Abstract
Bisulfite sequencing measures absolute levels of DNA methylation at single-nucleotide resolution, providing a robust platform for molecular diagnostics. Here, we optimize bisulfite sequencing for genome-scale analysis of clinical samples. Specifically, we outline how restriction digestion targets bisulfite sequencing to hotspots of epigenetic regulation; we show that 30ng of DNA are sufficient for genome-scale analysis; we demonstrate that our protocol works well on formalin-fixed, paraffin-embedded (FFPE) samples; and we describe a statistical method for assessing significance of altered DNA methylation patterns.
Keywords: Epigenome profiling, epigenetics, bisulfite sequencing, human disease samples, cancer, biomarker development, molecular diagnostics, FFPE
The role of DNA methylation in human diseases has sparked interest in genome-scale methods for DNA methylation profiling1. Among an array of protocols for measuring DNA methylation, bisulfite sequencing stands out for its ability to quantify the DNA methylation status of essentially all non-repetitive regions in the genome at single-nucleotide resolution2. We recently developed reduced representation bisulfite sequencing (RRBS) as an accurate yet cost-efficient method for genome-scale DNA methylation analysis3,4. Here, we show that RRBS is highly appropriate for DNA methylation profiling of human disease cohorts, and we address four obstacles that hamper epigenome mapping in clinical samples: (i) High input DNA requirements. Methods such as MeDIP-seq5, MBD-seq6, Methyl-seq7 and CHARM8 consume micrograms of genomic DNA, which is infeasible for many clinical samples such as tumors obtained by laser capture microdissection or rare stem cell populations. (ii) Inability to analyze FFPE samples. We are not aware of a genome-scale method for DNA methylation mapping that works well on formalin-fixed, paraffin-embedded (FFPE) clinical samples, rendering many of the best-annotated patient cohorts inaccessible for epigenome studies. (iii) Incomplete bisulfite conversion. Whole-genome bisulfite sequencing cannot use specific primers to enrich for fully converted DNA, such that incomplete bisulfite conversion is likely to result in measurement artifacts. (iv) Lack of data analysis tools. Few statistical methods or bioinformatic tools exist that would allow sensitive detection of DNA methylation alterations that distinguish disease case and control samples.
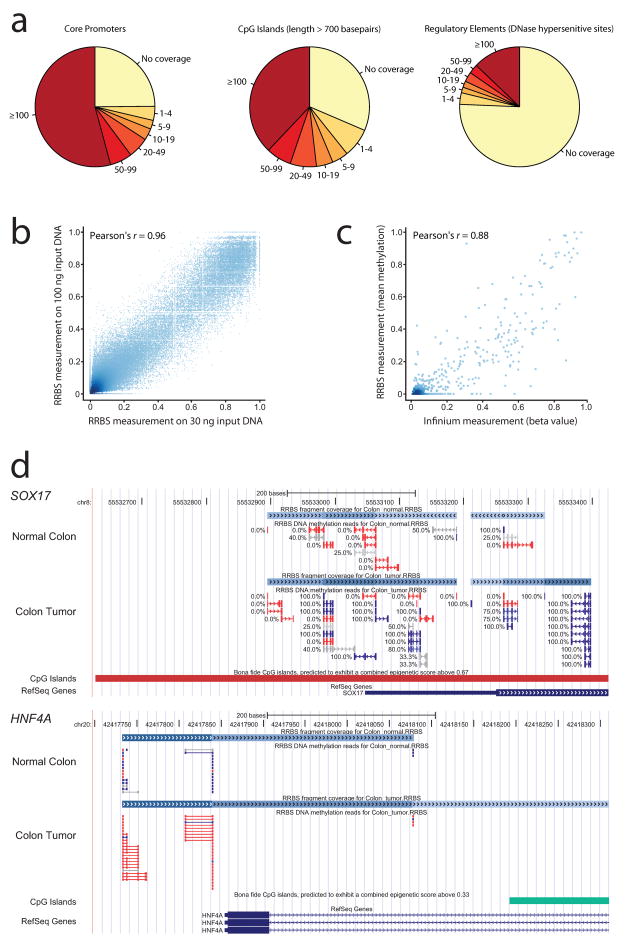
The RRBS protocol combines DNA digestion with a methylation-insensitive restriction enzyme and size selection to select a reproducible subset of the genome3,4. This ‘reduced representation’ is bisulfite-sequenced and its DNA methylation profile compared between disease cases and control samples. To translate the RRBS protocol from mouse to human, we initially performed in silico digestions, confirming that MspI digestion and a size selection of 40 basepairs to 220 basepairs enriches for CpG islands and promoter regions (data not