Abstract
The distinction between mycosis fungoides (MF) and inflammatory dermatoses (ID) by clinicopathologic criteria can be challenging. There is limited information regarding the performance characteristics and utility of TCRG and TCRB clonality assays in diagnosis of MF and ID from paraffin-embedded tissue sections. In this study, PCR tests were performed with both TCRG and TCRB BIOMED-2 clonality methods followed by capillary electrophoresis and Genescan analysis using DNA samples from 35 MF and 96 ID patients with 69 and 133 paraffin-embedded specimens, respectively. Performance characteristics were determined for each test individually and in combination. TCRG and TCRB tests demonstrated identical sensitivity (64%) and specificity (84%) when analyzed as individual assays. The positive predictive value, negative predictive value, and change of posttest MF probability over a range of MF pretest probabilities were obtained. These data were used to construct an algorithm for sequential use of TCRG and TCRB. As single tests, commercially available BIOMED-2 PCR-based TCRG and TCRB clonality tests on paraffin-embedded tissue have no significant difference in terms of sensitivity and specificity. Combined use of the two tests in patients with intermediate pretest probabilities as proposed in the algorithm could improve test utility.
Mycosis fungoides (MF) is the principle form of cutaneous T-cell lymphoma and accounts for nearly 50% of all primary cutaneous lymphomas.1 In a proportion of cases, especially during the early stages, it is difficult to distinguish MF from some reactive inflammatory dermatoses (ID) clinically and histopathologically, and thus a definitive diagnosis is often preceded by a variably long period. Based on the fact that the tumor cells of lymphomas harbor identically (clonally) rearranged T-cell receptor genes whereas reactive skin disorders consist of cells with polyclonal T-cell receptor genes,2 T-cell receptor clonality testing is commonly performed on cases of suspected MF as an ancillary study to provide additional evidence for diagnosis.
The BIOMED-2 collaborative study developed multiplex PCR assays for the detection of clonally rearranged TCR genes, which make interlaboratory comparison possible. Because of the restricted repertoire of V and J segments of TCRG locus and the existence of TCRG recombination in both TCRγδ and TCRαβ clonal T-cell proliferations as a result of the chronological order of TCR gene rearrangements in T-cell progenitors,3 PCR clonality assay of TCRG is the test most often performed in most clinical diagnostic laboratories.
The BIOMED-2 group reported an 89% rearrangement rate of the TCRG gene and 94% of the TCRB gene in T-cell malignancies in 2003.4 In 2006, Morgan et al used the BIOMED-2 PCR primers to examine 10 early-stage MF cases and 10 late-stage MF or Sezary syndrome cases with either fresh or paraffin-embedded tissue specimens. They reported a high frequency of clonally rearranged TCRG (17/20) and TCRB (15/20) genes.5 However, the currently available data on the sensitivity and specificity of BIOMED-2 TCRB clonality assay in testing paraffin-embedded skin tissue of MF is limited and from small datasets.
Rigorous test performance assessment is required to determine the utility of clonality testing in skin biopsies. Monoclonality is occasionally found in some benign cutaneous lymphocytic infiltrates and can be detected in only approximately 50% to 66.6% of early MF lesions.6,7 Langerak et al reported a 75% rate of polyclonality, 15% rate of oligoclonality, and 10% rate of monoclonality in frozen tissue specimens with reactive lymphoproliferations using BIOMED-2 Ig/TCR clonality assessment.8 Distinguishing early-stage MF from inflammatory dermatoses remains a major challenge in dermatopathology. Improving the accuracy of TCR clonality test and maximizing its utility in the differential diagnosis of MF and ID is still a topic under investigation. Several approaches have been proposed, including comparison of TCRG PCR results at several involved skin sites from the same patient or serial analysis of skin biopsies over the course of disease.9,10,11 An alternative approach is to test the TCRB gene.12 The BIOMED-2 group reported that the addition of TCRB rearrangements as PCR targets increased the clonality detection rate to 94% using frozen or fresh specimens of T-cell malignancies.13 Although the combined use of TCRG and TCRB clonality assessments seems a promising idea to help increasing test sensitivity, no large-scale comparison of the BIOMED-2 TCRG and TCRB clonality assays, which tests paraffin-embedded skin tissue from MF and ID patients, has been reported.
In this retrospective study, we aimed to (1) determine the performance characteristics of TCRG and TCRB clonality tests (as single test or combined tests) using BIOMED-2 primers in paraffin-embedded skin sections in terms of concordance, sensitivity, and specificity; (2) calculate positive predictive value as well as negative predictive value over a range of pretest probabilities; and (3) based on these results develop an evidence-based strategy for use of the two TCR clonality assays in paraffin embedded skin specimens.
Materials and Methods
Case Selection and Patient Follow-Up
A total of 202 paraffin-embedded skin biopsies from 131 patients (35 patients diagnosed as MF and 96 patients diagnosed as ID) were obtained from Stanford Hospital. Cases were included based on the availability of paraffin-embedded tissue for TCR clonality analysis during 1997 to 2006. This research was approved by Institutional Review Board of Stanford University. Both TCRG and TCRB clonality tests using BIOMED-2 primers were performed on each specimen. We were able to obtain clinical follow-up information for a total of 72 patients in the study, and the overall follow-up time ranged from 0.5 to 131 months with a median of 30 months. Of the 35 MF patients in the study, the follow-up information was available for 29 patients with the follow-up time ranging from 0.5 to 88 months and a median of 33 months. Clinical information was also obtained on the patients with inflammatory dermatoses to ensure that they did not ultimately develop mycosis fungoides. Of the 96 ID patients in the study, the follow-up information was available for 43 patients with the follow-up time ranging from 0.5 to 131 months and a median of 31 months.
