Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency, an X-linked inherited disease, is one of the most common enzymopathies and affects over 400 million people worldwide. In China at least 21 distinct point mutations have been identified so far. In this study high-resolution melting (HRM) analysis was used to screen for G6PD mutations in 260 unrelated Han Chinese individuals, and the rapidity and reliability of this method was investigated. The mutants were readily differentiated by using HRM analysis, which produced distinct melting curves for each tested mutation. Interestingly, G1388A and G1376T, the two most common variants accounting for 50% to 60% of G6PD deficiency mutations in the Chinese population, could be differentiated in a single reaction. Further, two G6PD mutations not previously reported in the Chinese population were identified in this study. One of these mutations, designated “G6PD Jiangxi G1340T,” involved a G1340T substitution in exon 11, predicting a Gly447Val change in the protein. The other mutation involved a C406T substitution in exon 5. The frequencies of the common polymorphism site C1311T/IVS (intervening sequence) XI t93c between patients with G6PD and healthy volunteers were not significantly different. Thus, HRM analysis will be a useful alternative for screening G6PD mutations.
Glucose-6-phosphate dehydrogenase (G6PD) is a key enzyme in the hexose monophosphate pathway, a unique NADPH-generating process in mature red blood cells.1,2 The coenzyme NADPH is essential for protection against and repair of oxidative damage. G6PD deficiency, an X-linked inherited disease, is one of the most common enzymopathies that affects over 400 million people worldwide.3 To date, more than 150 different G6PD mutations have been identified among different ethnic populations,4 and each ethnic population has a characteristic mutation profile. At least 21 distinct G6PD point mutations have been reported in China, and more than 90% of those are A95G, G392T, G487A, A493G, C592T, C1024T, C1360T, G1376T, or G1388A, spanning exons 2, 5, 6, 9, 11, and 12 of G6PD.5,6,7,8,9,10,11
Various methods have been developed to detect G6PD mutations; these include PCR-single strand conformational polymorphism analysis, DNA sequencing, amplification refractory mutation system, and denaturing high-performance liquid chromatography. However, these approaches are either expensive or technically challenging. Because various mutations may lead to different clinical presentations,5 a reliable and rapid genotyping method is necessary to assist both physicians and patients.
High-resolution melting (HRM) analysis is a new and rapid method for mutation scanning in which PCR and mutation scanning are performed simultaneously in a single procedure within 30 minutes.12 Sensitivity and specificity for mutation detection are extremely high, and this technique also has the advantages of cost and throughput.13 Recently, a growing number of potential causative mutations for different diseases have been measured by HRM analysis, which can identify hundreds of mutations in many different genes. The detection of human genetic diseases, specifically those that are autosomal dominant, recessive, or X chromosome-linked, has been one of the largest applications of HRM analysis. In addition, identification of somatic mutations acquired by human tumors has also been examined.13,14,15 For example, JAK2 exon 14 and exon 12 mutations in Philadelphia chromosome–negative myeloproliferative neoplasms were assessed, and HRM analyses of JAK2 exons 12 and 14 produced analytical sensitivities near 1%. The JAK2 exon 12 HRM results correlated well with those from sequencing analysis.14 Here an HRM assay for detecting G6PD mutations was developed, and the characteristic profile of G6PD-deficient variants in Han Chinese individuals was analyzed.
Materials and Methods
Samples
A total of 260 unrelated Han Chinese individuals with G6PD deficiency and their parents from 137 families were collected from Shanghai Newborn Screening Center in Shanghai Children's Hospital. Informed consent was obtained from each participant, and the study was approved by the ethics committee of Shanghai Children's Hospital. Normal peripheral blood samples were obtained from 57 healthy individuals as controls, and G6PD mutation status was determined. The enzyme activity of these individuals was measured by using the method recommended by the World Health Organization for measurement of the G6PD/6-phosphogluconate dehydrogenase (6PGD) ratio.16 Genomic DNA was extracted from peripheral blood by using the phenol/chloroform method.