shown). We tested this protocol on two fresh-frozen clinical samples, a colon tumor and adjacent normal tissue from the same patient. A total of 8.7 and 5.3 million high-quality aligned reads were obtained, yielding DNA methylation data for more than 1 million unique CpGs (Table 1). Highly quantitative data with more than 25 individual CpG measurements were obtained for 65% of core promoters, 50% of CpG islands and 17% of putative regulatory elements (Fig. 1a). Furthermore, we observed coverage of a sizable number of CpG island ‘shores’9, enhancers, exons, 3′ UTRs, and repetitive elements (see http://rrbs-techdev.computational-epigenetics.org for details). This constitutes a slight improvement compared to previously reported RRBS in mouse samples4.
Table 1.
Summary of reduced representation bisulfite sequencing experiments
| Run No. | Sample | Source material | Amount of input DNA | Bisulfite protocol | Bisulfite conversion rate | Number of lanes | Number of reads (total) | Number of reads (accepted) | Acceptance rate (reads) | Number of CpGs (total) | Number of CpGs (unique) | Mean CpG coverage |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Normal colon tissue | fresh-frozen tissue section | 1000 ng | 2×5 h | 99.3% | 4 | 32,072,287 | 5,344,273 | 16.7% | 10,190,227 | 1,134,963 | 9.0× |
| 2 | Colon tumor (matched pair with No. 1) | fresh-frozen tissue section | 1000 ng | 2×5 h | 99.5% | 4 | 40,015,958 | 8,747,180 | 21.9% | 16,891,325 | 1,297,296 | 13.0× |
| 3 | Normal colon tissue | FFPE tissue section | 1000 ng | 2×5 h | 98.7% | 2 | 25,635,110 | 7,099,198 | 27.7% | 19,040,643 | 1,375,696 | 13.8× |
| 4 | Colon tumor (matched pair with No. 3) | FFPE tissue section | 1000 ng | 2×5 h | 98.0% | 2 | 24,591,564 | 8,147,389 | 33.1% | 13,429,630 | 1,439,076 | 9.3× |
| 5 | Whole blood from healthy individual | fresh primary blood | 30 ng | 2×5 h | 99.5% | 1 | 6,761,136 | 3,151,765 | 46.6% | 8,145,565 | 840,860 | 9.7× |
| 6 | Whole blood from colon cancer patient | fresh primary blood | 30 ng | 2×5 h | 99.5% | 1 | 9,236,260 | 4,900,811 | 53.1% | 12,683,438 | 906,599 | 14.0× |
| 7 | In vitro methylated Jurkat cells | Cultured cell line | 250 ng | 2×5 h | 99.0% | 1 | 11,078,427 | 3,129,338 | 28.2% | 6,673,632 | 1,055,042 | 6.3× |
| 8 | In vitro methylated Jurkat cells | Cultured cell line | 250 ng | 14 h | 96.3% | 1 | 11,951,820 | 3,172,896 | 26.5% | 6,475,049 | 1,044,976 | 6.2× |
| 9 | In vitro methylated Jurkat cells | Cultured cell line | 250 ng | 5 h | 93.9% | 1 | 10,574,495 | 3,294,211 | 31.2% | 8,001,377 | 1,161,585 | 6.9× |
| 10 | 5-aza-cytidine demethylated Jurkat cells | Cultured cell line | 250 ng | 2×5 h | 99.2% | 1 | 15,241,239 | 3,900,421 | 25.6% | 8,051,251 | 1,192,898 | 6.7× |
| 11 | 5-aza-cytidine demethylated Jurkat cells | Cultured cell line | 250 ng | 14 h | 94.8% | 1 | 15,667,380 | 2,671,494 | 17.1% | 4,426,104 | 1,011,481 | 4.4× |
| 12 | 5-aza-cytidine demethylated Jurkat cells | Cultured cell line | 250 ng | 5 h | 92.0% | 1 | 16,371,085 | 4,383,094 | 26.8% | 8,387,319 | 1,229,928 | 6.8× |
