3 not only on atherogenic pathways, but on atherogenesis in the intact animal, Id3-/- mice were bred with atherosclerosis-prone ApoE-/- mice. The resulting Id3-/- ApoE-/- mice were placed on either a standard chow or Western diet and analyzed by en face analysis after 16 and 32 weeks of diet. While no significant amount of atherosclerosis was detected in chow-fed mice of either genotype after 16 weeks of chow diet, Id3-/- ApoE-/- mice had significantly more atherosclerosis than Id3+/+ApoE-/- mice after 32 weeks on a chow diet. Similarly, Id3-/- ApoE-/- mice had two-fold more atherosclerosis than Id3+/+ApoE-/- mice after 32 weeks of Western feeding (Figure 4). To confirm and extend these findings, cross-sectional analysis of the ascending aorta was also employed. In agreement with the results of the en face analysis, Id3-/- ApoE-/- mice had significantly more lesion in the ascending aorta than Id3+/+ ApoE-/- mice after 16 weeks of Western feeding (2.0-fold)(Figure 5A and 5B). Consistent with a role for Id3 in regulating genes in many pathways involved in atherogenesis, while the absolute amount of plaque was larger in the Id3-/- ApoE-/- mice, the gross characteristics of the plaques were similar. The proportion of cellular to acellular area is similar between the two groups (Figure 5C and 5D).
Figure 4. Id3-/- ApoE-/- mice have significantly more atherosclerosis than Id3+/+ ApoE-/- mice in the descending aorta.
Beginning at eight weeks of age, Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice were fed a chow or Western diet for 16 or 32 weeks. Aortas were perfused with paraformaldehyde, harvested, opened longitudinally and stained with Sudan IV. En face lesion area was quantitated using Image-Pro 5.0 software. A, Representative vessels from Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice. Regions shown are of the descending aorta from the bifurcation of the iliac arteries to the bifurcation of the left subclavian (left to right). B, Quantitation of lesion area by en face analysis in Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice after 16 weeks of chow diet, 32 weeks of chow diet or 32 weeks of Western diet. Each point represents one animal. *: p = 0.001. **: p = 0.005.
Figure 5. Id3-/- ApoE-/- mice have significantly more atherosclerosis than Id3+/+ ApoE-/- mice in the aortic arch.
Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice were fed a chow or Western diet for 16 weeks. Aortas were perfused with paraformaldehyde and the ascending portions from the aortic cusp to the bifurcation of the brachiocephalic artery were removed and paraffin embedded. Embedded tissue was sectioned into five μm thick intervals. Ten sections, 150 μm apart were stained by the Movat method and analyzed using Image-Pro 5.0 software. A, Representative cross sections from matched regions of the aortas of Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice after 16 weeks of Western feeding. Quantitation of: B atherosclerosis in Id3+/+ ApoE-/- and Id3-/- ApoE-/- mice after 16 weeks of Western diet for cross sectional lesion area (*:p = 0.0002), C, cellular vs. acellular content, and D, percent plaque that is cellular vs. acellular.
Atherosclerosis is known to be influenced by lipid levels, obesity and diabetic status. Western feeding induced the expected increase in total cholesterol, LDL cholesterol, glucose and insulin, but no significant differences were found in any of the lipid measures, insulin levels or body weights when Western or Chow fed Id3+/+ ApoE-/- were compared with the more atherogenic Id3-/- ApoE-/- mice (Supplementary Table IV). Interestingly, glucose levels were lower in the more atherogenic Western-fed Id3-/- ApoE-/- mice compared with Id3+/+ ApoE-/- mice but not in the Chow-fed group.
Discussion
The development of advanced technologies that allow sequencing and analysis of the human genome has led to the identification of a number of SNPs that are associated with disease; however a multitude of confounding factors hamper progress in identifying their functional significance. Few studies have been able to relate variants in candidate genes to a disease phenotype in an animal model or to understand how polymorphisms in the gene alter protein function and ultimately affect disease. In this study, we analyzed the functional significance of a human ID3 gene polymorphism at rs11574 that was associated with carotid IMT in DHS and found loss of a major functional property of Id3 when the specific polymorphic nucleotide at rs11574 was mutated from the major (Id3105A) to the risk (Id3105T) allele, and a marked increase in atherosclerosis with a loss of function approach in a mouse model of atherosclerosis. Together, these data suggest that Id3 may be an important upstream regulatory factor in atherogenesis.