Specimen Processing and DNA Extraction
The standard protocol of DNA extraction from paraffin-embedded tissue in the Stanford Molecular Pathology Lab was used in the study. Four to ten 20-μm sections were cut from each of the paraffin blocks and placed into 1.5-ml microcentrifuge tubes. Histoclear (National Diagnostics, Atlanta, GA) was added to each tube three times to remove the paraffin followed by two times of ethanol wash. The tissue pellets were dried completely on a 65°C heat block before Protein K buffer solution was added into each tube. The tissue specimens were incubated overnight at 62°C. The digestion was stopped by immersing the tubes in boiling water for 8 minutes. Agarose gel electrophoresis was performed to assess the DNA quality and concentration.
Multiplex PCR
TCRG and TCRB rearrangements were studied using the commercially available BIOMED-2 multiplex PCR assays manufactured by InVivoScribe Technologies (San Diego, CA) and the BIOMED-2 PCR protocol.4 The final 50-μl reaction volume includes 45-μl master mix (A, B, or C for TCRB; A or B for TCRG), 2U of Taq Gold (Applied Biosystems, Foster City, CA), and 100 to 200 ng of DNA. The PCR reactions were performed on ABI 9700 thermal cyclers. The amplification parameters were suggested by Invivoscribe assay as follows: initial denaturation for 7 minutes at 95°C, followed by 35 cycles of 45 seconds at 95°C, 45 seconds at 60°C, and 90 seconds at 72°C, with a final extension step of 10 minutes at 72°C.
Differential Fluorescence Detection on ABI 3100 (Genescan)
PCR product (1 μl) was added into the appropriate well of a 96-well PCR plate. Thaw Hi-Di formamide (11 μl, Applied Biosystems, Forster City, CA) and GeneScan-500 Rox size standard (1 μl) were prepared for each PCR product. The freshly prepared formamide with size standard was added into each well and mixed with PCR product. The plate was sealed with rubber plate septa and placed at 95°C for 2 minutes to denature, then the samples were placed on ice to snap chill for at least 5 minutes before being loaded to the analyzer.
Reaction Controls
DNA extracted from paraffin-embedded tissue specimens is frequently of moderate to poor quality and might contain PCR inhibitors. Consequently, it is essential to evaluate the integrity and amplifiability of DNA extracted from paraffin-embedded tissues.4 The multiplex control gene tube is included in both TCRG and TCRB assays with PCR products of 100, 200, 300, 400, and 600 bp. At least 300-bp control PCR products were required for a test to be included in the analysis. Besides the control tube for each sample, we set up four additional groups of controls:
-
1.
Positive control: DNA containing a clonal TCRG/TCRB gene rearrangement from a previously positive clinical sample or cell line. Two cell line DNA samples are provided with the clonality kit.
-
2.
Sensitivity control: Positive (clonal) control provided with the assay reagents (peer cell line), diluted to 2% to 5% with polyclonal DNA.
-
3.
Negative (polyclonal, normal) control: DNA extracted from nonclonal lymphoid-rich tissue such as tonsil, lymph node, or peripheral blood lymphocytes.
-
4.
Blank control: PCR water used in place of any template DNA.
Result Interpretation
Test results of all samples were independently interpreted by three pathologists who were blinded to the patients' histological or clinical information. Monoclonality was defined as one or two predominant peaks (equal or greater than two times the height of polyclonal background) within the appropriate size range. Oligoclonality was defined as three or more predominant peaks within the appropriate size range. Polyclonal pattern was considered to be negative.
Results
The 202 investigated paraffin-embedded specimens were collected from 131 patients who had a final diagnosis of either MF or ID. Among the 35 MF patients, there were 8 patients with 1 biopsy, 23 patients with 2, and 4 patients with 3 or more. Among the 96 ID patients, there were 66 patients with 1 biopsy, 24 patients with 2, and 6 patients with 3 or more. The reactive skin disorders included in the study and the numbers of their corresponding biopsies are listed in Table 1. The results of the two tests are summarized as a 3 × 3 contingency table in Table 2. The overall concordance rate of the two assays was 77.2%.
Table 1.
Reactive Skin Disorders Included in the Study
| Reactive skin disorder | Number of patients | Number of samples |
|---|---|---|
| Actinic reticuloid | 2 | 2 |
| Arthropod bite reaction | 2 | 3 |
| Gyrate erythema | 6 | 7 |
| Interface dermatitis | 4 | 4 |
| Lichen planus | 4 | 4 |
| Lichen sclerosus | 5 | 5 |
| Lichenoid dermatitis | 1 | 1 |
| Lichenoid drug eruption | 1 | 1 |
| Lupus | 2 | 4 |
| Morphea | 7 | 8 |
| Pigmented purpuric eryption | 9 | 12 |
| Pityriasis lichenoides et varioliformis acuta | 8 | 9 |
| Psoriasis | 14 | 27 |
| Spongiotic dermatitis | 15 | 15 |
| Superficial perivascular dermatitis | 5 | 5 |
| Dermatitis, NOS | 4 | 10 |
| Drug eruption | 1 | 2 |
| Small plaque parapsoriasis | 1 | 2 |
| Eczematous dermatitis | 3 | 8 |
| Hypersensitivity reaction | 1 | 2 |
| Erythema nodosum | 1 | 2 |
| Total | 96 | 133 |
Table 2.