Primer Design
Twelve PCR primer pairs that did not overlap with known single nucleotide polymorphisms were designed to detect known point mutations in G6PD. The primers are listed in Table 1.
Table 1.
Primers and Conditions for Screening the G6PD-Deficient Variants by HRM Analysis
| Mutation | Location | Primer name | Primer type | Sequence | Product length, bp |
|---|---|---|---|---|---|
| A95G | Exon 2 | 1A | Sense | 5′-GGCGATGCCTTCCATCAGTC-3′ | 109 |
| 1B | Antisense | 5′-AGGCATGGAGCAGGCACTTC-3′ | |||
| C274T | Exon 5 | 2A | Sense | 5′-TGTGTCTGTCTGTCCGTGTCTC-3′ | 108 |
| 2B | Antisense | 5′-AGGCTGCATCATCGTACTGG-3′ | |||
| G392T | Exon 5 | 3A | Sense | 5′-TACCAGCGCCTCAACAGC-3′ | 77 |
| C406T | 3B | Antisense | 5′-GGCAAGGCCAGGTAGAAGA-3′ | ||
| G487A | Exon 6 | 4A | Sense | 5′-GCAGCTCTGATCCTCACTCC-3′ | 137 |
| A493G | |||||
| T517C | 4B | Antisense | 5′-GGTTGGACAGCCGGTCA-3′ | ||
| C519G | |||||
| C592T | Exon 6 | 5A | Sense | 5′-TGTTCCGTGAGGACCAGATCTA-3′ | 64 |
| 5B | Antisense | 5′-AGGTTCTGCACCATCTCCTTG-3′ | |||
| A835G | Exon 8 | 6A | Sense | 5′-CGTGATGCAGAACCACCTACT-3′ | 92 |
| A835T | 6B | Antisense | 5′-CCTTCTCATCACGGACGTCA-3′ | ||
| G871A | Exon 9 | 7A | Sense | 5′-CCCAACTCAACACCCAAGGA-3′ | 86 |
| 7B | Antisense | 5′-TGGCCTGCACCTCTGAGAT-3′ | |||
| C1004T | Exon 9 | 8A | Sense | 5′-CCAAAGGGTACCTGGACGAC-3′ | 86 |
| 8B | Antisense | 5′-CTCATTCTCCACATAGAGGACGAC-3′ | |||
| C1024T | Exon 9 | 9A | Sense | 5′-CACTTTTGCAGCCGTCGT-3′ | 65 |
| 9B | Antisense | 5′-CTCGAAGGCATCACCTACCA-3′ | |||
| C1311T | Exon 11 | 10A | Sense | 5′-AGGCAGTGGCATCAGCAAG-3′ | 88 |
| 10B | Antisense | 5′-GCAGAAGACGTCCAGGATGAG-3′ | |||
| G1340T | Exon 11 | 11A | Sense | 5′-GCCTCATCCTGGACGTCTTC-3′ | 92 |
| G1360T | 11B | Antisense | 5′-CCCATAGCCCACAGGTATGC-3′ | ||
| t93c | Intron 11 | 12A | Sense | 5′-GCCCTCCCTCCCTGTGTG-3′ | 111 |
| G1376T | Exon 12 | ||||
| G1381A | Exon 12 | 12B | Antisense | 5′CAGCTCAATCTGGTGCAGCAGT-3′ | |
| G1388A | Exon 12 |
Note: Seven pairs of primers (1A/1B, 2A/2B, 5A/5B, 7A/7B, 8A/8B, 9A/9B, and 10A/10B) were individually used to amplify seven fragments containing the corresponding mutant sites (A95G, C274T, C592T, G871A, C1004T, C1024T, and C1311T), whereas the other five pairs of primers (3A/3B, 4A/4B, 6A/6B, 11A/11B, and 12A/12B) were applied to detect several mutations in one fragment.
