Abstract
JAK2 V617F is the most frequently found somatic mutation in polycythemia vera (PV). Among the cases negative for V617F, a significant fraction have a mutation in exon 12 of the JAK2 gene. Several groups have reported that the exon 12 mutations are present in only a small fraction of the blood cells in some patients. We have developed an assay to detect these mutations with an analytical sensitivity of 0.1% by using a “PCR clamp” to inhibit amplification of the normal sequence and enhance amplification of DNA containing a mutation in the clamp target sequence. The products of this reaction were analyzed by capillary electrophoresis to detect deletions, which are the most frequent type of exon 12 mutations, or by nucleotide sequencing to detect all of the mutations. In a survey of 34 specimens from patients with PV or idiopathic erythrocytosis who did not have a JAK2 V617F mutation, we found four with a mutation in exon 12, 3 of 10 with PV, and 1 of 24 with idiopathic erythrocytosis. In two cases the mutation was present in a small fraction of the cells and difficult to detect without the use of the clamp. The use of an assay with increased analytical sensitivity enhances the ability to identify patients with mutations in exon 12 of the JAK2 gene.
The discovery of the JAK2 V617F mutation has made a significant contribution to understanding the pathogenesis of myeloproliferative neoplasms and also eased the diagnosis of this group of diseases.1,2,3,4,5 JAK2 V617F is found in the myeloid lineage in more than 95% of patients with polycythemia vera (PV) as well as in about 50% of patients with essential thrombocytosis and myelofibrosis.6 Among the cases of PV negative for the V617F mutation, a mutation in exon 12 is found in a significant number of cases.7 JAK2 exon 12 mutations are included as a major criterion for diagnosis of PV in the most recent World Health Organization guidelines.8
More than 10 different sequence variations have been found in exon 12 of the JAK2 gene, most of which are in the region between codons 536 and 544.7,9,10,11,12,13,14,15,16,17,18 Many of these involve a deletion of three to six nucleotides, but some are 2-bp replacement mutations. Four duplications have also been reported.12,16,18,19 Several groups have reported that the exon 12 mutations are frequently present in only a small fraction of the granulocytes in some patients.7,11,14,17,20 Thus, the molecular diagnosis of mutations in exon 12 of the JAK2 gene is complicated by heterogeneity of the mutations and low allelic load. Our aim in this study was to establish an assay to detect JAK2 exon 12 mutations with a high degree of analytical sensitivity.
Materials and Methods
Patients
Thirty-four patients with JAK2 V617F-negative PV or idiopathic erythrocytosis referred to the Johns Hopkins Center for Chronic Myeloproliferative Disorders were studied, nine of whom were seen before May 2007 and reported previously.13,21 The remaining 25 patients were evaluated between December 2007 and September 2008 and were not previously reported. The diagnosis of PV was based on Polycythemia Vera Study Group criteria, which required an increased red blood cell mass, splenomegaly, and a normal oxygen saturation; thrombocytosis or leukocytosis also supported the diagnosis.22 A diagnosis of idiopathic erythrocytosis was supported by the presence of an increased red blood cell mass, a low or normal erythropoietin level, and the absence of a defining stimulus. All patients gave written informed consent by using a form approved by the Johns Hopkins Institutional Review Board.
DNA Preparation
DNA was extracted from density gradient purified granulocytes from EDTA-anticoagulated peripheral blood by using the Puregene Cell kit (Gentra Systems, Minneapolis, MN). The concentration of DNA was determined by specrophotometry using the Nanodrop ND-1000 spectrophotometer (Wilmington, DE).
PCR
The PCR was done in a volume of 10 μl with primers PCV-7 (5′-TGGTGTTTCTGATGTACCAACCTCAC-3′) and PCV-8 (5′-TCAAGAAAACAGATGTTGTTTTAAAAGGACAA-3′) and the clamp at a final concentration of 1 μmol/L each, 50 μmol/L of each deoxyribonucleotide triphosphate (dNTP), 1 mmol/L MgSO4, 1.25 U of Platinum Pfx DNA polymerase, and 2.5 μl of the 10X buffer provided by the enzyme manufacturer (Invitrogen, Carlsbad, CA). Template was added in a volume of 1 μl containing 50 to 100 ng of DNA. The sequence of the clamp was 5′-+T+C+A+C+A+A+AAT+C+A+GAA+A+T+G+AAG-3′, with the locked nucleic acid (LNA) positions preceded by a +. The clamp also had a 3′-terminal C3 linker to block extension during the PCR. The reaction was cycled 40 times between 94°C for 5 seconds, and 65°C for 45 seconds, preceded by 5 minutes at 94°C, and followed by 5 minutes at 68°C. When the reaction was followed by fragment analysis, the upstream primer was labeled with a 5′-fluorescein dye (6-FAM). The primers were purchased from Integrated DNA Technologies (Coralville, IA), and the LNA clamp was purchased from Sigma-Proligo (Woodlands, TX).