Correlation between TCRG and TCRB Test Results
|
TCRG |
||||
|---|---|---|---|---|
| Monoclonal | Polyclonal | Oligoclonal | Total | |
| TCRB | ||||
| Monoclonal | 43 | 22 | 1 | 66 |
| Polyclonal | 21 | 107 | 0 | 128 |
| Oligoclonal | 2 | 0 | 6 | 8 |
| Total | 66 | 129 | 7 | 202 |
Concordance rate of TCRG and TCRB assays = (43 + 107 + 6)/202 = 77.2%.
There are two ways to interpret results from the combined use of TCRG and TCRB clonality studies: requiring both tests to be monoclonal before calling a final positive result, or calling a positive result when either test is monoclonal. With the former approach, the test sensitivity for MF would drop to 49% and the specificity would increase to 93%; with the latter approach, the sensitivity would increase to 78% with the specificity dropping to 74%.
Table 3 shows the numbers of positive and negative results as well as test sensitivity and specificity by different ways of interpreting oligoclonal pattern. If an oligoclonal pattern is interpreted as negative for clonality, TCRG and TCRB tests when used alone, detected the same number of positive samples and thus had the same sensitivity (64%) and specificity (84%); although there was an overlap of the positive samples from the two tests, they were not identical. Alternatively, because the interpretation of oligoclonal pattern remains a gray area, we also calculated the sensitivity and specificity in Table 3 when oligoclonal pattern is interpreted as positive for clonality. In this cohort, oligoclonality was highly associated with MF, and interpreting oligoclonality as positive resulted in an approximately 10% increase in sensitivity with virtually no change in the specificity.
Table 3.
Test Results of TCRG, TCRB, and Combined Use of TCRG and TCRB When Interpreting Oligoclonality as Either Negative or Positive
| Classification of oligoclonal pattern | Clonality test(s) used | Test interpretation | Definitions of “positive” and “negative” | # of MF | # of ID | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| As negative | TCRG alone | Positive | Monoclonal | 44 | 22 | 64% | 84% |
| Negative | Oligoclonal or polyclonal | 25 | 111 | ||||
| TCRB alone | Positive | Monoclonal | 44 | 22 | 64% | 84% | |
| Negative | Oligoclonal or polyclonal | 25 | 111 | ||||
| TCRG and TCRB | Positive | Both tests are monoclonal | 34 | 9 | 49% | 93% | |
| Negative | At least one test is not monoclonal | 35 | 124 | ||||
| TCRG and TCRB | Positive | At least one test is monoclonal | 54 | 35 | 78% | 74% | |
| Negative | Neither test is monoclonal | 15 | 98 | ||||
| As positive | TCRG alone | Positive | Monoclonal or oligoclonal | 51 | 22 | 74% | 84% |
| Negative | Polyclonal | 18 | 111 | ||||
| TCRB alone | Positive | Monoclonal or oligoclonal | 51 | 23 | 74% | 83% | |
| Negative | Polyclonal | 18 | 110 | ||||
| TCRG and TCRB | Positive | Both tests are monoclonal or oligoclonal | 40 | 9 | 58% | 93% | |
| Negative | At least one test is polyclonal | 29 | 124 | ||||
| TCRG and TCRB | Positive | At least one test is monoclonal or oligoclonal | 62 | 36 | 90% | 73% | |
| Negative | Both tests are polyclonal | 7 | 97 | ||||
| Total # | 69 | 133 |
Results by T-Classification
Of the 69 MF samples, T-classification information (T-classification system: T1, patches/plaques <10% body surface area; T2, patches/plaques ≥10% body surface area; T3, tumors, rounded, or dome-shaped lesions >1 cm in diameter; T4, generalized erythroderma, >80% of body surface is affected) was available for 44 samples, as shown in Table 4. Among the 10 samples from T1 MF patients, 3 were detected by the TCRG test and 6 were detected by TCRB test. There was an overlap of two samples with positive results between the two tests, and thus a total of seven were detected with the combination of the two tests. Eleven samples were from patients with T2 MF, among which ten were monoclonal by TCRG and seven were monoclonal by TCRB. The seven samples detected as positive by TCRB were all detected as positive by TCRG, and the combined use of the two tests did not lead to a higher sensitivity. The T3 MF group included 15 samples, and TCRG and TCRB detected 11 and 9 samples, respectively. Again, the nine samples with positive TCRB results were all positive for TCRG so that the combined use of the two assays did not impact the sensitivity. For the eight samples from patients with T4 MF, TCRG and TCRB detected the same 6 samples, and the combined use of the two assays did not increase the sensitivity.
Table 4.