PCR and HRM Assay Conditions
PCR reactions were performed on the Rapid Cycler (Idaho Technology, Salt Lake City, UT). Approximately 50 ng of DNA was amplified in a total volume of 10 μl containing 200 nmol/L of each primer, 2 mmol/L MgCl2, 0.01% bovine serum albumin, and 1 μl LCGreen I dye (Idaho Technology). The reaction conditions were 94°C for 15 seconds, followed by 32 cycles of 94°C for 5 seconds, 63°C for 5 seconds, and 72°C for 15 seconds, with final extension at 72°C for 90 seconds. The cycling conditions were the same for both amplicons.
HRM analysis was performed over the range from 75°C to 95°C rising at 0.3°C per second with 100 acquisitions per degree. The melting curves were normalized for direct comparison among samples. Each of the genotypes detected by HRM was validated by DNA sequencing.
For comparison, three common G6PD mutations (A95G, G1376T, and G1388A) in the Han Chinese population were also detected by amplification refractory mutation system. Three pairs of allele-specific forward primers and three corresponding reverse primers were used as described.17 DNA samples were amplified simultaneously in two parallel reactions with each primer set to detect the mutant allele and the normal allele. The amplification products were detected by agarose gel electrophoresis.
Statistical Analysis
Frequencies of the common polymorphism C1311T/IVS (intervening sequence) XI t93c were compared between individuals with G6PD deficiency and healthy controls by using the χ2 test. Statistical analyses were performed by using the Statistical Package for the Social Sciences version 13.0. For all statistical tests, a P value less than or equal to 0.05 was considered statistically significant.
Results
Point Mutations Can be Identified Accurately and Rapidly by HRM Analysis
To identify the common point mutations in Han Chinese individuals by HRM, 12 pairs of PCR primers were designed (Table 1). Rapid-cycle PCR allowed amplification and screening of common point mutations in a closed-tube system within 20 minutes. The normalized melting curves of six G6PD mutations or polymorphisms are shown in Figure 1. The slope of the melting curve of heterozygous samples was markedly reduced, and heterozygotes could be easily distinguished from nonmutant samples based on differences in the melting curve shape. Sometimes, the melting characteristics of hemizygotes were identical to those obtained for wild-type samples, whereas the melting curves generated from A95G hemizygotes were shifted to the right as compared with those for wild-type samples (Figure 1A). In contrast, the melting curves from other hemizygous mutants, including A835T, G871A, C1004T, C1024T, and C1311T, were shifted to the left compared with wild-type curves (Figure 1B1C1D1E1F). Therefore, wild-type and hemizygous mutant samples generated similar melting curve profiles that were separated by 0.2°C to 0.6°C Temperature, based on the shape of the melting curves.
Figure 1.
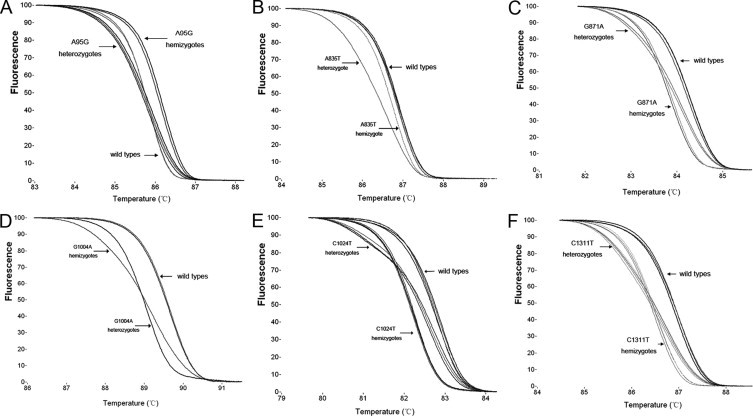
Screening for six G6PD mutations or polymorphisms containing A95G (A), A835T (B), G871A (C), C1004T (D), C1024T (E), and C1311T (F) in the Chinese population by high-resolution melting analysis. Differences in the melting curve shape can easily identify heterozygotes, hemizygotes, and wild-type alleles.
