Abstract
Using recently available mass sequencing and assembly technologies, we have been able to identify and quantify unique cell-free DNA motifs in the blood of patients with multiple sclerosis (MS). The most common MS clinical syndrome, relapsing-remitting MS (RRMS), is accompanied by a unique fingerprint of both inter- and intragenic cell-free circulating nucleic acids as specific DNA sequences that provide significant clinical sensitivity and specificity. Coding genes that are differentially represented in MS serum encode cytoskeletal proteins, brain-expressed regulators of growth, and receptors involved in nervous system signal transduction. Although coding genes distinguish RRMS and its clinical activity, several repeat sequences, such as the L1M family of LINE elements, are consistently different in all MS patients and clinical status versus the normal database. These data demonstrate that DNA motifs observed in serum are characteristic of RRMS and disease activity and are promising as a clinical tool in monitoring patient responses to treatment modalities.
Although multiple sclerosis (MS) remains a clinical diagnosis,1 the definitive standard for the confirmation of diagnosis and the clinical assessment of MS disease activity is T1-weighted gadolinium (Gd) enhanced magnetic resonance imaging (MRI). Gd-MRI supplies information about current disease activity by highlighting areas of breakdown in the blood-brain barrier that indicate inflammation.2 Areas of inflammation appear as active lesions. T1-weighted images also show “black holes,” which are thought to indicate areas of permanent damage. T2-weighted MRI scans are used to provide information about disease burden or lesion load. The high costs of randomized clinical trials in MS are directly associated with the requirement of frequent Gd-MRI scans to assess clinical activity as a function of pharmaceutical intervention and optimal dose assessment. Gadolinium carries significant risk for some patients. In 2007 the U.S. Food and Drug Administration issued a “black box” warning for the use of gadolinium (http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm142884.htm, last accessed October 13, 2009). This warning was based on research that linked the use of gadolinium as an image enhancement aid for MRI to the development of nephrogenic systemic fibrosis, a debilitating and potentially fatal disease.
The development of new therapies for MS is hindered by the lack of a low-cost, minimally invasive diagnostic assay to monitor disease activity. The high per patient cost of Gd-MRI prohibits the number of patients studied in randomized controlled clinical trials as well as the rate at which important questions can be tested. High per patient costs make it prohibitively expensive to study the comparative effectiveness of a treatment, prevention, or diagnostic regimen as it transitions from clinical trial to the larger venue of clinical practice. The cost for maximizing disease control in clinical practice has adverse economic consequences for the uninsured patient as well as societal effects on the health insurance industry and on local, state, and federal governments. The basic sequence data reported here from mass sequence and assembly (MSA) technology provides significant promise that the differential frequencies of specific DNA motifs in patients with relapsing-remitting MS (RRMS) can be translated into a rapid serum-based diagnostic assay for MS and assessment of its clinical activity.
Materials and Methods
Patients
Patients (n = 28) with RRMS from the Centro Sclerosi Multipla, Don Gnocchi Foundation (Milan, Italy) who satisfied the Poser criteria for the diagnosis of clinically definite MS were included in this study. All patients gave informed consent according to a protocol approved by the internal review board of the Don Gnocchi Foundation. Thirteen patients were in clinical relapse, and blood samples were obtained within 7 days of clinical relapse and before the initiation of therapy. These patients are classified as having relapsing MS and included 12 females and 1 male (median age 38 years, range 31−55 years) with median duration of MS of 12 years (range, 3 to 21 years) and a median Kurtzke Expanded Disability Status Scale score of 4.5 (range, 0.0 to 10.0). The Expanded Disability Status Scale ranks patients as a function of their physical disability. A median score of 4.5 signifies a relatively severe disability but requiring minimal assistance. These patients had not received immunomodulatory drug treatment for 1 year or more at the time of circulating nucleic acid (CNA) analysis. Fifteen RRMS patients with clinically stable disease (relapse-free for at least 6 months before CNA analysis) are classified for this study as stable MS. These included 12 females and 4 males (median age 39 years, range, 25 to 53 years) with a median disease duration of 12.5 years (range, 2–23 years) and a median Expanded Disability Status Scale score of 4.0. The diagnoses of relapsing MS and stable MS were confirmed by brain and spinal cord Gd-MRI. Enhancing lesions (dye-enhanced MRI plaques of acute inflammation)2 were present in all relapsing MS patients, but no areas of enhancement were seen at the time of enrollment in patients with stable MS. Serum from apparently healthy individuals (n = 50) was used as the control cohort.5
Sampling
Serum samples were collected, processed within 2 hours, and stored at −80°C. Frozen serum was thawed at 4°C, and cell debris was removed by brief centrifugation at 4000 × g for 20 minutes. Total nucleic acids were extracted from the supernatant using the High Pure Nucleic Acids Extraction Kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions.