| 13 | Normal colon tissue (same as No. 1) | fresh-frozen tissue section | 1000 ng | 14 h | 96.0% | 4 | 39,748,622 | 6,750,356 | 17.0% | 11,561,014 | 1,018,629 | 11.3× |
| 14 | Colon tumor (same as No. 2) | fresh-frozen tissue section | 1000 ng | 14 h | 98.1% | 4 | 43,324,210 | 8,292,609 | 19.1% | 14,171,997 | 1,068,498 | 13.3× |
| 15 | Mouse embryonic stem cells | Cultured cell line | 1000 ng | 14 h | 99.3% | 2 | 17,887,273 | 6,304,523 | 35.2% | 9,004,532 | 888,836 | 10.1× |
| 16 | Mouse embryonic stem cells | Cultured cell line | 300 ng | 14 h | 99.3% | 2 | 14,791,120 | 4,592,225 | 31.0% | 6,196,115 | 761,891 | 8.1× |
| 17 | Mouse embryonic stem cells | Cultured cell line | 100 ng | 14 h | 97.7% | 2 | 25,020,175 | 6,950,681 | 27.8% | 9,261,527 | 870,409 | 10.6× |
| 18 | Mouse embryonic stem cells | Cultured cell line | 30 ng | 14 h | 98.4% | 2 | 26,698,038 | 8,521,733 | 31.9% | 12,548,800 | 931,932 | 13.5× |
| TOTAL | 36 | 386,666,199 | 99,354,197 | 25.7% | 185,139,545 | 19,230,595 | 9.6× | |||||
Figure 1. Optimizing bisulfite sequencing for genome-scale profiling of human disease samples.
(a) Typical RRBS coverage of gene promoters (2-kilobase regions centered on RefSeq-annotated transcription start sites), CpG islands (annotated with a stringent version of the Gardiner-Garden criteria, requiring a minimum length of 700 basepairs) and putative regulatory elements (mapped by DNase hypersensitivity). All data are for colon tumor, run no. 1 in Table 1. The values refer to the number of individual CpG measurements in the region that pass quality control. Correlation between (b) RRBS performed on 30 ng and 100 ng of input DNA derived from mouse ES cells and (c) DNA methylation measurements for colon tumor, obtained by RRBS and Infinium, at 1,027 high confidence CpGs (RRBS: sequencing coverage ≥ 20, Infinium: detection p-value < 0.05). (d) Distribution of DNA methylation in the promoter regions of SOX17 (hypermethylated in the colon tumor) and HNF4A (hypomethylated). Unmethylated reads are shown in red, partially methylated reads in grey and methylated reads in blue.
For the analysis of clinical samples, three aspects of the RRBS protocol were specifically optimized. First, we minimized the input DNA requirement to be able to process minimal tissue samples and FACS-sorted cell populations (Fig. 1b). In two subsequent rounds of optimization we reduced the amount of input DNA from 1 μg to 300 ng and from 100 ng to 30 ng (Table 1), observing Pearson correlation coefficients of 0.97 and 0.96, respectively, calculated over all CpGs with at least 25-fold sequencing coverage. This analysis was performed on DNA from mouse ES cells rather than on human material to minimize the number of potential confounding factors. To confirm that the low-input protocol works well for human disease samples, we performed RRBS on two human blood samples using 30 ng of input DNA, and we observed a correlation of 0.96 between the two samples (Supplementary Table 1).