In this study additional mutations adjacent to certain primer pairs could be detected by HRM analysis within the same reaction tube (Table 1). For example, G392T is a common mutation in Han Chinese individuals, and the melting curve shapes in some cases were quite different from that of the G392T mutant (Figure 2A). We hypothesized that an unknown mutation was located near G392T. Indeed, sequence analysis confirmed a C to T substitution at nucleotide 406 (Figure 2B and 2C). G1340T, a new point mutation, was also identified by HRM analysis using the primer pair 11A/B (Figure 2D). This nucleotide substitution was also confirmed by DNA sequencing (Figure 2E and 2F).
Figure 2.
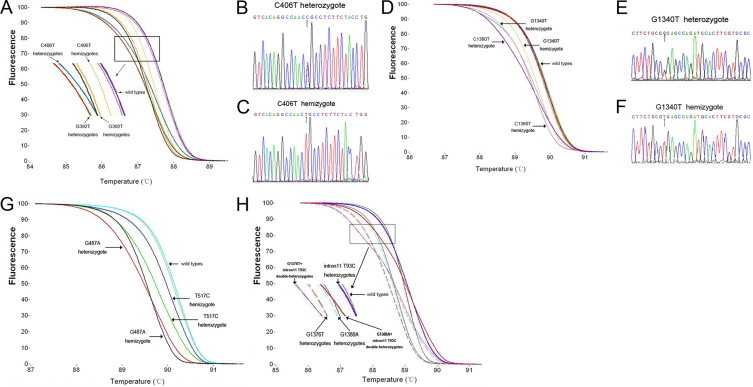
Detection of adjacent mutations with a pair of primers in the same reaction and identification of two new mutations in the Chinese population (as confirmed by DNA sequencing). Mutations G392T and C406T could be distinguished based on melting curve shape (A). The hemizygote and heterozygote of C406T were confirmed by DNA sequencing (B and C). Melting curve differences between C1360T and G1340T are shown (D); the hemizygote and heterozygote of G1340T were also confirmed by DNA sequencing (E and F). The hemizygote and heterozygote of G487A and T517C yielded clearly distinct melting curves (G). The double heterozygote samples such as G1376T/IVS XI t93c and G1388A/IVS XI t93c could also be distinguished by HRM analysis (H).
Importantly, double heterozygous mutants could be detected by HRM owing to the close proximity of mutations IVS XI t93c, G1376T, and G1388A. The melting curve of the 111-bp amplicon was biphasic in double heterozygous samples, G1376T/IVS XI t93c and G1388A/IVS XI t93c, suggesting two independent melting domains (Figure 2H).
Mutation Spectrum of G6PD Deficiency in the Han Chinese Population
A total of 16 point mutations, including 14 missense, one synonymous, and one intron polymorphism site, were identified from the 260 samples in this study (Table 2). Five common mutations, namely A95G, G871A, C1024T, G1376T, and G1388A, with a minimum frequency of 5% for each, accounted for approximately 80% of all G6PD mutations. The substitutions C274T, A493G, C519G, C592T, A835G, and G1381A, which were previously reported in the Chinese population, were not detected in these subjects. In addition, 6 male patients and 14 female patients had unknown mutations.
Table 2.