DNA Sequencing
When the PCR was done for preparation of template for sequencing, the primers were not labeled with a fluorescent dye. The amplicons were treated with ExoSap (Amersham Biosciences, Piscataway, NJ) to remove the primers and dNTPs, then 1 μl was sequenced by using the PCR primers as sequencing primers and Applied Biosystems (ABI, Foster City, CA) BigDye Terminator version 3.1 chemistry. The sequencing reactions were purified by using the CleanSeq system (Agencourt Bioscience, Beverly, MA) and then resolved by capillary electrophoresis on the ABI 3100 Prism Genetic Analyzer. The mutations were determined by comparison with the normal JAK2 sequence (accession NM-004972) and a normal control that was included in each run.
Fragment Analysis
After PCR the products were diluted 1:100 in water, then 1 μl was mixed with 11 μl ABI Hi-Di Formamide and size markers (ABI GeneScan 350 ROX), and resolved by capillary electrophoresis on an ABI Prism 3100 Genetic Analyzer. The results were evaluated and printed by using the computer program GeneMapper version 4.0 (ABI).
Results
Our aim was to design an assay to detect mutations in exon 12 of the JAK2 gene with a high degree of analytical sensitivity because previous work revealed several patients with a mutation in only a small fraction of the tested DNA.7,11,14,17,20 We based our assay on PCR followed by nucleotide sequencing because a fairly large number of different mutations have been found in this region, and an assay designed to detect only one mutation, such as JAK2 V617F, would not be adequate. To increase the analytical sensitivity of the assay, we used a “clamp” to suppress amplification of the normal sequence. Most of the mutations in exon 12 are deletions of three to six nucleotides. For this reason we were able to use fragment length analysis of a PCR product generated with a fluorescently labeled primer to speed up the optimization of the assay. The assay was optimized for thermostable polymerase, the concentration of the buffer concentrate, magnesium ion and clamp, and the annealing/extension temperature. The most serious problem during the development of this test was the finding of spurious mutations that appeared to result from preferential amplification of polymerase errors (data not shown). This was remedied by the use of a proofreading polymerase.
Figure 1 shows the effect of the clamp on the relative signal from normal and mutant DNA using the fragment analysis assay on a DNA specimen with a c.1624-1629delAATGAA (N542-E543del) mutation. The undiluted patient specimen DNA had a mutant signal a bit less abundant than the normal signal as seen in the trace in the upper left of Figure 1. Likely this shows that the mutation is heterozygous and the population of cells with the mutation constitutes most but not all of the granulocytes. When the PCR was done using the optimized assay, the LNA clamp suppressed the signal from the normal allele and only the mutation signal could be seen (Figure 1, upper right panel). Using this optimized assay, we were able to detect the mutant signal when the mutant genomic DNA was diluted down to 0.1% in normal DNA, whereas without the clamp the mutation was difficult to recognize at a 1:10 dilution (Figure 1). The effect of the LNA clamp was also evident in sequence analysis where only the mutant sequence can be seen when the genomic template was undiluted patient specimen DNA (Figure 2, top panel). The lack of overlapping normal sequence eases the interpretation of the sequence data, even when the mutant genomic template was diluted 10-fold in normal DNA (Figure 2, 10% mutant sequence electropherogram). The mutation is even visible in a 1000-fold dilution of mutant DNA in normal DNA, although it is not easy to read at that dilution (Figure 2, 0.1% mutant). Thus, the assay has a sensitivity of about 0.1%. The c.1611-1616delTCACAA mutation was also detectable at 1:1000 dilution of the patient specimen mutant DNA in normal DNA (data not shown).