Second, we optimized RRBS analysis for DNA extracted from FFPE tissue slices. Focusing on two matched colon samples that were stored in FFPE format since 2001, we observed the characteristic DNA degradation pattern of FFPE samples (Supplementary Fig. 1a). To avoid degradation products in the selected size range (40–220bp), we size-selected DNA fragments greater than 500 basepairs before digesting the genomic DNA with MspI. Our protocol resulted in high-quality RRBS libraries (Supplementary Fig. 1b), and the sequencing yield was comparable to fresh-frozen samples (Table 1). We also observed high overall agreement between the FFPE samples and the fresh-frozen samples in terms of genomic coverage and DNA methylation measurements (Supplementary Fig. 2). Specifically, the correlation of DNA methylation levels at CpGs with at least 25-fold sequencing coverage was 0.87 between the fresh-frozen and the FFPE colon tumor and 0.88 between the fresh-frozen and the FFPE normal colon tissues (Supplementary Table 1).
Third, we optimized bisulfite treatment in order to maximize conversion of unmethylated cytosines while minimizing loss of input DNA due to bisulfite-induced degradation. Across multiple experiments in clinical samples and mouse ES cells, we found a conversion protocol with two subsequent 5-hour bisulfite treatments10 was more effective than our previously used single-step 14-hour protocol (conversion rate >99% in all experiments). We also performed RRBS on in vitro methylated and in vivo demethylated DNA from a single cell line. This experiment confirmed that the overall level of DNA methylation does not have a visible effect on the bisulfite conversion rate (Table 1). Finally, we compared the DNA sequence properties (sequence composition, structural features, repeat content, etc.) between the regions that exhibited comparatively low vs. high levels of bisulfite conversion, using the EpiGRAPH web service11. No consistent correlation with the bisulfite conversion rates could be identified (data not shown), suggesting that systematic bisulfite conversion bias is not a problem when applying RRBS to human disease samples.
As an additional validation, we performed DNA methylation analysis of the fresh-frozen colon tumor sample using the Infinium HumanMethylation27 platform, which combines bisulfite conversion with a genotyping microarray to measure DNA methylation in promoter regions12. For 1,027 CpGs both methods yielded high-confidence measurements, and we observed a correlation of 0.88 between Infinium and RRBS (Fig. 1c). Furthermore, when we allowed for up to 100 basepairs distance between the CpGs assayed by Infinium and RRBS, the high-confidence overlap between both methods increased to 7,324 CpGs, while the correlation between the two assays remained high (Pearson’s r = 0.77). This observation is consistent with high autocorrelation of DNA methylation levels in the CpG-rich regions of the human genome13,14 and provides justification for measuring DNA methylation at a subset of indicator CpGs, rather than at every single CpG within a given region.
To complement the experimental optimizations described above, we developed a bioinformatic data analysis pipeline that is designed to identify subtle alterations of DNA methylation in genomic regions with putative gene-regulatory potential (Supplementary Note). This pipeline builds upon a comprehensive set of pre-annotated genomic regions (which includes promoters, CpG islands and many other genomic features). For each region it performs a statistical test for differential DNA methylation, and it calculates p-values without having to introduce any arbitrary threshold parameters. Multiple-testing correction is performed by controlling the false discovery rate. Importantly, restricting the analysis to a relevant subset of the genome increases the statistical power for detecting subtle alterations in gene-regulatory regions, because the p-values are not diluted by multiple-testing correction for regions that are a priori unlikely to be differentially methylated.
To illustrate the features of the bioinformatic analysis pipeline, we compared the DNA methylation profile of the colon tumor with matched normal colon tissue. We observed tumor-specific hypermethylation at 52 gene promoters, 114 CpG islands and hundreds of additional genomic regions. Affected genes include SOX17 (Fig. 1d) and GATA5 (Supplementary Fig. 3), which are known targets of hypermethylation in colon cancer15,16. However, classical targets such as APC and MGMT were unmethylated in this particular tumor. To corroborate the observation that the tumor exhibits hypermethylation at a relatively small number of genes, we assessed whether or not the tumor classifies as CpG island methylator phenotype (CIMP) based on a recently published biomarker17. CIMP is a characteristic property of a subset of colon cancers exhibiting widespread DNA methylation at a large number of CpG island promoters. We inspected the promoters of five genes that have been identified as predictive of CIMP17, and the RRBS data clearly denote the tumor as CIMP-negative. In addition to hypermethylation at a small but significant number of gene promoters, we also observe cases of tumor-specific hypomethylation. An example is HNF4A (Fig. 1d), a hepatic transcription factor that has an essential role in colon development18.