Generation of Random Circulating Nucleic Acid Libraries
One microliter of the total nucleic acid solution was subjected to a random primer DNA amplification protocol using the GenomePlex WGA4 kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Resulting circulating DNA preparations were molecular barcoded, pooled, and sequenced using a GS FLX high-throughput sequencer (Roche/454 Life Sciences, Branford, CT) according to the manufacturer's instructions. Raw sequences were trimmed for the adapters/primers used in library generation.
Sequence Analysis
Repetitive elements were detected using the RepeatMasker program (Smit AFA, Hubley R, Green P. RepeatMasker Open 3.0, http://www.repeatmasker.org, last accessed April 30, 2008), which makes use of Repbase (version 12.09; Genetic Information Research Institute, Mountain View, CA).3 The genomic origin of the nonrepetitive circulating DNA was investigated by local alignment analyses using the BLAST program with high stringent parameters.4 Each sequence was subjected to sequential BLAST analyses querying databases of known bacterial genomes, viral genomes, and the human genome (reference genome build 36.2). For each query fragment and each database search, the highest scoring Blast hit with a length of >40 bp was recorded in a SQL database with the start and stop positions for query and hit and the description of the respective matching subject. With use of the genome annotation file as obtained from National Center for Biotechnology Information (NCBI) the position of each hit was evaluated, leading to the hit counts and corresponding hit lengths within annotated genes, pseudogenes, the transcribed parts of a gene (denoted as RNA), the coding sequences and untranslated regions (UTRs). The genome annotation file was used to calculate the expected values for the respective features as percentages of the total genome length.
Statistics
Nucleotides matching to the different chromosomes were normalized by all nucleotides matching to the genome. Nucleotides matching to repetitive elements were normalized by all nucleotides matching to repetitive elements. A database from 50 apparently healthy humans was used as a normal control group.5 Comparisons between the healthy and diseased groups were evaluated using the C value (index) of receiver operator characteristic (ROC) curves,6 which was calculated as area under the curve by applying the linear trapezoidal rule. All elements showing at least an area under the curve value of 0.65 were considered for further evaluation. Fisher's exact P values were calculated at the point of best separation (as defined by the maximum of the product of sensitivity and specificity). Multivariate regression models were estimated using the stepwise-in approach. All statistical evaluations were calculated using Microsoft Excel.
Results
We sequenced serum DNA from 28 patients with definite RRMS using MSA. Thirteen patients were diagnosed as undergoing clinical relapse. Blood was drawn within 1 week of the diagnosis and before any therapy was initiated. The remaining 15 patients were in a remission for at least 6 months before sampling. A total of 2.1 × 105 high-quality sequences and 3.75 × 107 nucleotides were generated. The average number of sequences per sample was 7.6 × 103, averaging 178 nucleotides in length. Of all nucleotides obtained from relapsing MS and stable MS patients, 82% produced significant hits on one of the public databases, of which >99% aligned to the human genome. The relative amount of genes, pseudogenes, transcribed, and untranslated regions (annotated as RNA and UTR) and coding sequences was calculated and compared with the relative amounts observed in the circulating DNA pool of control samples obtained from 50 apparently healthy individuals.6 The representation of coding sequences, genes, RNAs, UTRs, and pseudogenes in the circulating DNA of RRMS patients was comparable with the representation for the circulating DNA (P > 0.1) from healthy control subjects.
The relative abundance of serum DNA fragments matching to specific genes was compared between the different patient groups. Comparison of the relapsing MS patients with the healthy control group yielded fragment matches to 20 genes, which are differentially represented and yielded c values of ≥0.65 in the ROC statistics (Supplemental Table S1, see http://jmd.amjpathol.org). Interestingly, 12 of the 20 genes that yielded a c value ≥0.65 in the individual analysis are either exclusively or predominantly expressed in the central nervous system (CNS). The combination of the differential representation of fragments matching to 7 of these genes in a multivariate regression (MVR) model is sufficient to separate the relapsing MS patients from the normal control group with a combined c value of 0.98 (P < 0.0001) (Table 1). Four of these genes are expressed predominantly in the CNS (GRID2, CHL1, CNTNAP5, and NELL2). The remaining three genes (PLCB1, DTNB, and FHOD3) of the MVR model are ubiquitous in their tissue expression (Table 2). All of the gene fragments identified in the MVR analysis are underrepresented relative to the normal control subjects with the exception of CHL1 and CNTNAP5. Of the genes that were differentially represented between relapsing MS patients and normal control subjects only GRID2 was also underrepresented in the stable MS group compared with the normal control group. Contrary to the relapsing MS group, comparison of the stable MS group with the normal control group yielded only a few genes with predominantly CNS expression (Table 1, Supplemental Table S2, see http://jmd.amjpathol.org). Similar to the relapsing MS group, MVR yielded seven genes that are sufficient to separate stable MS patients from the normal control group (c value = 0.98; P < 0.0001). Differentiation between relapsing MS and stable MS patients (Supplemental Table S3, see http://jmd.amjpathol.org) can be achieved by combining three genes (SATB2, ELAVL2, and CUGBP2) in a MVR model (c value = 1; P < 0.0001). Finally, 18 genes showed significantly different representation in the serum of all MS patients compared with the normal control group (Table 1, Supplemental Table S4, see http://jmd.amjpathol.org). A multivariate regression model using the representation of seven genes results in a C value of 0.98 (P < 0.00001).