Genotypes of rs11574 were associated with a 9% change in IMT between individuals who were homozygous for G versus homozygous for A (Table 2). Intima-media thickness of the carotid artery is a surrogate quantitative measure of atherosclerosis that has a graded, predictive relationship to overt CVD24. IMT serves as an indicator of atherosclerotic burden25, and a predictor of subsequent cardiovascular events26. IMT changes of the magnitude of those seen in association with ID3 polymorphism have been associated with significant increases in the relative risk of cardiovascular events24, 27, suggesting that in addition to being statistically significant, the percent change in IMT associated with ID3 polymorphism is clinically significant. A large percentage of individuals in the DHS study were on lipid-lowering therapy (44.7%) which is associated with reduced IMT in humans. The average LDL cholesterol in the studied population was only 104.1 with a SD of 32.4. It is possible that an even greater reduction in IMT could be seen in those without the disease allele in a group with high lipids. While lipid values (LDL and HDL) were adjusted for in the covariate analysis and the association of rs11574 with IMT was independent of these variables (p<0.005), the present study does not have sufficient patients with high lipid levels to test this hypothesis directly. Confirmation of the association of ID3 polymorphism with IMT and other indices of atherosclerotic burden in cohorts with a broad range of cholesterol levels and with and without diabetes needs to be performed. Nonetheless, results from the DHS population provided support for Id3 as a candidate gene modulating atherosclerosis and raised the interesting possibility that polymorphism in ID3 at rs11574 alters Id3 function.
Polymorphism at rs11574 results in an amino acid substitution in the C-terminus of Id3, a region of the protein that has been shown to have functional significance. For example, a variant of human Id3, which has an alternate C-terminus generated by retention of an intron (Id3L), was unable to abrogate binding of E-protein to its consensus E-box site in a DNA mobility shift assay28. Consistent with these data, here we have shown that a SNP in this essential domain attenuated E-protein interaction (Figure 2B). Further support for a loss of function phenotype for rs11574 was demonstrated by promoter-reporter assays in which Id3105T had a diminished ability to inhibit E12-mediated promoter activation, confirming that this substitution alters the ability of Id3 to regulate transcription (Figure 3). Interestingly, the inhibition of luciferase activity with the highest concentration of Id3105T appears greater than what might be predicted based on co-IP results (Figure 2B and 2C). Our luciferase results suggest that at high Id3:E12 ratios, Id3105T has some capacity, although significantly less than Id3105A, to inhibit E12 activation of the SMaA promoter (Figure 3). It is possible that the alteration in Id3:E12 interaction for Id3105T is not an all or none phenomenon. The amino acid change at Id3105 may alter affinity for E12, may change the interaction of Id3 with an obligate co-factor or result in a change in partnering with other bHLH factors in the cell. Further detailed biochemical characterization of binding constants and competition assays will be needed to determine the mechanism behind this effect.