The RRBS method’s deep coverage of gene promoters plus selective sampling of all other types of genomic regions makes it most useful for detecting novel epigenetic alterations, for example in the context of biomarker discovery19. Compared to truly genome-wide bisulfite sequencing, its focus on a reduced representation of the genome translates into a substantial cost advantage and the ability to screen larger patient cohorts. On the other hand, padlock-targeted bisulfite sequencing and epigenotyping microarrays currently achieve substantially lower genomic coverage, making these technologies more suitable for validating findings than for initial discovery. In terms of sample quality and input DNA requirements, RRBS is more forgiving than any other method for epigenome profiling that we are aware of. It is thus possible to run RRBS as an add-on for essentially all ongoing tumor genomics initiatives, and to generate genome-wide methylation profiles of some of the most interesting and best-annotated sample collections. Finally, with ever-decreasing sequencing costs RRBS will readily scale to more comprehensive genomic coverage, for example, by using additional restriction enzymes or widening the size-selection window.
Online Methods
Sample origin and DNA extraction
DNA for a primary colon tumor and adjacent normal colon tissue was purchased from BioChain (lot number A704198). Both samples came from the same donor, an 81-year-old male patient diagnosed with moderately differentiated adenocarcinoma. In vitro methylated and 5-aza-cytidine demethylated Jurkat genomic DNA samples were obtained from New England Biolabs (NEB). Formalin fixed, and paraffin-embedded (FFPE) colon carcinoma and matching normal tissue blocks were purchased from OriGene Technical Inc. These samples were derived from an 89-year-old male patient and FFPE-processed in 2001. Genomic DNA was isolated using a RecoverALL total nucleic acid isolation kit (Applied Biosystems/Ambion) according to the manufacture’s recommendation. After purification, degraded genomic DNA was size-selected on a 0.8% agarose gel. DNA fragments larger than 500 basepairs were extracted using a QIAGEN gel purification kit. Genomic DNA from human blood cells was extracted as described previously20. To obtain comparable, high-quality DNA and to remove residual cellular proteins, all commercial DNA samples were further purified using a standard phenol:chloroform:isoamyl alcohol (25:24:1) protocol20. Mouse ES cells were cultured according to established protocols4. All cells were grown on 0.2% gelatin for at least two passages before isolation of DNA. Mouse genomic DNA was extracted as described previously20.
RRBS library construction
30 ng to 1 μg of human or mouse genomic DNA was digested with 5 to 20 units of MspI (NEB) in a 20 μl reaction for 16 to 20 hours at 37 °C. Digested DNA was phenol:chloroform:isoamyl alcohol purified as described above and DNA pellets were resolved in 10 μl EB buffer for end-repair. Digested DNA was filled in and adenylated in a 50 μl reaction containing 10 units of Klenow fragment (3′ → 5′ exo−, NEB), 40 μM dGTP, 40 μM 5′ methylated dCTP (Roche), 400 μM dATP and 1× NEB buffer 2. The reaction was incubated 20 min at 30 °C followed by 20 min at 37 °C. Purified adenylated DNA fragments were ligated with pre-annealed synthetic 5-methylcytosine-containing Illumina adapters in a 50 μl reaction consisting of 2,000 cohesive end units of T4 ligase (NEB), 10 μl adenylated DNA and 0.5 to 1.0 μM of the adapter for 16 to 20 hours at 16 °C. Before size selection, the adapter-ligated DNA was purified following the standard phenol:chloroform:isoamyl alcohol protocol. Size selection was conducted as described previously20. Briefly, purified DNA was run on a 3% NuSieve 3:1 agarose gel