Table 1.
Serum DNA Signatures Predictive of RRMS and Disease Activity Obtained by MVR Analysis
| Normal* | c statistic | Stable | c statistic | |
|---|---|---|---|---|
| Genes† | ||||
| All MS | PARN(+), COMMD10(+), TMEM117(+), SPAG17(−), NEB(−), PTPRR(−), TRIO(−) | 0.97 (0.85–0.99) | ||
| Relapsing MS | GRID2(−), CHL1(+), PLCB1(−), DTNB(−), CNTNAP5(+), NELL2(−), FHOD3(−) | 0.98 (0.81–0.98) | SATB2(−), ELAVL2(−), CUGBP2(−) | 1.0 (0.90–1.0) |
| Stable MS | FRMD3(+), PLD5(+), GPR158(−), HPSE2(−), PPM1H(−), ABLIM1(−), RHBDD1(−) | 0.98 (0.90–1.0) | ||
| Repeats‡ | ||||
| All MS | (TG)n(+), AT_rich(−), L1MA4(−), L1M5(−), L1MD(−), (GGTTG)n(+), L1MC(−) | 0.98 (0.93–1.0) | ||
| Relapsing MS | AT_rich(−), L1M5(−), Hs17(+), L1MD(−), Satellite(−), HERVK-int(−), L1MA4(−) | 1.0 (0.98–1.0) | (CAA)n(−), LTR16B1(−), Satellite(−), MER21-int(+), (TCTCTG)n(−), L1ME4a(−) | 1.0 (0.90–1.0) |
| Stable MS | (TG)n(+), AT_rich(−), CER(+), L1MC(−), (TC)n(+), L1MD(−), LTR16A(+) | 1.0 (0.98–1.0) |
(+) signifies an excess compared with the normal database; (−) signifies a reduction compared with the normal database.
Derived from Supplemental Tables S1 to S4 (see http://jmd.amjpathol.org).
Derived from Supplemental Tables S5 to S8 (see http://jmd.amjpathol.org).
Table 2.
Gene Functions of Dysregulated CNAs from Patients with RRMS Versus Healthy Control Subjects
| Relapsing MS versus normal | Stable MS versus normal | Relapsing MS versus normal | Relapsing MS versus stable MS | Total | |
|---|---|---|---|---|---|
| Cytoskeleton/cell morphology | NEBL, DTNB, KLHL1, FHOD3, ARHGAP15 | ABLIM1, FRMD3, FARP1, CLASP1 | SNTG2, SPAG17, NEB, SGCZ | NEBL, FHOD3, UBR4, FARP1, SORBS1 | 18 |
| Neuronal growth factors/growth regulators/differentiation regulators | NELL2, MDGA2, CHL1, CNTN5, CNTNAP5, NAV3, FAM19A2 | SDK1 | TRIO, PDGFD, PTPRR | CNTNAP5, CHL1, ELAVL2, NEGR1 | 15 |
| Phosphatases/phospholipases/kinases | DGKI, MAST4, ITPK1, PLCB1 | PPM1H, PLD5, KSR2 | ITPK1, PRKCB1, KSR2, PLD5 | 11 | |
| Nervous signal transduction | GRID2, GRIA1 | GRID2, SLC44A5 | GRID2, STXBP6 | 6 | |
| Ion channels/ion transporters/ion channel interacting | NKAIN3 | KCNMA1 | SFXN5 | PDZD2 | 4 |
| RNA processing | SRPK2, PARN | CUGBP2 | 3 | ||
| Polysaccharide metabolism | HPSE | CHSY2 | 2 | ||
| G protein-coupled receptors | GPR158 | GPR158 | 2 | ||
| Apoptosis | WWOX, ELMO1 | 2 | |||
| Cell cycle regulation | MCC | 1 | |||
| Proteases | RHBDD1 | 1 | |||
| Inflammation | COMMD10 | 1 | |||
| Lipid rafts | RFTN1 | 1 | |||
| Unclassified | MDS1 | PLEKHA5, SPATA5, TMEM117 | CUBN, SATB2, CCDC60, SERGEF, GSG1L, MDS1, MUC16, EBF1, TBC1D22A, SSBP2 | 14 | |
| Total | 20 | 15 | 18 | 28 | 81 |
Genes with exclusive or predominant expression in nervous tissues are in bold; genes included in MVR(C) are underlined.