The multifactorial nature of atherosclerosis suggests that, for a polymorphism in a single gene to be associated with a change in IMT of the magnitude seen in our study, it must regulate multiple pathways or cell types. Ids exert their dominant negative effect by inhibiting bHLH factors such as the E-proteins from binding to their cognate consensus sites (E-boxes)29. bHLH factors are broadly expressed in many cell types and regulate a wide range of genes that play a role in atherosclerosis and T2D, including C-reactive protein 30, cholesterol synthesis genes31, 32, p2133, plasminogen activator inhibitor type-1 (PAI-1)34, fatty acid synthase35, 36 and insulin37. Recently, we have demonstrated that E-proteins interact with SREBP-1c to activate the adiponectin promoter and by sequestering E-proteins, Id3 can inhibit SREBP-1c activation of transcription8. SREBP-1c has been implicated in the development of obesity, T2D, dyslipidemia and atherosclerosis in humans (reviewed in 38), thus Id3 regulation of SREBP-1c may contribute to atherogenesis. In the present study, pathway analysis of microarray data from VSMC derived from WT and Id3-/- mice reveal that loss of Id3 results in significant modulation of multiple genes within atherogenic pathways (Table 3 and Supplementary Tables II and III). In addition to potential upstream regulation of many VSMC genes involved in atherogenesis, Id3 may regulated atherogenic genes in other cell types known to be important in atherosclerosis, including T cells39, B cells40 and endothelial cells41. Thus, the potential of Id3 to regulate many genes in many cell types may amplify the effects of the rs11574 SNP. Identification of E12 target genes in cells involved in atherogenesis may provide valuable insights into the pathways whereby ID3 polymorphisms and loss of function lead to vascular disease phenotypes.
Deletion of the Id3 gene in ApoE-/- mice, a more dramatic phenotype than attenuated Id3 function due to a SNP, provides further evidence that loss of Id3 function increases atherosclerosis development. Id3-/- ApoE-/- mice were found to have significantly more atherosclerotic lesion than Id3+/+ApoE-/- mice after 16 or 32 weeks of Western feeding. Furthermore, even in the setting of less hyperlipidemia, the Id3-/- ApoE-/- mice maintained on a chow diet for 32 weeks also developed more atherosclerosis than the Id3+/+ ApoE-/- mice (Figure 4). Id3+/+ApoE-/- and Id3-/- ApoE-/- animals had similar lipid values whether chow fed or Western fed despite significantly different amounts of atherosclerosis, providing evidence that the impact of Id3 on atherogenesis is not mediated by alterations in lipid values (Supplementary Table IV). As en face analysis does not provide information on depth or composition of the lesion, cross-sectional lesion area analysis was also performed. Consistent with our en face findings, Id3-/- ApoE-/- mice were found to have significantly more cross-sectional lesion area than Id3+/+ApoE-/- mice in the proximal aorta after 16 weeks of Western diet (Figure 5A and 5B). While the overall size of the lesions was significantly greater in the Id3-/- ApoE-/- mice, the gross lesion composition was similar between groups (Figure 5C and D). The finding that the size of the lesion, not lesion characteristics, were increased in mice null for Id3 is consistent with our array findings that Id3 is an upstream regulator of multiple processes and pathways involved in atherogenesis and consistent with the finding that a single SNP in a single gene could be associated with IMT in humans.
These data are the first to demonstrate that loss of Id3 increases atherosclerosis in mice. Moreover, results provide the first report of a SNP in the ID3 gene associated with IMT in humans that leads to the expression of an Id3 protein with attenuated function. Taken together, results underscore the importance of identifying pathway regulators like Id3 and their partner proteins that may be important therapeutic targets in the prevention of atherogenesis.
Novelty and Significance.
What is known?
1p34-36, a susceptibility locus for premature myocardial infarction in humans, and Athsq1, a murine atherosclerosis susceptibility locus, both contain the gene encoding the helix-loop-helix factor Id3.
Id3 has been shown in vitro to regulate many pathways in many cell types implicated in atherogenesis, but the impact of modulating the expression or function of Id3 on atherogenesis in vivo is unknown.
What new information does this article contribute?
The present study is the first to identify a single nucleotide polymorphism in the ID3 gene (at rs11574) that is associated with carotid intima-media thickness (a surrogate marker of atherosclerotic plaque burden and clinical cardiovascular events) in humans.
The Id3 protein encoded by the minor (disease-associated) allele has attenuated function as an inhibitor of gene expression.
Consistent with these findings, ApoE-/- mice null for Id3 have a significant increase in atherosclerosis.