Figure 1.

Fragment analysis showing the effect of the LNA clamp on the relative level of signal from normal DNA and a 6-bp deletion mutant. The genomic DNA was heterozygous for a 6-bp deletion, N542-E543del. As indicated on the left of the figure (% Mutant) serial 10-fold dilutions of mutant DNA in normal DNA (no JAK2 exon 12 mutation) were prepared and used for template. The PCR was done with and without the LNA clamp as described at the top of each column of fragment analysis results. At the bottom of the figure, the positions of the fragments corresponding to the normal (WT) and mutant (Del) are indicated. In each graph, the y axis represents relative fluorescence units and the x axis represents molecular weight.
Figure 2.
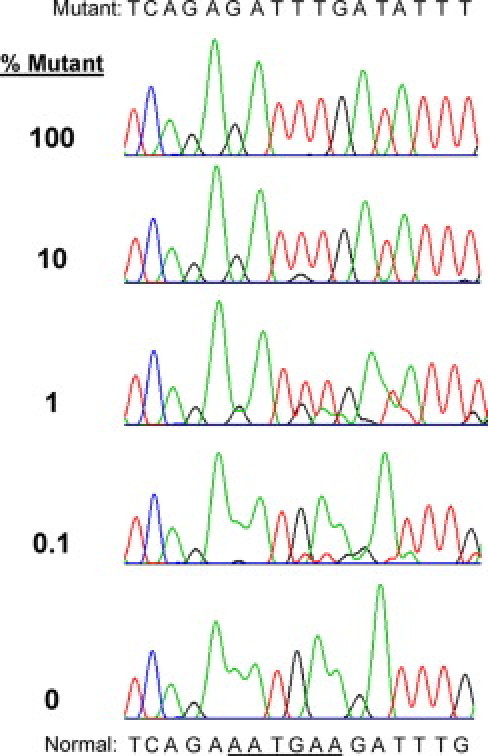
Sequence analysis showing the effect of the LNA clamp on the relative level of signal from normal DNA and a 6-bp deletion mutant. The genomic DNAs were the same serially diluted template DNAs that were used in Figure 1. The genomic DNA mixtures were amplified with the LNA clamp and subjected to dideoxy nucleotide sequencing. The sequence of the mutant DNA is written at the top of the figure, and the normal sequence is written at the bottom with the six nucleotides deleted in the mutant underlined. The percentage of mutant DNA specimen in the template is indicated to the left of the sequence traces. The genomic DNA used for the sequence electropherogram at the bottom was pure normal.
Using this assay, we surveyed a group of DNAs from purified granulocytes from 10 patients with PV and 24 with idiopathic erythrocytosis, all of whom lacked the JAK2 V617F mutation. Three of the patients with PV had mutations, including c.1611-1616delTCACAA (F537-K539delinsL), c.1622-1627delGAAATG (R541-E543delinsK), and c1627-1632delGAAGAT (E543-D544del), and one of the patients with idiopathic erythrocytosis had c.1624-1629delAATGAA (N542-E543del). All four mutations have been reported previously. The E543-D544del mutation was clearly demonstrated with the assay enhanced with the clamp using both fragment analysis (Figure 3A) and sequencing (Figure 3B). Without the clamp the signal from the mutant DNA was very low and hard to distinguish from the normal, although visible. With the clamp the mutation is obvious in both assays and easy to decipher by nucleotide sequencing (Figure 3).
Figure 3.

Analysis of an E543–D544del mutation present at low allelic load using fragment analysis (A) and sequence analysis (B). The top panels in both parts of the figure show a normal control. The middle and bottom panels used the patient specimen genomic DNA with the PCR done with and without the LNA clamp as indicated. In A the positions of the deletion mutant and normal fragments are indicated at the top. In each graph, the y axis represents relative fluorescence units and the x axis molecular weight. In B the normal sequence is written at the top of the figure with the six nucleotides deleted in the mutant underlined, and the mutant sequence is written between the lower two sequence electropherograms.