TCR Clonality Results and T-Classification of MF
| T-classification | TCRG (positive/total) | TCRB (positive/total) | TCRG and TCRB* (positive/total) | TCRG or TCRB† (positive/total) |
|---|---|---|---|---|
| T1 | 3/10 (30%) | 6/10 (60%) | 2/10 (20%) | 7/10 (70%) |
| T2 | 10/11 (90.9%) | 7/11 (63.6%) | 7/11 (63.6%) | 10/11 (90.9%) |
| T3 | 11/15 (73.3%) | 9/15 (60%) | 9/15 (60%) | 11/15 (73.3%) |
| T4 | 6/8 (75%) | 6/8 (75%) | 6/8 (75%) | 6/8 (75%) |
| MF (total) | 30/44 (68.2%) | 28/44 (63.6%) | 24/44 (54.5%) | 34/44 (77.3%) |
T-classification system: T1, patches/plaques <10% body surface area; T2, patches/plaques ≥10% body surface area; T3, tumors (rounded or dome-shaped lesions >1 cm in diameter); T4, generalized erythroderma (>80% of body surface is affected).
A final positive test result was called for a certain sample when both TCRG and TCRB tests were positive, and a final negative test result was called when at least one test was negative.
A final positive test result was called for a certain sample when at least one test was positive, and a final negative test result was called when both tests were negative.
Test Concordance
Using TCRG as the initial test and TCRB as an additional test showed that TCRB contributed equally in accurate and inaccurate results (see Supplemental Table S1 at http://jmd.amjpathol.org). In the MF group, among the 44 samples with positive TCRG results, 34 (77%) also had positive TCRB results, whereas 10 (23%) had negative TCRB results; among the 25 samples of MF with negative TCRG results, 15 (60%) also had negative TCRB results, whereas 10 (40%) were positive by TCRB analysis. In the ID group, among the 22 samples with positive TCRG results, 9 (41%) were also positive for TCRB, whereas 13 (59%) were TCRB negative; among the 111 ID samples with negative TCRG results, 98 (73.7%) were also negative by TCRB, whereas 13 (9.8%) had positive TCRB results.
Test Utility/Predictive Value
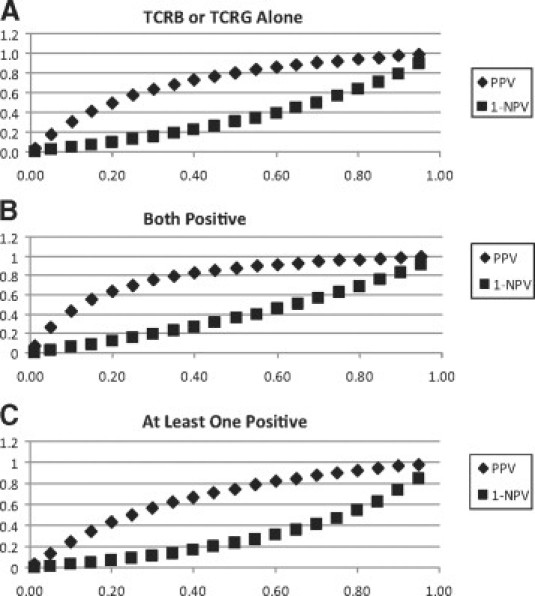
The positive predictive value (PPV) and 1 − negative predictive value (1-NPV) were determined under different pretest probabilities as presented in supplemental Table S2 (http://jmd.amjpathol.org). The calculations of PPV and NPV combined three estimates of test sensitivity, test specificity, and the patients' pretest probability as described in previous literature.14,15 For each pretest probability, three ways of test interpretation were listed: calling positive when using TCRG or TCRB as a single test, calling positive only when both tests are positive, and calling positive when at least one test is positive. Both the PPV and 1-NPV (posterior probability of disease given a negative result) values increase as pretest probability increases in all three ways of interpretation, with “both positive” as the highest value and “at least one positive” as the lowest value. The relationships between pretest MF probability and PPV/1-NPV are plotted in Figure 1A1B1C.
Figure 1.
Plots showing relationships between pretest MF probability and PPV/1-NPV when TCRG and TCRB tests are used alone and together with different interpretations. x axis: Pretest probability of MF; y axis: PPV or (1-NPV). A:TCRG and TCRB tests when used alone had the same PPV and 1-NPV, so they are presented as the same plots here. B: Combined use of TCRG and TCRB: positive result was called only when both tests were positive, and a negative result when either test was negative. C: Combined use of TCRG and TCRB: positive result was called when at least one test was positive, and a negative result when both tests were negative.
The relative changes of MF probability when TCRG and TCRB tests are used alone and together with different diagnostic cut-offs are shown in Figure 2. The three curves plot the change in MF probability as a function of the pretest probability and test result. Over the range of pretest probability from 0.01 to 0.95, the “both positive” testing method increases MF probability most dramatically, the “TCRG or TCRB alone” method comes next, and the “at least one positive” method causes the least increase in MF probability. It can also be observed from Figure 2 that the largest increase in MF probability after a positive test result occurs when pretest probability ranges from 0.15 to 0.50. The largest decrease in MF probability after a negative test result occurs when pretest probability ranges from 0.50 to 0.75. These results suggest that when pretest probability is in the moderately low range (0.15 to 0.50), the more stringent diagnostic cut-off (requiring both tests to be positive) will provide the most clinically informative test performance, whereas when the pretest probability is in the moderately high range (0.50 to 0.75), the less stringent (at least one positive) diagnostic cut-off will provide the most clinically informative test performance.