The majority of differentially represented genes can be grouped into functional clusters as shown in Table 2. Four main functional groups were identified. The first group consists of proteins forming the cytoskeleton or cell morphology. The second group comprises regulators of growth and differentiation, the majority of which are explicitly expressed in the brain. The third major group comprises phosphatases, phospholipases, and kinases. The fourth group consists of receptors and carrier proteins involved in nervous system signal transduction. Differential serum representation of neuronal growth regulators is especially pronounced in the relapsing MS patient group in whom significant differences from the normal group were detected for five genes (MDGA2, CNTNAP5, CHL1, CNTN5, and NAV3) that are involved in axon guidance, nervous system cell development, and regeneration. Furthermore, we have found differential serum representations for the neural epidermal growth factor gene (NELL2) and FAM19A2 related sequences. NELL2 is involved in cell growth regulation and differentiation, whereas the FAM19A2 protein possibly acts as a regulator of immune and nervous system cells. Furthermore, four neuronal growth factor/regulator genes showed clear differences in representation in the serum of relapsing MS and stable MS patients (Table 2).
MS patient DNA motifs are further distinguished from those of normal subjects by alterations in the frequencies of repetitive elements and fragments derived from specific human chromosomes. An MVR model that uses the repetitive elements, (TG)n, AT_rich, L1MA4, L1M5, L1MD, (GGTTG)n, and L1MC, distinguishes MS patients from normal control subjects (Table 1, Supplemental Table S5, see http://jmd.amjpathol.org) with high statistical reliability (c value = 1, P < 0.0001). A second MVR model was calculated using families of repetitive element DNA sequences and sequences derived from different chromosomes. The combination of sequences matching to chromosome 17 (Hs17), LINE1 elements belonging to the L1M family, chromosome 4 (Hs4), and the mammalian apparent long terminal repeat-retrotransposon family (LTR/MaLR) yields a similar highly significant relative risk P value for MS patients compared with the normal control group. The repetitive elements, repetitive element families, and chromosomes with individual c values of >0.65 are given in Supplemental Table S5 (see http://jmd.amjpathol.org). Eight of the 16 individual repetitive elements with high c values are microsatellites and 6 are different LINE1 elements. All LINE elements are significantly reduced compared with normal control LINE element DNA motifs, whereas (TG)n, (CACAA)n, (CAACC)n, (CCCCAA)n, and (GGTTG)n repeats are overrepresented.
Differentially represented repetitive elements in MS DNA motifs provided additional evidence that distinguishes active from stable disease (Table 1). An MVR model using the repetitive elements (CAA)n, LTR16B1, satellite, MER21, (TCTCTG)n, and L1ME4a yielded a c value of 1 (P < 0.0001) between the two groups (Table 1, Supplemental Table S6, see http://jmd.amjpathol.org). Stable MS patients can be distinguished from healthy control subjects by combining the down-regulation of L1MC, L1MD, and AT_rich small repeats, and up-regulation of CER, (TG)n, and LTR16A sequences (P < 0.0001) (Table 1, Supplemental Table S7, see http://jmd.amjpathol.org. An MVR model using the single repetitive elements AT_rich, L1M5, Hs17, L1MD, satellite, HervK-int (endogenous retrovirus), L1MA4, and chromosome-specific nonrepetitive elements of chromosome Hs17 provided highly significant differences (c value = 1, P < 0.0001) that distinguish relapsing MS patients from healthy control subjects (Table 1, Supplemental Table S8, see http://jmd.amjpathol.org). An MVR model using the repetitive element families L1M, satellite, and DNA/hAT and nonrepetitive sequences derived from chromosomes Hs17 and Hs4 yielded a similar highly significant differentiation (Supplemental Table S8, see http://jmd.amjpathol.org). The differentially represented Hs17-specific DNA motifs are derived from three hot spots of overrepresentation (Figure 1) that occur at approximate nucleotide positions 2.5 × 107, 5.3 × 107, and 7.0 × 107.
Figure 1.
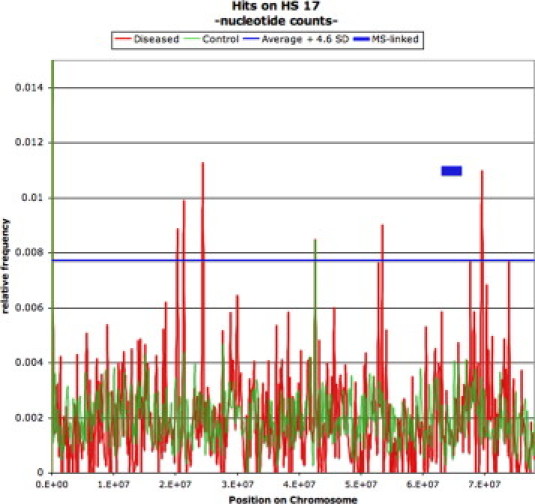
Relative representation of DNA sequences assignable to homo sapiens chromosome 17 as a function of distribution over the chromosome, based on a 0.5-kb window. The green line shows distribution in 50 normal subjects, and the red line represents patients with relapsing MS. The blue horizontal line depicts the border of significance adopted for patients versus normal control subjects, which is equivalent to the mean + 4.6 SD (equals P = 0.01 after Bonferroni correction). The blue box indicates the genomic region linked to MS.4
Our results demonstrate that serum DNA can be shown to have profiles that are distinct from those of genomic DNA when MSA procedures are used. Figure 2A2B2C2D, illustrates ROC curves for acute and stable RRMS patients as a function of differential frequencies of coding genes and repeat elements versus those in normal patients. These profiles are highly statistically different in the serum DNA from MS patients compared with serum DNA profiles of healthy normal subjects. Relative frequencies of genes and repetitive elements can be used to distinguish MS patients from normal control subjects by serum-based analysis as well as distinguish disease activity. A known MS locus is present on Hs17.7 In our analysis Hs17 is significantly overrepresented as a source of DNA motifs in relapsing MS patients versus control subjects with three distinct chromosomal hot spots. These results confirm the value of MSA technologies to detect informative serum DNA biomarkers that we have reported recently in experimental neurodegenerative diseases of cattle8,9 and elk.9
Figure 2.