Distribution of G6PD Variants from G6PD-Deficient Individuals in Shanghai
| Nucleotide change | Amino acid change | Heterozygote | Hemizygote | Homozygote | Total | Frequency, % |
|---|---|---|---|---|---|---|
| A95G | Arg 32 His | 12 | 11 | 0 | 23 | 8.8 |
| A95G/C1311T/IVS XI t93c | Arg 32 His | 1 | 0 | 0 | 1 | 0.38 |
| G392T | Gly 131 Val | 4 | 5 | 0 | 9 | 3.5 |
| C406T | Arg 136 Cys | 1 | 2 | 0 | 3 | 1.2 |
| C406T/C1311T/IVS XI t93c | Arg 136 Cys | 1 | 1 | 0 | 2 | 0.77 |
| G487A | Gly 163 Ser | 1 | 2 | 0 | 3 | 1.2 |
| G487A/C1311T/IVS XI t93c | Gly 163 Ser | 1 | 0 | 0 | 1 | 0.38 |
| T517C | Phe 173 Leu | 1 | 1 | 0 | 2 | 0.77 |
| A835T | Thr 279 Ser | 1 | 1 | 0 | 2 | 0.77 |
| G871A/C1311T/IVS XI t93c | Val 291 Met | 11 | 7 | 2 | 20 | 7.7 |
| C1004T | Ala 334 Phe | 1 | 0 | 0 | 1 | 0.38 |
| C1004T/IVS V 638 (-t) | Ala 334 Phe | 0 | 1 | 0 | 1 | 0.38 |
| C1004T/C1311T/IVS XI t93c | Ala 334 Phe | 1 | 1 | 0 | 2 | 0.77 |
| C1024T | Leu 342 Phe | 9 | 13 | 0 | 22 | 8.5 |
| C1024T/C1311T/IVS XI t93c | Leu 342 Phe | 2 | 0 | 0 | 2 | 0.77 |
| C1311T/IVS XI t93c | 1 | 0 | 0 | 1 | 0.77 | |
| G1340T | Gly 447 Val | 1 | 1 | 0 | 2 | 0.77 |
| G1360T/C1311T/IVS XI t93c | Arg 454 Cys | 1 | 1 | 0 | 2 | 0.77 |
| G1376T | Arg 459 Leu | 31 | 34 | 2 | 67 | 25.8 |
| G1376T/C1311T/IVS XI t93c | Arg 459 Leu | 6 | 0 | 0 | 6 | 2.3 |
| G1388A | Arg 463 His | 28 | 36 | 0 | 64 | 24.6 |
| G1388A/C1311T/IVS XI t93c | Arg 463 His | 2 | 0 | 0 | 2 | 0.77 |
| A95G/G1388A | Arg 32 His/Arg 463 His | 0 | 1 | 0 | 1 | 0.38 |
| G871A/G1376T/C1311T/IVS XI t93c | Val 291 Met/Arg 459 Leu | 1 | 0 | 0 | 1 | 0.38 |
| Mutation unknown | Unknown | 20 | 7.7 |
Interestingly, two G6PD mutations that had not been reported in the Chinese population were identified. One of these mutations, designated as G6PD Jiangxi 1340T, involved a G→T substitution at nucleotide 1340 in exon 11, yielding a Gly447Val change in the protein. The other mutation involved a C→T change at nucleotide 406 in exon 5, resulting in an Arg136Cys change in the protein. This G6PD mutation was detected in five individuals from three unrelated families. This is the first report of the C406T mutation in the Chinese population.
The C1311T and IVS-XI t93c Polymorphism in Han Chinese Individuals
The frequency of the common polymorphism C1311T/IVS XI t93c in the Han Chinese population is shown in Table 3. Linkage disequilibrium between the mutation 871A and the polymorphic variants 1311T and IVS XI + 93c was observed in all tested individuals with G6PD Viangchan G871A, and the data were consistent with previous reports on Asian populations.18,19,20 In some cases, the mutations A95G, C406T, G487A, C1004T, C1024T, G1360T, G1376T, and G1388A were located in cis with a C1311T/IVS XI t93c silent polymorphism.
Table 3.
The Frequency of Common Polymorphism Site C1311T/IVS XI t93c in the Chinese Han Population
| Nucleotide change | A95G | C406T | G487A | G871A | C1004T | C1024T | G1360T | G1376T | G1388A | Total | Healthy volunteers |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive individuals | 1 | 3 | 1 | 21 | 2 | 2 | 2 | 6 | 2 | 40 | 9 |
| Frequency, % | 4.2 | 60 | 25 | 100 | 50 | 8.3 | 100 | 8.1 | 3.0 | 15.4 | 15.8 |
The frequency of the common polymorphism variant 1311T/IVS XI + 93c among the 260 tested individuals was 15.8%. The frequency of this mutation among 57 healthy individuals was 15.4% (Table 3), indicating no statistical difference between these two groups.