Figure 2.
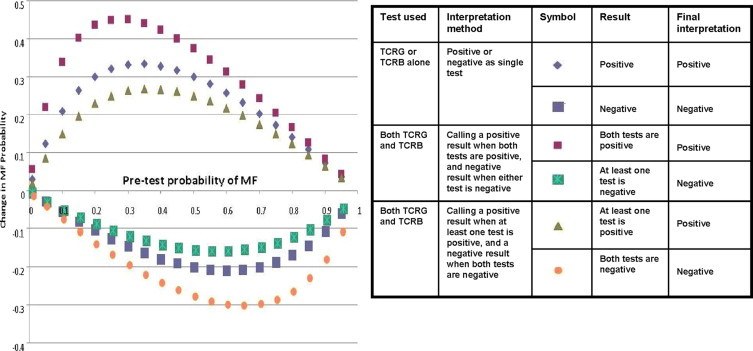
Plots showing changes of MF probability before and after TCR clonality tests (with different testing methods) in terms of positive and negative results (final interpretation). Change of MF probability when the test result was positive = PPV − pretest probability; Change of MF probability when test result was negative = 1 − NPV − pretest probability.
Discussion
The combined use of PCR-based TCRG and TCRB clonality tests to assist in the diagnosis of lymphoproliferative diseases has been proposed as a means to increase clonality testing sensitivity.4,13,16 With the advent of the BIOMED-2 concerted action BHM4-CT98-3936, standardized TCRG and TCRB clonality assays have become commercially available. Some validation work has been reported involving the combined use of the two assays.13,16 However, prior work studied several categories of T-cell malignancies and the data on MF was limited. Most published data are based on fresh or frozen tissue, whereas in clinical practice these tests are commonly performed using paraffin embedded tissue specimens. We assessed the performance characteristics of BIOMED-2 TCRG and TCRB clonality tests on paraffin embedded skin tissues diagnosed as MF or ID.
In our study, the concordance rate between the two tests was 77.2%. In theory, TCRG and TCRB tests can complement each other in detecting clonal rearrangements. The circumstances when TCRG and TCRB have concordant results, yet were at odds with the clinical diagnosis, are worthy of comment. For example, one 70-year-old patient with MF T4 classification was biopsied when the disease appeared to be worsening. However, both TCRG and TCRB tests on all three biopsies were negative. Another 48-year-old patient diagnosed as T1 MF was also tested negative with both TCRG and TCRB in three biopsies. Two other patients, diagnosed as either MF T3 or MF T4, had oligoclonal results from TCRG and TCRB tests in each of the 2 biopsies. The false negative results in these cases could have been attributable to technical limitations such as amount or quality of DNA or amplification efficiency, incorrect clinical diagnosis (especially in cases with short follow-up times), as well as defects in primer design (there is no primer for the Jγ1.2 [JγP] segment, which is used in a small percentage of T-cell lymphomas, particularly in the skin). Some other possible explanations for the false negative results of the T1 and T4 cases include the number of neoplastic cells present being below the threshold of sensitivity for PCR analysis, the primers did not anneal properly, there were partial or incomplete rearrangements of the TCR, or translocation involving the TCR region.9 Of perhaps greater concern are the false positive clonality results in patients with ID, leading to a risk of being incorrectly diagnosed and treated. These results emphasize the necessity of interpreting the results of clonality data in the context of all of the available clinical and pathological information, and being aware of the performance characteristics of T cell clonality studies using paraffin embedded tissue.
Sensitivity and Specificity
Before the BIOMED-2 assays became available, Klemke et al studied 41 patients with either early-stage MF or parapsoriasis using their own TCRG primers and protocol, as a result, a monoclonal T-cell infiltrate was demonstrated in paraffin-embedded lesional skin specimens in 19.2% of parapsoriasis cases and in 66.6% of early-stage MF cases.7 Van Dongen et al reported in 2003 that the combined usage of TCRB and TCRG tubes detected virtually all clonal T-cell populations,4 and Bruggemann et al reported in 2007 that the combined use of the two revealed two or more clonal signals in 95% of all TCR clonal cases (T-prolymphocytic leukemia, T-large granular lymphocytic leukemia, angioimmunoblastic T-cell lymphoma, anaplastic large cell lymphoma, peripheral T-cell lymphoma, unspecified) with fresh or frozen specimens.13 Nevertheless, as shown in Table 3, with paraffin-embedded T1 to T4 skin specimens from MF patients and when interpreting oligoclonal pattern as negative in the conventional way, we detected equal sensitivity of the two clonality tests at 64%; and the combined usage of TCRG and TCRB could increase the sensitivity to 78%. Specificity is also the same for TCRG and TCRB as single tests at 84% and can be maximized by calling clonal support only when both tests are positive. On the other hand, it is known that especially early MF can present with an oligoclonal pattern of T cells,11 thus we also looked at the effect on test characteristics by classifying oligoclonal pattern as positive. In this study cohort, the seven specimens with oligoclonal TCRG results were all in the MF group, among which six had known T-classification information (one T2, three T3, and two T4). Seven of the eight specimens with oligoclonal TCRB results were in the MF group (two T2, three T3, and two T4), and the remaining case was in the ID group. Because of the high association of oligoclonality with MF (although not specific to early MF) in this cohort, sensitivity is improved whereas the specificity is slightly decreased by the reclassification of oligoclonal pattern. Taken together, TCRG and TCRB showed no significant difference in either sensitivity or specificity as single tests, and the combined usage of the two leads to the question of how to appropriately interpret the results when there is discordance.