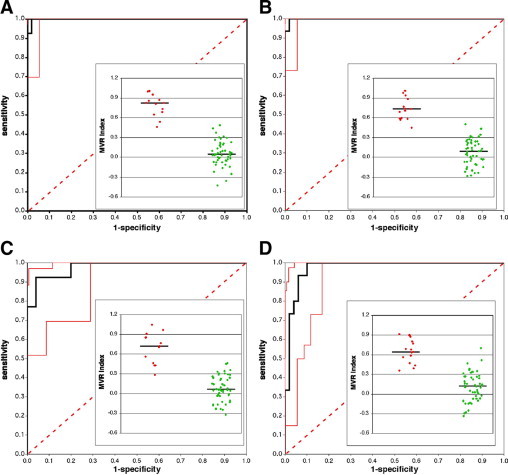
MVR index distribution (inset) and resulting ROC curves of chromosome (Hs) and repetitive element (RE) representation as well as intragenic representation in serum DNA from RRMS patients versus normal healthy control serum DNA. A: Representation of Hs and RE in relapsing MS patients versus normal control subjects. B: Representation of Hs and RE in stable MS patients versus normal control subjects. C: Representation of genes in relapsing MS patients versus normal control subjects. D: Representation of genes in stable MS patients versus normal control subjects. Main figures (ROC curves): the black line is the actual ROC curve, the solid red lines indicate the 95% confidence interval (2.5 to 97.5% limits) of the ROC curves, and the dashed line represents a line of no separation (c value = 0.5). Data insets: Red dots depict either relapsing MS or stable MS values, green dots are normal control values; the solid line represents the group median.
Discussion
Progressive MS is a physically devastating human demyelinating disease of the CNS expressed in a complex genetic and immunological milieu and is widely considered to be a disease of autoimmunity against the components of myelin.10,11,12 Autoimmunity to myelin components can be demonstrated in patients with MS and experimentally induced murine models (experimental autoimmune encephalomyelitis). Immune responses against these components provide supportive experimental evidence for this hypothesis. Alternatively, a variety of intracellular pathogens have been proposed as etiological agents and include human herpesvirus-6,13 endogenous retroviruses,14 and Chlamydophila pneumoniae 15,16. Intrathecal immunoglobulins observed as oligoclonal bands in the cerebrospinal fluid are most commonly observed in chronic infections of the CNS.17 Antibodies to C. pneumoniae as components of the oligoclonal bands in MS18 provide further supportive evidence for the role of this pathogen in MS. Molecular mimicry to antigens of C. pneumoniae19,20 has been demonstrated in murine models of MS. The severity of experimental autoimmune encephalomyelitis is increased with systemic infection with C. pneumoniae that disseminates to the CNS.19 In the rat experimental autoimmune encephalomyelitis model immunization with a 20-mer protein homologous with a C. pneumoniae protein that shares a critical seven-amino acid motif with myelin basic protein results in a severe experimental autoimmune encephalomyelitis response.20 Our mass sequence analysis of serum DNA provides additional evidence of an infectious process. The ecotropic viral integration site, EVI1, is predictive of the progressive spongiform encephalopathy observed in chronic wasting disease of elk.9 Other endogenous retroviral elements have been similarly observed in bovine spongiform encephalopathy.8,9 EVI1 is recognized also as the CCR7 chemokine receptor gene. The EVI1/CCR7 gene is important in that human endogenous retrovirus (HERV) peptides in the expression of type 1 cytokine profiles in vitro of peripheral blood mononuclear cells from patients in relapsing phases of MS but not from stable MS patients have been reported.21 Moreover, LTR16B1 of ERV3 was significantly decreased in serum of patients with relapsing MS. Down-regulation of the MS-associated retrovirus has similarly been positively associated in 11 patients with response to treatment with recombinant interferon-β.22 Our MSA and comparative analysis of serum DNA has expanded the observation of HERV sequences in MS to include the underexpression of AT_rich repeats and LINE elements L1M5, L1MA4, L1MD, and L1MC compared with the normal database. An overexpression of (TG)n and (GGTTG)n was found in the all-inclusive MS group compared with the normal database.