Discussion
HRM Analysis is a Powerful Approach to High-Throughput Screening for G6PD Deficiency
HRM was applied to screen 260 unrelated individuals for G6PD mutations in the present study, and 92.3% (240 of 260) of the samples resulted in positive mutations that were confirmed by DNA sequencing. The mutation detection rate was higher than the denaturing high-performance liquid chromatography approach (only ∼86%) in other reports.5,11 Simultaneously, the amplification refractory mutation system was used to detect three common G6PD mutations (A95G, G1388A, and G1376T) in this study. Of the 171 tested samples, 12 (∼7%) of them resulted in false negative or positive results. Accordingly, HRM appears to be more convenient, sensitive, and accurate than other methods for identifying G6PD mutants.
Like other PCR-based methods, HRM cannot detect deletions encompassing the whole gene or entire exons. Nevertheless, performing HRM as an initial screen for potential mutations will considerably reduce the number of samples that need to be sequenced as well as costs and labor. Furthermore, HRM is the least expensive of the currently used screening methods.
The effective amplicon design is an important consideration for achieving robust and reproducible results in HRM analysis. Amplicon length may also influence the sensitivity of genotyping because shorter amplicons generally allow better discrimination of small sequence variations such as single base differences.21,22 As amplicon size decreases, the Tm differences among the genotypes increases, thus allowing better differentiation between mutant and nonmutant samples.23 In this study the lengths of amplicons were limited within 150 bp because most exons in G6PD are not longer than 200 bp. Moreover, the amplicons were designed to have similar Tm values to facilitate amplification under the same PCR conditions. Based on melting curve shape, the heterozygotes and hemizygotes carrying the most common G6PD mutations could be easily distinguished.
Adjacent Mutations Can be Determined Using the Same Primer Pair
More than 90% of individuals with G6PD deficiency in the Chinese population have mutations that span exons 2, 5, 6, 9, 11, and 12 of G6PD, and some point mutations are located in close proximity to one another. Therefore, it was convenient for us to amplify one fragment containing several mutations. For example, the primer pair 3A/3B is usually used to detect the G392T mutant (Figure 2A). However, the shapes of melting curves generated in some G392T mutant samples were shifted to the right. Based on this result, it was speculated that an unknown mutation may be located adjacent to G392T. Indeed, sequencing data revealed the presence of a different mutation, C406T (Figure 2B and 2C), which was not previously reported in the Chinese population. This result shows the power of HRM to detect both novel and known mutations.
The G1388A and G1376T G6PD variants are the most common in the Chinese population, whereas IVS XI t93c, in cis with C1311T, is the most frequent polymorphism in different ethnic populations around the world. The results showed that the frequencies of these two mutations reached 53.8%, and the C1311T/IVS XI t93c haplotype was present in 15.8% of the individuals with G6PD deficiency. As such, a single reaction that distinguishes among G1388A, G1376T, and IVS XI t93c would be beneficial for genetic screening. In this study the primer pair 12A/12B was designed to amplify the fragment that included these mutations in the same PCR product. The melting curves of hemizygotes generated from G1376T, G1388A, and IVS XI t93c mutants shifted to the right (Figure 2H), and we were able to differentiate among them based on their melting curves in one single reaction. Based on the data, the two most common variants, which account for 50% to 60% of mutations related to G6PD deficiency in Han Chinese individuals, could be differentiated in a single reaction. In addition, two common double heterozygote mutants, G1376T/IVS XI t93c and G1388A/IVS XI t93c, could also be distinguished based on different melting curves generated with the 12A/12B primers (Figure 2H). Overall, the present study developed an efficient and accurate HRM method for screening more than 90% of G6PD mutations in the Chinese population.
Identification of New G6PD Mutations in the Chinese Han Population by HRM Analysis
HRM analysis predicted a new G6PD mutation, G1340T, which results in the replacement of the smallest amino acid, Glycine (Gly), with the larger residue, Valine (Val). This amino acid change could result in altered enzyme activity. The result revealed a G6PD/6PGD ratio of 0.2 for the patient with this mutation, thus identifying the molecular basis for the severe clinical symptoms presented by this patient.