Discussion
In a recent review, Patnaik and Tefferi23 recommend testing for mutations in exon 12 of the JAK2 gene in patients with erythrocytosis and low erythropoietin who are negative for the V617F mutation. Nucleotide sequencing is a good way to accomplish this considering the wide variety of mutations that have been discovered in JAK2 exon 12.15 However, several authors have reported patients with a mutation in a small fraction of the isolated DNA, which may not be detected by using standard dideoxy nucleotide sequencing.7,14,17,20 Our approach was to use an oligo clamp to suppress amplification of the normal sequence and enhance amplification of mutant DNA. This approach has been called clamped PCR or wild-type blocking PCR.24,25 We previously used such a strategy to improve the analytical sensitivity of an assay for NPM1 mutations in acute myeloid leukemia.26 For the JAK2 exon 12 mutations, we aimed for and achieved a higher level of analytical sensitivity mostly by increasing the concentration of the clamp. Using the clamp at a concentration of 1 μmol/L, we were able to detect the JAK2 exon 12 mutations N542-E543del and F537-K539delinsL when the heterozygous patient specimen DNA was diluted in a 1000-fold excess of normal DNA (Figures 1 and 2; data not shown). It is likely that the analytical sensitivity for detection of all of the deletion mutations, which account for about 86% of the reported exon 12 mutations,15 will be similar. We have not tested the analytical sensitivity of this method for the K539L mutation, which is caused by two A>T substitutions and accounts for about 8% of the exon 12 mutations in the JAK2 gene.15 Clamp-based methods have been used to achieve a 0.1% or better analytical sensitivity for single bp substitutions,27 and it is likely that the JAK2 K539L mutation will be detectable with this assay with a similar level of sensitivity, although in the absence of evidence we cannot exclude the need for further optimization.
The LNA clamp hybridizes to a region that overlaps at least part of all of the JAK2 exon 12 mutations that have been described to date, except for a set of duplications that have been seen in four patients.12,16,19 The analytical sensitivity for detection of these duplications would not be enhanced over that of sequence analysis alone because the discontinuity in the nucleotide sequence is not in the clamp target region. However, these mutations are in the region sequenced and would be detected with the standard analytical sensitivity of nucleotide sequencing.
The main problem we encountered in developing this assay was the generation of pseudo-mutations that were likely due to the preferential amplification of polymerase errors that happened in early cycles (data not shown). This phenomenon was reported recently by Gilje et al27 in the context of a clamped PCR assay for KRAS mutations. We solved this problem, as suggested by Gilje et al,27 by using a high-fidelity thermal stable polymerase. We were successful using two different high-fidelity thermostable DNA polymerases, Phusion HS (New England Biolabs, Ipswich, MA), which was used by Gilje et al27, and Platinum Pfx (Invitrogen). The results for the latter enzyme are shown in this article.
Several assays have been published that can detect most or all JAK2 exon 12 mutations with an analytical sensitivity of 1 to 10%.17,19,21,28 Assays based on allele-specific PCR are much more sensitive, but are limited to detecting only known mutations and usually only one mutation per assay.7,14 Other methods to increase the analytical sensitivity of the assay include using DNA purified from plasma and using RNA as a starting material rather than DNA.18,29 Another approach to increased analytical sensitivity is to select cells for analysis that are more likely to harbor the mutation. Several of the reports of JAK2 exon 12 mutations that were present in only a small minority of blood cells studied DNA purified from endogenous erythroid colonies.7,20 Although analysis of endogenous erythroid colonies is not a common practice for detection of JAK2 mutations, many molecular diagnostic laboratories purify granulocytes before JAK2 mutation analysis. Interestingly, one case has been reported in which the JAK2 V617F mutation was present in the erythroid lineage and not in granulocytes.30 The use of a test with a high degree of analytical sensitivity may allow reliable detection of JAK2 exon 12 mutations in total blood cell DNA without enrichment of particular lineages or prior growth of endogenous erythroid colonies. Our assay, with an analytical sensitivity capable of detecting 0.1% cells with a heterozygous deletion mutation, is likely to be able to detect a JAK2 exon 12 mutation in patients that would be missed by many of the other assays.
However, the clinical significance of finding a mutation in such a small fraction of the cells is unknown. Two studies of randomly collected blood samples using very sensitive assays for the JAK2 V617F mutation detected individuals with a mutation that did not correlate with the clinical status.31,32 Similar to the situation with oncogenic fusion genes, there is likely to be a portion of the population that carries an oncogenic mutation in a small proportion of cells with unknown clinical significance.33,34,35 Thus, finding a mutation in a small proportion of cells must be interpreted with caution.