Genomics has emerged as a powerful method with the potential to identify gene loci and gene variants (such as single nucleotide polymorphisms or SNPs) that may provide novel pathophysiological insights and serve as useful biomarkers of complex diseases. Recent studies have identified gene loci and SNPs associated with atherosclerosis in humans, yet little is known about the impact of these genes on atherosclerosis or if these SNPs alter function of the protein encoded by the gene variant. The present study is the first to report a SNP in the ID3 gene (at rs11574) associated with carotid intima-media thickness (IMT) in humans. Moreover, our results demonstrate that the protein encoded by this disease-associated ID3 variant has significantly attenuated function as an inhibitor of gene activation. Loss of Id3 resulted in modulation of many genes in pathways involved in atherogenesis, highlighting the potential magnitude of the impact of altered Id3 function. Consistent with these findings, Id3 gene deletion in ApoE-/- mice resulted in a marked increase in atherosclerosis formation. Taken together, these results provide evidence for a pathophysiological role of attenuated Id3 function in promoting atherogenesis and suggest that ID3 polymorphism at rs11574 may be a biomarker of atherosclerotic burden in humans.
Supplementary Material
Acknowledgments
The authors wish to thank Hong Pei and Yongde Bao for excellent technical assistance.
Sources of Funding: This work was supported by NIH RO1 HL62522, NIH P01 HL55798 (C.A.M.), NIH 5-T32 HL007355-29 (M.J.L.), NIH F32 HL085989 (A.B.L.), an American Heart Association Pre-Doctoral Fellowship (A.C.D.), M01 RR07122 (WFU GCRC), and NIH R01 HL67348 (D.W.B.).
Non Standard Abbreviations and Acronyms
- ApoE
Apolipoprotein E
- bHLH
Basic helix-loop-helix
- CAT
Chloramphenicol acetyltransferase
- Co-IP
Co-immunoprecipitation
- CVD
Cardiovascular disease
- DHS
Diabetes Heart Study
- Id3
Inhibitor of Differentiation-3
- IMT
Intima-media thickness
- LDLR
LDL receptor
- MI
Myocardial infarction
- oxLDL
Oxidized LDL
- QTL
Quantitative trait locus
- PREST
Pedigree Relationship Statistical Test
- SmaA
Smooth muscle alpha actin
- T2D
Type 2 diabetes
- tagSNP
Tagging SNPs
- VSMC
Vascular smooth muscle cell
Footnotes
Disclosures: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geng YJ, Libby P. Progression of atheroma: a struggle between death and procreation. Arteriosclerosis, thrombosis, and vascular biology. 2002;22:1370–1380. doi: 10.1161/01.atv.0000031341.84618.a4. [DOI] [PubMed] [Google Scholar]
- 3.Stary HC. Natural history and histological classification of atherosclerotic lesions: an update. Arteriosclerosis, thrombosis, and vascular biology. 2000;20:1177–1178. doi: 10.1161/01.atv.20.5.1177. [DOI] [PubMed] [Google Scholar]
- 4.Berenson GS, Srinivasan SR, Bao W, Newman WP, 3rd, Tracy RE, Wattigney WA. Association between multiple cardiovascular risk factors and atherosclerosis in children and young adults. The Bogalusa Heart Study. N Engl J Med. 1998;338:1650–1656. doi: 10.1056/NEJM199806043382302. [DOI] [PubMed] [Google Scholar]
- 5.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Rao S, Shen GQ, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, Topol EJ. Premature myocardial infarction novel susceptibility locus on chromosome 1P34-36 identified by genomewide linkage analysis. Am J Hum Genet. 2004;74:262–271. doi: 10.1086/381560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, Liang CP, Barlow C, Dansky H, Breslow JL, Tall AR. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc Natl Acad Sci U S A. 2001;98:7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doran AC, Meller N, Cutchins A, Deliri H, Slayton RP, Oldham SN, Kim JB, Keller SR, McNamara CA. The helix-loop-helix factors Id3 and E47 are novel regulators of adiponectin. Circulation research. 2008;103:624–634. doi: 10.1161/CIRCRESAHA.108.175893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar MS, Hendrix JA, Johnson AD, Owens GK. Smooth muscle alpha-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circulation research. 2003;92:840–847. doi: 10.1161/01.RES.0000069031.55281.7C. [DOI] [PubMed] [Google Scholar]