Correlation of TCR Clonality Tests and T-Classification of MF
Sandberg et al reported the equal clonality detection rates of TCRG and TCRB in both early stage (stage I to II, 5/8) and late stage (stage III to IV, 4/6) MF cases using BIOMED-2 primers and GeneScan analysis.17 With T-classification, our results showed that TCRB detected clonal expansion in six of ten T1 specimens, whereas TCRG only detected three of ten; however, TCRG performed better in T2 and T3 specimens than TCRB, and the two had the same detection rate in T4 specimens. Based on this dataset, adding TCRB test to TCRG test increased the clonal detection rate in T1 MF, but did not add value to T2 to T4 MF.
PPV, NPV, and Pretest Probability
While sensitivity and specificity characteristics are frequently used to assess test performance, PPV and NPV are also important and informative characteristics for clinicians. A previous Swiss study of 263 general practitioners showed that clinicians often confuse test sensitivity with positive predictive value, and as a result, tend to overvalue positive test result and undervalue clinical history. However, when they were given likelihood ratios, they correctly interpreted PPV.18 Pretest probability in this study refers to the probability that the patient under investigation has MF before TCR clonality test; PPV is the probability that a patient has MF given a positive test result; NPV is the probability that a patient does not have MF given a negative test result. The PPV and 1-NPV (posterior probability of disease given a negative result) over a range of pretest probabilities are visualized as plots in Figure 1. As the pretest probability increases, PPV and 1-NPV values gradually increase and form two curves. To further evaluate the change of MF probability before and after TCR clonality test given either positive or negative result with different pretest probabilities, the corresponding data were plotted in Figure 2. Figure 2 (as well as supplemental Table S2 at http://jmd.amjpathol.org) can serve as references for clinicians to estimate the change of MF probability by TCRG/TCRB clonality tests before ordering the test based on certain pretest probability, and accordingly, the value of performing the test.
With the information of change of MF probability by TCR clonality test in hand, we are able to look into the data for the answers to the following questions: Under what pretest probabilities will a TCR clonality test be most helpful, and which testing method will be most optimal to aid diagnosis in clinical settings? Based on our analysis, some recommended pretest probability cut-offs and optimal TCRG and/or TCRB testing methods are listed in Table 5.
Table 5.
Recommended Pretest Probability Cut-Offs and Optimal TCRG and/or TCRB Testing Methods
| Pretest MF probability | Recommended use of TCR clonality test(s) | Rationale |
|---|---|---|
| <0.15 | Of limited utility | Increment of PPV would not be adequate to change patient management |
| Little clinical significance | ||
| 0.15 to 0.50 | Combined use of TCRG and TCRB tests | Low pretest probability, specificity is more important |
| Both tests need to be positive for the TCR test to be reported as supporting clonal process | Maximize PPV and minimize possibility of false positive results (1-PPV) | |
| 0.50 to 0.75 | Combined use of TCRG and TCRB tests | Higher pretest probability, sensitivity is more important |
| At least one test needs to be positive for the TCR test to be reported as supporting clonal process | Maximize NPV and minimize possibility of false negative results (1-NPV) | |
| >0.75 | Of limited utility | Change of MF probability would not change patient management |
| Little clinical significance |
For those patients with intermediate pretest MF probabilities, TCR testing can significantly change the MF likelihood as shown in Figure 1. When pretest probability is in the moderately low range (15% to 50%), requiring both TCRG and TCRB to be positive may increase the PPV of the clonality study to a more clinically useful level (55% to 88%) than either test alone (41% to 80%) or than requiring at least one test to be positive (35% to 75%). When the pretest probability is in the moderately high range (50% to 75%), requiring at least one test to be positive may reduce the probability of false negatives to a more clinically useful level (23% to 47%) as compared with either test alone (30% to 56%) or requiring both tests to be positive (35% to 62%).
TCR clonality test is of limited utility when the pretest MF probability is very low or very high. When pretest MF probability is very low (<0.15), TCR clonality test, by any testing and interpretation method we analyzed, is of limited utility in terms of influencing clinical management of patients (Figure 1 and Supplemental Table S2 at http://jmd.amjpathol.org). For example, when pretest MF probability is 0.01, obtaining a positive test result from TCRG or TCRB alone can increase the probability to 0.04, and getting positive results from both can increase the probability to 0.07; however, a 3% or 6% increase in disease probability is of questionable clinical utility. When the pretest probability is less than 15%, the PPV of each of the three algorithms is less than 50%, and therefore, the majority of patients with positive studies will not have MF. If the pretest probability is greater than 75%, our data indicate that clonality testing will not result in a clinically significant change in probability of MF.