Note Added in Proof
Scott Reading, Ph.D. (ARUP Laboratories) presented a very similar strategy for detection of JAK2 exon 12 mutations at the 2009 Annual Meeting of the Association for Molecular Pathology.
Footnotes
Supported by grants from the National Institutes of HealthRO1HL082995 to A.R.M. and K12HL087169-0 to B.L.S.
References
- 1.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 2.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJP, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D'Andrea A, Fröhling S, Döhner K, Marynen P, Vandenberghe P, Mesa M, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Todd R, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 3.James C, Ugo V, Le Couédic J-P, Staerk J, Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kralovics R, Passamonti F, Buser AS, Teo S-S, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7:673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
- 7.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF, Green AR. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the WHO classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Avilés L, Besses C, Alvarez-Larrán A, Cervantes F, Hernández-Boluda JC, Bellosillo B. JAK2 exon 12 mutations in polycythemia vera or idiopathic erythrocytosis. Haematologica. 2007;92:1717–1718. doi: 10.3324/haematol.12011. [DOI] [PubMed] [Google Scholar]
- 10.Pardanani A, Lasho TL, Finke C, Hanson CA, Tefferi A. Prevalence and clinicopathologic correlates of JAK2 exon 12 mutations in JAK2V617F-negative polycythemia vera. Leukemia. 2007;21:1960–1963. doi: 10.1038/sj.leu.2404810. [DOI] [PubMed] [Google Scholar]
- 11.Percy MJ, Scott LM, Erber WN, Harrison CN, Reilly JT, Jones FGC, Green AR, McMullin MF. The frequency of JAK2 exon 12 mutations in idiopathic erythrocytosis patients with low serum erythropoietin levels. Haematologica. 2007;92:1607–1614. doi: 10.3324/haematol.11643. [DOI] [PubMed] [Google Scholar]
- 12.Albiero E, Madeo D, Ruggeri M, Bernardi M, Giorgetti A, Rodeghiero F. Loss of the JAK2 intramolecular auto-inhibition mechanism is predicted by structural modelling of a novel exon 12 insertion mutation in a case of idiopathic erythrocytosis. Br J Hematol. 2008;142:986–990. doi: 10.1111/j.1365-2141.2008.07180.x. [DOI] [PubMed] [Google Scholar]
- 13.Williams DM, Kim AH, Rogers O, Spivak JL, Moliterno AR. Phenotypic variations and new mutations in JAK2 V617F-negative polycythemia vera, erythrocytosis, and idiopathic myelofibrosis. Exp Hematol. 2007;35:1641–1646. doi: 10.1016/j.exphem.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouroupi E, Zoi K, Parquet N, Zoi C, Kiladjian JJ, Grigoraki V, Vainchenker W, Lellouche F, Marzac C, Schlageter MH, Dosquet C, Scott LM, Fenaux P, Loukopoulos D, Chomienne C, Cassinat B. Mutations in exon 12 of JAK2 are mainly found in JAK2 V617F-negative polycythaemia vera patients. Br J Hematol. 2008;142:676–679. doi: 10.1111/j.1365-2141.2008.07223.x. [DOI] [PubMed] [Google Scholar]
- 15.Bernardi M, Ruggeri M, Albiero E, Madeo D, Rodeghiero F. Isolated erythrocytosis in V617F negative patients with JAK2 exon 12 mutations: report of a new mutation. Am J Hematol. 2009;84:238–260. doi: 10.1002/ajh.21357. [DOI] [PubMed] [Google Scholar]
- 16.Pietra D, Li S, Brisci A, Passamonti F, Rumi E, Theocharides A, Ferrari M, Gisslinger H, Kralovics R, Cremonesi L, Skoda R, Cazzola M. Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)-negative myeloproliferative disorders. Blood. 2008;111:1686–1689. doi: 10.1182/blood-2007-07-101576. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Kralovics R, De Libero G, Theocharides A, Gisslinger H, Skoda RC. Clonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2-V617F mutations. Blood. 2008;111:3863–3866. doi: 10.1182/blood-2007-09-111971. [DOI] [PubMed] [Google Scholar]