- 10.Murre C. Helix-loop-helix proteins and lymphocyte development. Nature immunology. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 11.Forrest ST, Barringhaus KG, Perlegas D, Hammarskjold ML, McNamara CA. Intron retention generates a novel Id3 isoform that inhibits vascular lesion formation. The Journal of biological chemistry. 2004;279:32897–32903. doi: 10.1074/jbc.M404882200. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura ME, Li F, Berthoux L, Wei B, Lobe DR, Jeon C, Hammarskjold ML, McNamara CA. Vascular injury induces posttranscriptional regulation of the Id3 gene: cloning of a novel Id3 isoform expressed during vascular lesion formation in rat and human atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2001;21:752–758. doi: 10.1161/01.atv.21.5.752. [DOI] [PubMed] [Google Scholar]
- 13.Taylor AM, Li F, Thimmalapura P, Gerrity RG, Sarembock IJ, Forrest S, Rutherford S, McNamara CA. Hyperlipemia and oxidation of LDL induce vascular smooth muscle cell growth: an effect mediated by the HLH factor Id3. Journal of vascular research. 2006;43:123–130. doi: 10.1159/000090131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura ME, Lobe DR, McNamara CA. Contribution of the helix-loop-helix factor Id2 to regulation of vascular smooth muscle cell proliferation. The Journal of biological chemistry. 2002;277:7293–7297. doi: 10.1074/jbc.M108986200. [DOI] [PubMed] [Google Scholar]
- 15.Mueller C, Baudler S, Welzel H, Bohm M, Nickenig G. Identification of a novel redox-sensitive gene, Id3, which mediates angiotensin II-induced cell growth. Circulation. 2002;105:2423–2428. doi: 10.1161/01.cir.0000016047.19488.91. [DOI] [PubMed] [Google Scholar]
- 16.Nickenig G, Baudler S, Muller C, Werner C, Werner N, Welzel H, Strehlow K, Bohm M. Redox-sensitive vascular smooth muscle cell proliferation is mediated by GKLF and Id3 in vitro and in vivo. FASEB J. 2002;16:1077–1086. doi: 10.1096/fj.01-0570com. [DOI] [PubMed] [Google Scholar]
- 17.Kuhel DG, Zhu B, Witte DP, Hui DY. Distinction in genetic determinants for injury-induced neointimal hyperplasia and diet-induced atherosclerosis in inbred mice. Arteriosclerosis, thrombosis, and vascular biology. 2002;22:955–960. doi: 10.1161/01.atv.0000017994.77066.75. [DOI] [PubMed] [Google Scholar]
- 18.Orford JL, Selwyn AP, Ganz P, Popma JJ, Rogers C. The comparative pathobiology of atherosclerosis and restenosis. The American journal of cardiology. 2000;86:6H–11H. doi: 10.1016/s0002-9149(00)01094-8. [DOI] [PubMed] [Google Scholar]
- 19.Bowden DW, Rudock M, Ziegler J, Lehtinen AB, Xu J, Wagenknecht LE, Herrington D, Rich SS, Freedman BI, Carr JJ, Langefeld CD. Coincident linkage of type 2 diabetes, metabolic syndrome, and measures of cardiovascular disease in a genome scan of the diabetes heart study. Diabetes. 2006;55:1985–1994. doi: 10.2337/db06-0003. [DOI] [PubMed] [Google Scholar]
- 20.Lange LA, Bowden DW, Langefeld CD, Wagenknecht LE, Carr JJ, Rich SS, Riley WA, Freedman BI. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke; a journal of cerebral circulation. 2002;33:1876–1881. doi: 10.1161/01.str.0000019909.71547.aa. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Chen B, Han BH, Sun XH, Lim RW. Inhibition of muscle-specific gene expression by Id3: requirement of the C-terminal region of the protein for stable expression and function. Nucleic Acids Res. 1997;25:423–430. doi: 10.1093/nar/25.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung F, Johnson AD, Kumar MS, Wei B, Hautmann M, Owens GK, McNamara C. Characterization of an E-box-dependent cis element in the smooth muscle alpha-actin promoter. Arteriosclerosis, thrombosis, and vascular biology. 1999;19:2591–2599. doi: 10.1161/01.atv.19.11.2591. [DOI] [PubMed] [Google Scholar]