An MS susceptibility locus occurs on human chromosome 17q24 flanked by long segmental, highly homologous, intrachromosomal duplications.7 We have observed DNA fragments traceable to three hot spots on Hs 17 in MS patients. Cell-free DNA in the blood is associated with histones (nucleosomes) protected from nuclease digestion.23 Epigenetic modifications such as DNA methylation as well as acetylation and methylation of histones result in nucleosome positioning alterations and chromatin order.24 Extensive hypomethylation of LINE elements and endogenous and endogenous retroviruses have been reported in several cancers.25 The duplications flanking chromosome 17q24 may affect the biological activity of the regional genes.7 The observed differential frequencies of serum repetitive element DNA in the blood of MS patients versus healthy controls may similarly be a reflection of DNA plasticity. Alternatively, differential repetitive element representation in the CNA pool may originate from different processes by which DNA is released from cells. Several forms of cell death other than apoptosis (eg, necrosis, autophagy, or mitotic catastrophe) as well as an active release of newly synthesized DNA have been hypothesized as possible sources of blood CNA.
L1 elements (LINES) and HERV encode reverse transcriptase that provides an RNA intermediate for new chromosomal integration sites. L1s are the most active autonomous elements of the human genome26 and are estimated to be present in >500,000 copies.27 Because of 5′ truncation-associated RT dissociation from its RNA template and various disruptive mutations, only 30 to 60 LINES are active transposing elements. Based on the similarity of LINE-generated pseudogenes and the HERV-W family of retroviruses, it has been postulated that they are derived from LINE-mediated retrotransposition of retroviral mRNA.28 Nevertheless, the number of complete pseudogenes is greater than expected.29 A significant question arises from the results of our total sequencing approach of serum DNAs: What role do L1 retroelements and microsatellite repeats play in MS? The importance of this question is related to whether or not these repeat elements should be considered for pharmaceutical targeting because L1 retrotransposons have been demonstrated to be regulatory-sensitive to small interfering RNAs.30 Further, these newly identified biomarkers might have some role in the dynamic equilibrium of autoreactive T lymphocytes that play a pivotal role in the prevention of autoimmune diseases such as MS.31
Gene expression profiles derived from peripheral blood mononuclear cells have provided evidence that a small number of genes are down-regulated in MS,32 similar to the reduction in both genes and repetitive element circulating DNA that is observed in the present study, especially in the relapsing disease stage. Translation of the observed microchip data to a complex ratio of gene expression profiles in peripheral blood mononuclear cells using quantitative RT-PCR has demonstrated the potential utility in clinical diagnosis.33 On the basis of the data presented here, it should be possible to develop an inexpensive serum-based quantitative multiplex PCR assay to diagnose MS without the significant expense of mRNA preservation in peripheral blood mononuclear cells and a reverse transcription step. In addition, our mass sequencing data allow a simple laboratory analysis of disease activity. Our ROC MVR analysis suggests that the repetitive elements will yield the best separation of true positive results from the normal population with predicted extraordinary sensitivity and specificity (Figure 2). Table 3 summarizes the various repetitive elements that distinguish RRMS and its clinical activity from normal individuals. Current list prices for hospital-based Gd-MRI average approximately $3500 (https://www.mymedicalcosts.com/Pricing.aspx, last accessed October 18, 2009). With the ever-increasing efforts to reduce the costs of medical care, serum-based assays (estimated to be $100 to $250) with appropriate validation could replace Gd-MRI as the primary standard for diagnosis as well as disease monitoring as a function of drug therapy. The data further suggest that circulating nucleic acids in MS may provide additional tools to address pathogenesis.
Table 3.
Prevalence of Repetitive Circulating DNA in Multiple Sclerosis Patients Versus Healthy Control Subjects
| Statistically significant MS dysregulation versus healthy CNA |
|||||||
|---|---|---|---|---|---|---|---|
| Repetitive element | MS | P | AMS | P | SMS | P | Comments |
| LINE | All LINE elements decreased in MS regardless of disease activity | ||||||
| L1MA4 | ↓ | 0.0003 | ↓ | 0.0021 | |||
| L1M5 | ↓ | <0.0001 | ↓ | <0.0001 | |||
| L1MC | ↓ | 0.0004 | ↓ | 0.0002 | |||
| L1MD | ↓ | <0.0001 | ↓ | 0.0008 | ↓ | 0.0008 | |
| Small repeats | Mixed levels of the small repeat repetitive elements characterize disease activity; decrease of AT rich versus healthy controls equivalent in both relapsing and remitting MS | ||||||
| (TG)n | ↑ | <0.0001 | ↑ | <0.0001 | |||
| (TC)n | ↑ | 0.0025 | |||||
| AT_rich | ↓ | <0.0001 | ↓ | <0.0001 | ↓ | <0.0001 | |
| (GGTTG)n | ↑ | <0.0001 | |||||
| LTRs | LTR elements clearly distinguish between relapsing and remitting MS | ||||||
| CER | ↑ | 0.0007 | |||||
| LTR16A | ↑ | 0.0084 | |||||
| HervK-int | ↓ | 0.0015 | |||||
| Satellite | ↓ | 0.0009 | Decreased levels during acute disease activity | ||||
| MVR analysis | 0.0001 | 0.0001 | 0.0001 | All MVR analyses using the indicated repetitive elements are highly significant | |||
Repetitive elements were used in the predictive algorithms for each category (all MS, relapsing MS, and stable MS). Complete quantitative data on all repetitive data and MVR formulas are available in Supplemental Tables S5 to S8 (see http://jmd.amjpathol.org).