- 18.Ma W, Kantarjian H, Zhang X, Yeh CH, Zhang ZJ, Verstovsek S, Albitar M. Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias. J Mol Diagn. 2009;11:49–53. doi: 10.2353/jmoldx.2009.080114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnittger S, Bacher U, Haferlach C, Geer T, Müller P, Mittermüller J, Petrides P, Schlag R, Sandner R, Selbach J, Slawik HR, Tessen HW, Wehmeyer J, Kern W, Haferlach T. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica. 2009;94:414–418. doi: 10.3324/haematol.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher CM, Hahn U, To LB, Gecz J, Wilkins EJ, Scott HS, Bardy PG, D'Andrea RJ. Two novel JAK2 exon 12 mutations in JAK2V617F-negative polycythaemia vera patients. Leukemia. 2007;22:870–873. doi: 10.1038/sj.leu.2404971. [DOI] [PubMed] [Google Scholar]
- 21.Jones AV, Cross NC, White HE, Green AR, Scott LM. Rapid identification of JAK2 exon 12 mutations using high resolution melting analysis. Haematologica. 2008;93:1560–1564. doi: 10.3324/haematol.12883. [DOI] [PubMed] [Google Scholar]
- 22.Berk PD, Goldberg JD, Donovan PB, Fruchtman SM, Berlin NI, Wasserman LR. Therapeutic recommendations in polycythemia vera based on Polycythemia Vera Study Group protocols. Semin Hematol. 1986;23:132–143. [PubMed] [Google Scholar]
- 23.Patnaik MM, Tefferi A. The complete evaluation of erythrocytosis: congenital and acquired. Leukemia. 2009;23:834–844. doi: 10.1038/leu.2009.54. [DOI] [PubMed] [Google Scholar]
- 24.Orum H, Nielsen PE, Egholm M, Berg RH, Buchardt O, Stanley C. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acid Res. 1993;21:5332–5336. doi: 10.1093/nar/21.23.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez PL, Kolodney MS. Wild-type blocking polymerase chain reaction for detection of single nucleotide minority mutations from clinical specimens. Oncogene. 2005;24:6830–6834. doi: 10.1038/sj.onc.1208832. [DOI] [PubMed] [Google Scholar]
- 26.Laughlin TS, Becker MW, Liesveld JL, Mulford DA, Abboud CN, Brown P, Rothberg PG. Rapid method for detection of mutations in the nucleophosmin gene in acute myeloid leukemia. J Mol Diagn. 2008;10:338–345. doi: 10.2353/jmoldx.2008.070175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilje B, Heikkilä R, Oltedal S, Tjensvoll K, Nordgård O. High-fidelity DNA polymerase enhances the sensitivity of a peptide nucleic acid clamp PCR assay for K-ras mutations. J Mol Diagn. 2008;10:325–331. doi: 10.2353/jmoldx.2008.070183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapado I, Grande S, Albizua E, Ayala R, Hernández JA, Gallardo M, Gilsanz F, Martinez-Lopez J. High resolution melting analysis for JAK2 Exon 14 and Exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J Mol Diagn. 2009;11:155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma W, Kantarjian H, Zhang X, Sun W, Buller AM, Jilani I, Schwartz JG, Giles F, Albitar M. Higher detection rate of JAK2 mutation using plasma. Blood. 2008;111:3906–3907. doi: 10.1182/blood-2008-02-139188. [DOI] [PubMed] [Google Scholar]
- 30.Zehentner BK, Loken MR, Wells DA. JAK2V617F mutation can occur exclusively in the erythroid lineage and be absent in granulocytes and progenitor cells in classic myeloproliferative disorders. Am J Hematol. 2006;81:806–807. doi: 10.1002/ajh.20663. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, Zhang Q, Luo J, Xing S, Li Q, Krantz SB, Fu X, Zhao Z. JAK2(V617F): prevalence in a large Chinese hospital population. Blood. 2007;109:339–342. doi: 10.1182/blood-2006-03-009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidon P, El Housni H, Dessars B, Heimann P. The JAK2V617F mutation is detectable at very low level in peripheral blood of healthy donors. Leukemia. 2006;20:1622. doi: 10.1038/sj.leu.2404292. [DOI] [PubMed] [Google Scholar]
- 33.Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, Hows JM, Navarrete C, Greaves M. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 35.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]