until the bromophenol blue within the loading dye had run for 4 to 5 cm. To obtain 40 to 120 basepair and 120 to 220 basepair MspI digested genomic DNA fragments, we excised adapter-ligated fragments that run at 150 to 230 basepair and 230 to 330 basepair, respectively. (Due to special characteristics of the Illumina adapters, the lengths of the final DNA fragments are not directly additive.) To generate the 30 ng input RRBS library, 50 ng sheared and dephosphorylated Escherichia coli K12 genomic DNA was used as a carrier for gel size-selection and subsequent bisulfite conversion. Size-selected DNA was bisulfite-treated using EpiTect Bisulfite Kit (Qiagen). To validate and improve bisulfite conversion conditions of human genomic DNA, we tested different bisulfite conversion protocols: (i) the manufacturer’s standard 5-hour conversion protocol (99 °C for 5 min, 60 °C for 25 min, 99 °C for 5 min, 60 °C for 85 min, 99 °C for 5 min, 60 °C for 175 min); (ii) two rounds of the standard conversion10; (iii) an approximately 14-hour phase conversion which included three additional cycles of 5 min of denaturation at 95 °C followed by 3 h at 60 °C after the 5-hour phase standard conversion4,20. Bisulfite-converted DNA was eluted twice from the EpiTect spin column with 20 μl pre-heated EB buffer per elution. Analytical (10 μl) PCR reactions containing 0.5 μl of bisulfite-treated DNA, 0.2 μM each of Illumina PCR primers LPX1.1 and 2.1 and 0.5 U PfuTurbo Cx Hotstart DNA polymerase (Stratagene) were set up to determine the minimum number of PCR cycles. Reactions were performed under the following thermocycler conditions: 5 min at 95 °C, n × (20 s at 95 °C, 30 s at 65 °C, 30 s at 72 °C), followed by 7 min at 72 °C, with n ranging from 12 to 18 cycles. The final libraries were generated by large-scale amplification (8 × 25 μl) with each 25 μl aliquot containing 2 to 4 μl of bisulfite-converted template, 1.25 U PfuTurbo Cx Hotstart polymerase, and 0.2 μM each of Illumina LPX1.1 as well as 2.1 PCR primers. PCR was performed using the same PCR profile as in the analytical protocol. QIAquick-purified PCR products were subjected to a final size-selection step on a 3% NuSieve 3:1 agarose gel. SYBR-green-stained gel slices containing adaptor-ligated fragments were excised. RRBS library material was recovered from the gel (QIAquick) and quantified by a Quant-iT (Invitrogen) assay.
Sequencing and alignment
RRBS libraries were sequenced on the Genome Analyzer II (Illumina) using the established single-end sequencing protocol. Genomic alignment was performed as described previously4. Briefly, two reference sequences of size-selected MspI fragments are constructed in silico, one in which the genomic sequence is maintained as is, and one in which all Cs are replaced by Ts (the latter reflects complete bisulfite conversion of cytosines into thymines). During the alignment, residual Cs within each read are also converted into Ts and aligned against the reference sequence that consists of all converted MspI fragments. The alignment itself uses a straightforward seed-and-extension algorithm, which identifies all perfect 12 bp alignments and extends without gaps from either end of the established seed. Mismatches are counted and used as a quality measure. However, C-to-T conversions between the genomic DNA sequence and the read are not counted as mismatches but marked for downstream methylation calling. The best alignment is kept only in cases where the second-best alignment has at least three more mismatches, while reads that do not meet this stringency criterion are discarded. Bisulfite conversion rates are calculated as the number of genomic cytosines outside a CpG context that are unconverted, divided by the total number of cytosines outside a CpG context.