AMS, relapsing (acute) MS; SMS, remitting (stable) MS.
Acknowledgements
We thank Sara Henneke, Stefan Balzer, and Carsten Müller for their skillful assistance, and Sascha Glinka and Birgit Ottenwälder at Eurofins Medigenomix GmbH for performing the GS FLX/454 sequencing. Access to the MS serum bank maintained by the Don C. Gnocchi ONLUS Foundation IRCCS is gratefully acknowledged.
Footnotes
Supported by the EMPRO (European Microbicides Project) and AVIP (AIDS Vaccine Integrated Project) European Commission WP6 Projects, the nGIN (Next Generation HIV-1 Immunogens inducing broadly reactive Neutralising antibodies) and GISHEAL (Genetic and Immunological Studies of European and African HIV-1+ Long Term Non Progressors) WP7 Projects, the Japan Health Science Foundation, and 2007 Ricerca Finalizzata (Italian Ministry of Health).
None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
H.B.U., J.B., and E.S. are employees of Chronix Biomedical. W.M.M. is an independent board of directors member. M.C. and W.M.M. serve on the Chronix Medical Advisory Committee.
Web Extra Material
Relapsing MS Serum DNA Variance from Healthy Controls
Stable MS Serum DNA Variance from Healthy Controls (Genes)
Relapsing MS Serum DNA Variance from Stable MS (Genes)
MS (Relapsing MS + Stable MS) Serum DNA Variance from Healthy Controls (Genes)
MS (Relapsing MS + Stable MS) Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)
Relapsing MS Serum DNA Variance from Stable MS (HS and Repetitive Elements)
Stable MS Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)
Relapsing MS Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)
References
- 1.Birnbaum G. Making the diagnosis of multiple sclerosis. Adv Neurol. 2006;98:111–124. [PubMed] [Google Scholar]
- 2.Traboulsee A, Li DK. The role of MRI in the diagnosis of multiple sclerosis. Adv Neurol. 2006;98:125–146. [PubMed] [Google Scholar]
- 3.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase; Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck J, Urnovitz HB, Riggert J, Clerici M, Schütz E. Profile of the circulating DNA in healthy individuals. Clin Chem. 2009;55:730–738. doi: 10.1373/clinchem.2008.113597. [DOI] [PubMed] [Google Scholar]
- 6.Kestler HA. ROC with confidence—a Perl program for receiver operator characteristic curves. Comput Methods Programs Biomed. 2001;64:133–136. doi: 10.1016/s0169-2607(00)00098-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen DC, Saarela J, Clark RA, Miettinen T, Chi A, Eichler EE, Peltonen L, Palotie A. Segmental duplications flank the multiple sclerosis locus on chromosome 17q. Genome Res. 2004;14:1483–1492. doi: 10.1101/gr.2340804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck J, Urnovitz H, Grochup M, Ziegler U, Brenig B, Schütz E. Serum nucleic acids in an experimental bovine transmissible spongiform encephalopathy model. Zoonoses Public Health. 2009;56:384–390. doi: 10.1111/j.1863-2378.2009.01260.x. [DOI] [PubMed] [Google Scholar]
- 9.Gordon PM, Schütz E, Beck J, Urnovitz HB, Graham C, Clark R, Dudas S, Czub S, Sensen M, Brenig B, Groschup MH, Church RB, Sensen CW. Disease-specific motifs can be identified in circulating nucleic acids from live elk and cattle infected with transmissible spongiform encephalopathies. Nucleic Acids Res. 2009;37:550–556. doi: 10.1093/nar/gkn963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. Multiple sclerosis. N Engl J Med. 2000;343:938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- 11.Hartung HP, Kieseier BC, Hemmer B. Purely systemically active anti-inflammatory treatments are adequate to control multiple sclerosis. J Neurol. 2005;252(Suppl 5):v30–v37. doi: 10.1007/s00415-005-5006-3. [DOI] [PubMed] [Google Scholar]
- 12.Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 2008;31:247–269. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- 13.Fotheringham J, Jacobson S. Human herpesvirus 6 and multiple sclerosis: potential mechanisms for virus-induced disease. Herpes. 2005;12:4–9. [PubMed] [Google Scholar]
- 14.Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol. 2006;176:7636–7644. doi: 10.4049/jimmunol.176.12.7636. [DOI] [PubMed] [Google Scholar]
- 15.Sriram S, Stratton CW, Yao S, Tharp A, Ding L, Bannan JD, Mitchell WM. Chlamydia pneumoniae infection of the central nervous system in multiple sclerosis. Ann Neurol. 1999;46:6–14. [PubMed] [Google Scholar]