Strategy for Optimal Use of TCRG and TCRB Clonality Tests for Evaluation of Possibility of MF in Paraffin-Embedded Skin Sections
An international consensus panel has previously emphasized the importance of integrating clinical, immunological, and molecular pathology data for diagnosis of early mycosis fungoides.19 PCR methods have also evolved greatly since the publication of this consensus. Therefore, it is important to rigorously define the performance characteristics of the BIOMED-2 primer sets and high-resolution capillary electrophoresis methods for TCRG and TCRB tests. A general guideline of using BIOMED-2 protocols for PCR-based clonality diagnostic tests of suspected lymphoproliferations with an inconclusive diagnosis or with unusual histology, immunophenotype, or clinical presentation was proposed by van Krieken et al in 2007.16 This guideline covered the strategy of both B-cell and T-cell proliferations and suggested using TCRB and TCRG tests in parallel for suspected T-cell proliferations. In this study, based on the performance characteristics of TCRG and TCRB tests in the well-characterized cohorts, we calculated the PPV, 1-NPV, and the change of MF probability over a range of pretest probabilities from 0.01 to 0.95 (Figures 1 and 2). An algorithm demonstrating an application of these data is proposed in Figure 3—when a patient's pretest MF probability is very low (<0.15) or very high (>0.75), TCR clonality test is not recommended because the test will not impact clinical likelihood of disease; however, the test will provide useful information for patients with intermediate pretest MF probabilities.
Figure 3.
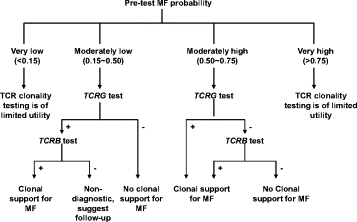
Strategy for optimal use of TCRG and TCRB clonality tests for evaluation of possibility of MF in paraffin-embedded skin sections.
When the patient's pretest probability for MF is moderately low (0.15 to 0.50), TCRG test would be performed as the primary test. Under this circumstance, if the TCRG test result is negative (polyclonal or oligoclonal), TCRB test is not necessary and an interpretation of no support for clonal process can be made; if the TCRG test result is positive (monoclonal), TCRB is suggested as a secondary test. A positive TCRB result in this setting would support the existence of clonal TCR rearrangement and clinical correlation is suggested, whereas a negative TCRB would be nondiagnostic and close follow-up is suggested.
When the patient's pretest probability for MF is moderately high (0.50 to 0.75), TCRG test is performed first. If the TCRG result is positive (monoclonal), then the TCRB test is not necessary, and the molecular test is considered to support the diagnosis of MF; if the TCRG result is negative (polyclonal or oligoclonal), TCRB testing is suggested as a secondary test. A clonal TCRB in this setting would be a molecular support for the diagnosis of MF, whereas a negative TCRB would mean no clonal support for MF.
While the algorithm requires prospective clinical validation, it integrates clinical information into molecular testing by maximizing sensitivity of the testing when the pretest probability is moderately high and maximizing specificity when the pretest probability is moderately low.
Conclusions
As single tests, PCR-based TCRG and TCRB clonality tests have no significant difference in terms of sensitivity and specificity in testing paraffin-embedded skin tissue specimens from MF and ID patients using commercially available BIOMED-2 assays. In our proposed algorithm, which integrates clinical information into molecular testing, the combined use of TCRG and TCRB clonality tests can maximize NPV when clinical suspicion is moderately high and maximize PPV when the pretest probability is moderately low.
Footnotes
Supported in part by NIH grant 5PO1CA49605 (to J.L.Z.) and the Departments of Pathology and Dermatology, Stanford University School of Medicine.
None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Web Extra Material
Clonality test results when using TCRG as the initial test and TCRB as an additional test.
Relation between pre-test probability, PPV, and 1-NPV with different TCRG and TCRB testing methods based on Bayes' theorem.
References
- 1.Florell SR, Cessna M, Lundell RB, Boucher KM, Bowen GM, Harris RM, Petersen MJ, Zone JJ, Tripp S, Perkins SL. Usefulness (or lack thereof) of immunophenotyping in atypical cutaneous T-cell infiltrates. Am J Clin Pathol. 2006;125:727–736. doi: 10.1309/3JK2-H6Y9-88NU-AY37. [DOI] [PubMed] [Google Scholar]
- 2.Hummel M, Stein H. Clonality and malignancy. PCR assays for the diagnosis of clonal B- and T-cell proliferations: potentials and pitfalls. Verh Dtsch Ges Pathol. 2003;87:102–108. [PubMed] [Google Scholar]
- 3.Cozzio A, French LE. T-cell clonality assays: how do they compare? J Invest Dematol. 2008;128:771–773. doi: 10.1038/jid.2008.49. [DOI] [PubMed] [Google Scholar]