Bioinformatic analysis
Disease-specific epigenetic alterations are typically more subtle than tissue-specific differences and changes related to cell differentiation, to which RRBS was originally applied4. We therefore developed a bioinformatic pipeline that scores epigenetic alterations according to strength and significance, and links them to potentially affected genes. To that end, we collected a comprehensive set of regions of interest, which includes promoters, CpG islands, CpG island shores, enhancers, exons, introns, and repetitive elements. For each of these regions, the number of methylated and unmethylated CpG observations is determined, and a p-value is assigned using Fisher’s exact test. Once all p-values are calculated, multiple-testing correction is performed separately for each region type using the q-value method21, which controls the false discovery rate to be below a user-specified threshold (typically 10%). A simple power calculation shows that – for a given candidate region with coverage of 50 individual CpG measurements (Fig. 1a) – the power to detect a difference of 0% methylation vs. 20% methylation is 95% (significance level of 0.05). The software pipeline is implemented in Python (alignment processing module) and R (statistical analysis module). The source code package with documentation and demonstration data is available online (Supplementary Note). Future updates will be posted on http://rrbs-techdev.computational-epigenetics.org.
Supplementary website
Using the software described in the previous section, we generated a comprehensive analysis of the RRBS experiments described in this paper. The supplementary website hosts all relevant results, including genome browser tracks visualizing the raw DNA methylation data; coverage statistics, mean methylation boxplots and clustering diagrams that provide an initial overview; and scatterplots, pairwise correlations as well as statistical significance calls that facilitate in-depth analysis. The supplementary website is available from the following URL: http://rrbs-techdev.computational-epigenetics.org.
Supplementary Material
Supplementary Figure 1. Library generation from FFPE samples
Supplementary Figure 2. DNA methylation in fresh-frozen and FFPE colon samples
Supplementary Table 1. Pairwise correlation of DNA methylation among the patient samples
Supplementary Note. Source code of Epigenome pipeline package with documentation and demonstration data
Acknowledgments
We thank K. Halachev (Max Planck Institute for Informatics) for the provision of genome annotation files and H. Cedar (The Hebrew University of Jerusalem) for providing the human blood DNA samples. C. Bock is supported by a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation. A. Meissner is supported by the Massachusetts Life Science Foundation and the Pew Charitable Trusts. The described work was in part funded by NIH grants R01HG004401, U54HG03067 and U01ES017155.
Footnotes
Author contributions
Conceived and designed the experiments: H. Gu, C. Bock, A. Gnirke, E. Lander, A. Meissner. Performed the experiments: H. Gu, E. Tomazou. Analyzed the data: C. Bock. Contributed reagents/materials/analysis tools: T. Mikkelsen, Z. D. Smith, N. Jäger. Wrote the paper: H. Gu, C. Bock, A. Meissner.
Competing interests
The authors declare no competing financial interests.
References
- 1.Esteller M. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 2.Lister R, Ecker JR. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meissner A, Gnirke A, Bell GW, et al. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meissner A, Mikkelsen TS, Gu H, et al. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Down TA, Rakyan VK, Turner DJ, et al. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serre D, Lee BH, Ting AH. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp992. published online November 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunner AL, Johnson DS, Kim SW, et al. Genome Res. 2009;19:1044–1156. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry RA, Ladd-Acosta C, Wen B, et al. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R, O’Malley RC, Tonti-Filippini J, et al. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock C, Halachev K, Büch J, et al. Genome Biol. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Le J, Barnes B, et al. Epigenomics. 2009;1:177. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 13.Bock C, Walter J, Paulsen M, et al. Nucleic Acids Res. 2008;36:e55- . doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckhardt F, Lewin J, Cortese R, et al. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellebrekers DM, Lentjes MH, van den Bosch SM, et al. Clin Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, Glockner SC, Guo M, et al. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberger DJ, Siegmund KD, Campan M, et al. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 18.Garrison WD, Battle MA, Yang C, et al. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bock C. Epigenomics. 2009;1:99. doi: 10.2217/epi.09.6. [DOI] [PubMed] [Google Scholar]
- 20.Smith ZD, Gu H, Bock C, et al. Methods. 2009 doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storey JD, Tibshirani R. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Library generation from FFPE samples
Supplementary Figure 2. DNA methylation in fresh-frozen and FFPE colon samples
Supplementary Table 1. Pairwise correlation of DNA methylation among the patient samples
Supplementary Note. Source code of Epigenome pipeline package with documentation and demonstration data