- 16.Tang YW, Sriram S, Li H, Yao SY, Meng S, Mitchell WM, Stratton CW. Qualitative and quantitative detection of Chlamydophila pneumoniae DNA in cerebrospinal fluid from multiple sclerosis patients and controls. PLoS ONE. 2009;4:e5200. doi: 10.1371/journal.pone.0005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stratton CW, Wheldon DB. Multiple sclerosis: an infectious syndrome involving Chlamydophila pneumoniae. Trends Microbiol. 2006;14:474–479. doi: 10.1016/j.tim.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Yao SY, Stratton CW, Mitchell WM, Sriram S. CSF oligoclonal bands in MS include antibodies against Chlamydophila antigens. Neurology. 2001;56:1168–1176. doi: 10.1212/wnl.56.9.1168. [DOI] [PubMed] [Google Scholar]
- 19.Du C, Yao SY, Ljunggren-Rose A, Sriram S. Chlamydia pneumoniae infection of the central nervous system worsens experimental allergic encephalitis. J Exp Med. 2002;196:1639–1644. doi: 10.1084/jem.20020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz DC, Lu L, Conant SB, Wolf NA, Gérard HC, Whittum-Hudson JA, Hudson AP, Swanborg RH. A Chlamydia pneumoniae-specific peptide induces experimental autoimmune encephalomyelitis in rats. J Immunol. 2001;167:1803–1808. doi: 10.4049/jimmunol.167.3.1803. [DOI] [PubMed] [Google Scholar]
- 21.Clerici M, Fusi ML, Caputo D, Guerini FR, Trabattoni D, Salvaggio A, Cazzullo CL, Arienti D, Villa ML, Urnovitz HB, Ferrante P. Immune responses to antigens of human endogenous retroviruses in patients with acute or stable multiple sclerosis. J Neuroimmunol. 1999;99:173–182. doi: 10.1016/s0165-5728(99)00123-x. [DOI] [PubMed] [Google Scholar]
- 22.Mameli G, Serra C, Astone V, Castellazzi M, Poddighe L, Fainardi E, Neri W, Granieri E, Dolei A. Inhibition of multiple-sclerosis-associated retrovirus as biomarker of interferon therapy. J Neurovirol. 2008;14:73–77. doi: 10.1080/13550280701801107. [DOI] [PubMed] [Google Scholar]
- 23.Holdenrieder S, Nagel D, Schalhorn A, Heinemann V, Wilkowski R, von Pawel J, Raith H, Feldmann K, Kremer AE, Müller S, Geiger S, Hamann GF, Seidel D, Stieber P. Clinical relevance of circulating nucleosomes in cancer. Ann NY Acad Sci. 2008;1137:180–189. doi: 10.1196/annals.1448.012. [DOI] [PubMed] [Google Scholar]
- 24.Ballestar E, Esteller M. The impact of chromatin in human cancer: linking DNA methylation to gene silencing. Carcinogenesis. 2002;23:1103–1109. doi: 10.1093/carcin/23.7.1103. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 26.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 27.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Pavlícek A, Paces J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlícek A, Paces J, Zíka R, Hejnar J. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: implications for retrotransposition and pseudogene detection. Gene. 2002;300:189–194. doi: 10.1016/s0378-1119(02)01047-8. [DOI] [PubMed] [Google Scholar]
- 30.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 31.Saresella M, Marventano I, Guerini FR, Zanzottera M, Delbue S, Marchioni E, Maserati R, Longhi R, Ferrante P, Clerici M. Myelin basic protein-specific T lymphocytes proliferation and programmed cell death in demyelinating diseases. Clin Immunol. 2008;129:509–517. doi: 10.1016/j.clim.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, Aune TM. Cutting edge: molecular portrait of human autoimmune disease. J Immunol. 2002;169:5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 33.Fossey SC, Vnencak-Jones CL, Olsen NJ, Sriram S, Garrison G, Deng X, Crooke PS, 3rd, Aune TM. Identification of molecular biomarkers for multiple sclerosis. J Mol Diagn. 2007;9:197–204. doi: 10.2353/jmoldx.2007.060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relapsing MS Serum DNA Variance from Healthy Controls
Stable MS Serum DNA Variance from Healthy Controls (Genes)
Relapsing MS Serum DNA Variance from Stable MS (Genes)
MS (Relapsing MS + Stable MS) Serum DNA Variance from Healthy Controls (Genes)
MS (Relapsing MS + Stable MS) Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)
Relapsing MS Serum DNA Variance from Stable MS (HS and Repetitive Elements)
Stable MS Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)
Relapsing MS Serum DNA Variance from Healthy Controls (HS and Repetitive Elements)