- 4.van Dongen JJM, Langerak AW, Bruggemann M, Evans PAS, Jummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JHJM, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 5.Morgan SM, Hodges E, Mitchell TJ, Harris S, Whittaker SJ, Smith JL. Molecular analysis of T-cell receptor β genes in cutaneous T-cell lymphoma reveals Jβ1 bias. J Invest Dematol. 2006;126:1893–1899. doi: 10.1038/sj.jid.5700304. [DOI] [PubMed] [Google Scholar]
- 6.Cerroni L, Gatter K, Kerl H. An illustrated guide to skin lymphoma. Second edition. Wileyx-Blackwell; 2004. p. 21. [Google Scholar]
- 7.Klemke C-D, Dippel E, Dembinski A, Ponitz N, Assaf C, Hummel M, Stein H, Goerdt S. Clonal T cell receptor γ-chain gene rearrangement by PCR-based GeneScan analysis in the skin and blood of patients with parapsoriasis and early-stage mycosis fungoides. J Pathol. 2002;197:348–354. doi: 10.1002/path.1133. [DOI] [PubMed] [Google Scholar]
- 8.Langerak AW, Molina TJ, Lavender FL, Pearson D, Flohr T, Sambade C, Schuuring E, Saati TA, van Dongen JJM, van Krieken JHJM. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls. A report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia. 2007;21:222–229. doi: 10.1038/sj.leu.2404482. [DOI] [PubMed] [Google Scholar]
- 9.Thurber SE, Zhang B, Kim YH, Schrijver I, Zehnder J, Kohler S. T-cell clonality analysis in biopsy specimens from two different skin sites shows high specificity in the diagnosis of patients with suggested mycosis fungoides. J Am Acad Dermatol. 2007;57:782–790. doi: 10.1016/j.jaad.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Ponti R, Fierro MT, Quaglino P, Lisa B, Paola FdC, Michela O, Paolo F, Comessatti A, Novelli M, Bernengo MG. TCRγ-chain gene rearrangement by PCR-based GeneScan: diagnostic accuracy improvement and clonal heterogeneity analysis in multiple cutaneous T-cell lymphoma samples. J Invest Dematol. 2008;128:1030–1038. doi: 10.1038/sj.jid.5701109. [DOI] [PubMed] [Google Scholar]
- 11.Vega F, Luthra R, Medeiros LJ, Dunmire V, Lee S-J, Duvic M, Jones D. Clonal heterogeneity in mycosis fungoides and its relationship to clinical course. Blood. 2002;100:3369–3373. doi: 10.1182/blood.V100.9.3369. [DOI] [PubMed] [Google Scholar]
- 12.Jones D, Duvic M. The current state and future of clonality studies in mycosis fungoides. J Invest Dematol. 2003;121:ix. doi: 10.1046/j.1523-1747.2003.12458.x. [DOI] [PubMed] [Google Scholar]
- 13.Bruggemann M, White H, Gaulard P, Garcia-Sanz R, Gameiro P, Oeschger S, Jasani B, Ott M, Delsol G, Orfao A, Tiemann M, Herbst H, Langerak AW, Spaargaren M, Moreau E, Groenen PJTA, Sambade C, Foroni L, Carter GI, Hummel M, Bastard C, Davi F, Delfau-Larue M-H, Kneba M, van Dongen JJM, Beldjord K, Molina TJ. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies report of the BIOMED-2 concerted action BHM4-CT98-3936. Leukemia. 2007;21:215–221. doi: 10.1038/sj.leu.2404481. [DOI] [PubMed] [Google Scholar]
- 14.Soon SL, McCall CO, Chen SC. Computerized digital dermoscopy: sensitivity and specificity aren't enough. J Invest Dermatol. 2003;121:214–215. doi: 10.1046/j.1523-1747.2003.12316.x. [DOI] [PubMed] [Google Scholar]
- 15.Hagen M. How good is that test? Prim Care. 1995;22:213–223. [PubMed] [Google Scholar]
- 16.van Krieken JHJM, Langerak AW, Macintyre EA, Macintyre EA, Kneba M, Hodges E, Sanz RG, Morgan GJ, Parreira A, Molina TJ, Cabecadas J, Gaulard P, Jasani B, Garcia JF, Ott M, Hannsmann ML, Berger F, Hummel M, Davi F, Bruggemann M, Lavender FL, Schuuring E, Evans PAS, White H, Salles G, Groenen PJTA, Gameiro P, Pott C, van Dongen JJM. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 concerted action BHM4-CT98-3936. Leukemia. 2007;21:201–206. doi: 10.1038/sj.leu.2404467. [DOI] [PubMed] [Google Scholar]
- 17.Sandberg Y, Heule F, Lam K, Lugtenburg PJ, Wolvers-Tettero ILM, van Dongen JJM, Langerak AW. Molecular immunoglobulin/T-cell receptor clonality analysis in cutaneous lymphoproliferations. Experience with the BIOMED-2 standardized polymerase chain reaction protocol. Haematologica. 2003;88:659–670. [PubMed] [Google Scholar]
- 18.Steurer J, Fischer JE, Bachmann LM, Koller M, ter Riet G. Communicating accuracy of tests to general practitioners: a controlled study. BMJ. 2002;324:824–826. doi: 10.1136/bmj.324.7341.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pimpinelli N, Olsen EA, Santucci M, Vonderheid E, Haeffner AC, Stevens S, Burg G, Cerroni L, Dreno B, Glusac E, Guitart J, Heald PW, Kempf W, Knobler R, Lessin S, Sander C, Smoller BS, Telang G, Whittaker S, Iwatsuki K, Obitz E, Takigawa M, Turner ML, Wood GS, International Society for Cutaneous Lymphoma Defining early mycosis fungoides. J Am Acad Dermatol. 2005;53:1053–1063. doi: 10.1016/j.jaad.2005.08.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clonality test results when using TCRG as the initial test and TCRB as an additional test.
Relation between pre-test probability, PPV, and 1-NPV with different TCRG and TCRB testing methods based on Bayes' theorem.