The C406T variant, resulting in an Arginine (Arg) to Cysteine (Cys) substitution at residue 136, was originally reported in a Spanish man.24 This mutation was first identified in three unrelated Chinese families. We suggested that the C406T mutation might have evolved in the Chinese population independently rather than from gene flow.
The C1311T and IVS-XI t93c Polymorphism Might Not be the Cause of Reduced G6PD Activity
C1311T is a common polymorphic variant that does not result in an amino acid change. The haplotype C1311T/IVS XI t93c is common among different ethnic groups, and its frequency varies significantly among different populations. The result showed that the frequency was 15% in Han Chinese individuals, similar to that reported in Mediterranean populations,25 but this frequency is higher than previous reports in the Chinese population. Jiang et al5 showed that this haplotype was found at a frequency of less than 7% in the Chinese population, and the percentage in ethnic Hans was higher than that in other ethnic groups. In this study all subjects were ethnically Han, indicating that the haplotype C1311T/IVS XI t93c is more frequent in ethnic Hans than in other Chinese ethnic populations.
This study detected the haplotype C1311T/IVS XI t93c among healthy controls as well as individuals with G6PD deficiency, and the frequencies between these two groups were not statistically different (P > 0.05). The G6PD/6PGD ratio data also suggested that certain cis-compound mutations such as G1376T/C1311T/IVS XI t93c do not have cumulative effects on G6PD enzyme activity (data not shown). The G6PD activity of such mutants did not differ from that of individuals with corresponding single mutations, indicating that the C1311T/IVS XI t93c haplotype cannot account for the reduced G6PD activity observed in mutants.
Acknowledgements
We are grateful to the patients and their families. We thank Professor Bao-zhong Lu for critical reading of this article.
Footnotes
Supported by grants from National Basic Research “973” Program of China (2007CB511904), National “863” Program of China (2007AA02Z400), Key Project from Shanghai Municipality (08JC1413000) and Shanghai Shen-Kang Hospital Developmental Center (SHDC12007101), Joint-Key Project from Shanghai Health Bureau (2008ZD004), Project from Shanghai Municipal Education Commission and State (06BZ055), and Shanghai Leading Academic Discipline (B204).
Contributor Information
Fanyi Zeng, Email: ytzeng@stn.sh.cn.
Shu-zhen Huang, Email: szhuang1@yahoo.com.
References
- 1.Levy HR. Glucose-6-phosphate dehydrogenases. Adv Enzymol. 1979;48:97–192. doi: 10.1002/9780470122938.ch3. [DOI] [PubMed] [Google Scholar]
- 2.Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose-6-phosphate dehydrogenase: G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler E. G6PD deficiency. Blood. 1994;84:3613–3636. [PubMed] [Google Scholar]
- 4.Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- 5.Jiang W, Yu G, Liu P, Geng Q, Chen L, Lin Q, Ren X, Ye W, He Y, Guo Y, Duan S, Wen J, Li H, Qi Y, Jiang C, Zheng Y, Liu C, Si E, Zhang Q, Tian Q, Du C. Structure and function of glucose-6-phosphate dehydrogenase-deficient variants in Chinese population. Hum Genet. 2006;119:463–478. doi: 10.1007/s00439-005-0126-5. [DOI] [PubMed] [Google Scholar]
- 6.Chiu DT, Zuo L, Chao L, Chen E, Louie E, Lubin B, Liu TZ, Du CS. Molecular characterization of glucose-6-phosphate dehydrogenase (G6PD) deficiency in patients of Chinese descent and identification of new base substitutions in the human G6PD gene. Blood. 1993;81:2150–2154. [PubMed] [Google Scholar]