- 24.O'Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK., Jr Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. Cardiovascular Health Study Collaborative Research Group. N Engl J Med. 1999;340:14–22. doi: 10.1056/NEJM199901073400103. [DOI] [PubMed] [Google Scholar]
- 25.Persson J, Stavenow L, Wikstrand J, Israelsson B, Formgren J, Berglund G. Noninvasive quantification of atherosclerotic lesions. Reproducibility of ultrasonographic measurement of arterial wall thickness and plaque size. Arterioscler Thromb. 1992;12:261–266. doi: 10.1161/01.atv.12.2.261. [DOI] [PubMed] [Google Scholar]
- 26.Hodis HN, Mack WJ, LaBree L, Selzer RH, Liu CR, Liu CH, Azen SP. The role of carotid arterial intima-media thickness in predicting clinical coronary events. Annals of internal medicine. 1998;128:262–269. doi: 10.7326/0003-4819-128-4-199802150-00002. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz MW, von Kegler S, Steinmetz H, Markus HS, Sitzer M. Carotid intima-media thickening indicates a higher vascular risk across a wide age range: prospective data from the Carotid Atherosclerosis Progression Study (CAPS) Stroke; a journal of cerebral circulation. 2006;37:87–92. doi: 10.1161/01.STR.0000196964.24024.ea. [DOI] [PubMed] [Google Scholar]
- 28.Deed RW, Jasiok M, Norton JD. Attenuated function of a variant form of the helix-loop-helix protein, Id-3, generated by an alternative splicing mechanism. FEBS Lett. 1996;393:113–116. doi: 10.1016/0014-5793(96)00868-x. [DOI] [PubMed] [Google Scholar]
- 29.Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 30.Szalai AJ, Wu J, Lange EM, McCrory MA, Langefeld CD, Williams A, Zakharkin SO, George V, Allison DB, Cooper GS, Xie F, Fan Z, Edberg JC, Kimberly RP. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J Mol Med. 2005;83:440–447. doi: 10.1007/s00109-005-0658-0. [DOI] [PubMed] [Google Scholar]
- 31.Salero E, Gimenez C, Zafra F. Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. Biochem J. 2003;370:979–986. doi: 10.1042/BJ20021142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrest ST, Taylor AM, Sarembock IJ, Perlegas D, McNamara CA. Phosphorylation regulates Id3 function in vascular smooth muscle cells. Circulation research. 2004;95:557–559. doi: 10.1161/01.RES.0000142735.67542.5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Providence KM, White LA, Tang J, Gonclaves J, Staiano-Coico L, Higgins PJ. Epithelial monolayer wounding stimulates binding of USF-1 to an E-box motif in the plasminogen activator inhibitor type 1 gene. J Cell Sci. 2002;115:3767–3777. doi: 10.1242/jcs.00051. [DOI] [PubMed] [Google Scholar]
- 35.Moldes M, Boizard M, Liepvre XL, Feve B, Dugail I, Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem J. 1999;344(Pt 3):873–880. [PMC free article] [PubMed] [Google Scholar]
- 36.Towle HC, Kaytor EN, Shih HM. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 37.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 38.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 40.Sugai M, Gonda H, Nambu Y, Yokota Y, Shimizu A. Role of Id proteins in B lymphocyte activation: new insights from knockout mouse studies. J Mol Med. 2004;82:592–599. doi: 10.1007/s00109-004-0562-z. [DOI] [PubMed] [Google Scholar]
- 41.Benezra R, Rafii S, Lyden D. The Id proteins and angiogenesis. Oncogene. 2001;20:8334–8341. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.