- 7.Du CS, Xu YK, Hua XY, Wu QL, Liu LB. Glucose-6-phosphate dehydrogenase variants and their frequency in Guangdong, China. Hum Genet. 1988;80:385–388. doi: 10.1007/BF00273657. [DOI] [PubMed] [Google Scholar]
- 8.Chiang SH, Wu SJ, Wu KF, Hsiao KJ. Neonatal screening for glucose-6- phosphate dehydrogenase deficiency in Taiwan. Southeast Asian J Trop Med Public Health. 1999;30 Suppl 2:72–74. [PubMed] [Google Scholar]
- 9.Tang TK, Huang WY, Tang CJ, Hsu M, Chen TA, Chen KH. Molecular basis of glucose-6-phosphate dehydrogenase (G6PD) deficiency in three Taiwan aboriginal tribes. Hum Genet. 1995;95:630–632. doi: 10.1007/BF00209477. [DOI] [PubMed] [Google Scholar]
- 10.Du CS, Liu LB, Liu B, Tokunaga K, Omoto K. Glucose-6-phosphate dehydrogenase deficiency among three national minorities in Hainan Island, China. Gene Geogr. 1988;2:71–74. [PubMed] [Google Scholar]
- 11.Yan T, Cai R, Mo O, Zhu D, Ouyang H, Huang L, Zhao M, Huang F, Li L, Liang X, Xu X. Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: description of four novel mutations. Haematologica. 2006;91:1321–1328. [PubMed] [Google Scholar]
- 12.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 13.Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans. 2009;37:433–437. doi: 10.1042/BST0370433. [DOI] [PubMed] [Google Scholar]
- 14.Rapado I, Grande S, Albizua E, Ayala R, Hernández JA, Gallardo M, Gilsanz F, Martinez-Lopez J. High resolution melting analysis for JAK2 Exon 14 and Exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J Mol Diagn. 2009;11:155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, Guelly C, Bauernhofer T, Samonigg H, Hoefler G, Dandachi N. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11:140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DA, Wang XY, Wang Z, Zhou DF, Cai WW. Molecular characterization of 71 cases of glucose-6-phosphate dehydrogenase deficiency in Hainan province. Zhonghua Xue Ye Xue Za Zhi. 2007;28:250–254. [PubMed] [Google Scholar]
- 17.Beutler E, Blume KG, Kaplan JC, Löhr GW, Ramot B, Valentine WN. International Committee for Standardization in Haematology: recommended screening test for glucose-6-phosphate dehydrogenase (G6PD) deficiency. Br J Haematol. 1979;43:465–467. doi: 10.1111/j.1365-2141.1979.tb03774.x. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Westwood B, Bartscocas CS, Malcorra-Azpiazu JJ, Indrak K, Beutler E. Glucose-6 phosphate dehydrogenase mutations and haplotypes in various ethnic groups. Blood. 1995;85:257–263. [PubMed] [Google Scholar]
- 19.Tang TK, Liu TH, Tang CJ, Tam KB. Glucose-6-phosphate dehydrogenase (G6PD) mutations associated with F8C/G6PD haplotypes in Chinese. Blood. 1995;85:3767–3768. [PubMed] [Google Scholar]
- 20.Beutler E, Westwood B, Kuhl W. Definition of the mutations of G6PD Wayne, G6PD Viangchan, G6PD Jammu, and G6PD ‘LeJeune.’. Acta Haematol. 1991;86:179–182. doi: 10.1159/000204830. [DOI] [PubMed] [Google Scholar]
- 21.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: kRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 23.Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85:50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarza R, Pujades A, Rovira A, Saavedra R, Fernandez J, Aymerich M, Corrons JLV. Two new mutations of the glucose-6-phosphate dehydrogenase (G6PD) gene associated with haemolytic anaemia: clinical, biochemical and molecular relationships. Br J Haematol. 1997;98:578–582. doi: 10.1046/j.1365-2141.1997.2563071.x. [DOI] [PubMed] [Google Scholar]
- 25.Cikes V, Abaza I, Krzelj V, Terzić IM, Tafra R, Trlaja A, Marusić E, Terzić J. Prevalence of factor V Leiden and G6PD 1311 silent mutations in Dalmatian population. Arch Med Res. 2004;35:546–548. doi: 10.1016/j.arcmed.2004.07.005. [DOI] [PubMed] [Google Scholar]


