Summary
We report that, in a simple, static culture system, wild-type Vibrio cholerae El Tor forms a three-dimensional biofilm with characteristic water channels and pillars of bacteria. Furthermore, we have isolated and characterized transposon insertion mutants of V. cholerae that are defective in biofilm development. The transposons were localized to genes involved in (i) the biosynthesis and secretion of the mannose-sensitive haemagglutinin type IV pilus (MSHA); (ii) the synthesis of exopolysaccharide; and (iii) flagellar motility. The phenotypes of these three groups suggest that the type IV pilus and flagellum accelerate attachment to the abiotic surface, the flagellum mediates spread along the abiotic surface, and exopolysaccharide is involved in the formation of three-dimensional biofilm architecture.
Introduction
Bacteria in aquatic environments are rarely found in the planktonic or free-swimming phase (Costerton et al., 1987). Rather, they are found in association with a solid surface. Attached bacteria may take the form of a dispersed monolayer of cells on a surface, they may be clustered on the surface in microcolonies, or they may be organized into a three-dimensional biofilm (Costerton et al., 1995).
It has been suggested that attachment to surfaces and formation of a biofilm may provide an adaptive advantage for aquatic organisms. For example, less soluble and less easily metabolizable large organic compounds (i.e. humic acids), which are found adsorbed to aquatic surfaces, may provide nutrients for attached bacteria (Mills and Powelson, 1996). Alternatively, attachment to surfaces, such as the exoskeletons of crustaceans or insects, which are made out of chitin, may provide nutrition directly to organisms, such as Vibrio cholerae, which are able to metabolize chitin (Fletcher, 1996). Formation of a biofilm may also provide protection from toxic compounds, such as antibiotics, which are present in the environment (Anwar et al., 1992; Vess et al., 1993). Biofilm formation may therefore be a survival mechanism for bacteria and other microbes living in an aquatic environment.
Biofilm development is a multistep process. Bacteria approach the surface, attach and then become immobilized on the surface. In order to form microcolonies, the bacteria move along the surface and associate with one another. Finally, an ordered three-dimensional structure is formed. This structure is composed of pillars of bacteria surrounded by water channels that allow nutrients to reach biofilm-associated bacteria and allow toxic metabolites to diffuse out of the biofilm (Costerton et al., 1995).
The genetic basis of colonization of abiotic surfaces has been studied in Escherichia coli (Pratt and Kolter, 1998) and Pseudomonas aeruginosa (O’Toole and Kolter, 1998a). In E. coli, both type I pili and flagella are important for attachment to the surface, while flagella alone are important for bacterial movement on a surface. In contrast, in P. aeruginosa, flagella are important for initial attachment to the surface, while microcolony formation does not occur in the absence of type IV pili, presumably because of a lack of twitching motility. Twitching is a type of surface motility that is thought to occur through anchoring of a pilus to a fixed surface, followed by retraction of the pilus against the surface (Wall and Kaiser, 1999).
Biofilm formation has been described as a developmental process (Costerton et al., 1995; O’Toole and Kolter, 1998). In support of this, Davies et al. (1998) have shown that an acylated homoserine lactone functions as a developmental signal in the formation of the characteristic three-dimensional biofilm architecture in P. aeruginosa. Furthermore, the expression of alginate, an exopolysaccharide present as an extracellular matrix in P. aeruginosa biofilms, is induced upon contact with a surface (Davies et al., 1993; Hoyle et al., 1993).
We are interested in understanding the impact that the ability to form a biofilm has on the emergence of human pathogens that are also natural inhabitants of the aquatic environment. V. cholerae, a Gram-negative rod responsible for the severe diarrhoeal disease known as cholera, is a prototype of such pathogens. During recorded history, two biotypes of V. cholerae have devastated large regions of the world in seven pandemics (Colwell, 1996). The classical biotype is thought to have caused the first six pandemics. Currently, V. cholerae El Tor is the dominant biotype of V. cholerae worldwide and is responsible for the seventh pandemic of cholera. It has been hypothesized that the adaptation of V. cholerae to survival in estuarine and freshwater aquatic environments is essential to its persistence as a pandemic pathogen (Colwell and Spira, 1992). Because attachment of V. cholerae to aquatic surfaces is believed to be adaptive, and many studies have demonstrated attachment of V. cholerae to environmental surfaces such as plants, filamentous green algae, zooplankton, crustaceans and insects (Huq et al., 1986; Tamplin et al., 1990; Shukla et al., 1995), we hypothesize that biofilm formation may be an important factor in the survival of V. cholerae in the aquatic environment.
A study of the genetic basis of biofilm development by V. cholerae El Tor is necessary to begin to unravel the role played by biofilm formation in the emergence of this pathogen. Recently, the type IV pilus, MSHA, which is responsible for mannose-sensitive haemagglutination by V. cholerae El Tor, has been implicated in biofilm formation on non-nutritive, abiotic surfaces (Watnick et al., 1999). In addition, studies have clearly demonstrated that the colony morphology of the rugose form of V. cholerae El Tor, which forms thicker biofilms than non-rugose El Tor, results from an exopolysaccharide (EPS) encoded by the vps locus (Yildiz and Schoolnik, 1999). The steps in the development of large, three-dimensional structures of V. cholerae on abiotic surfaces, however, have not been defined. In this work, we undertake a genetic and microscopic analysis of the development of a V. cholerae El Tor biofilm.
Results
A simple experimental system for the development of three-dimensional biofilms
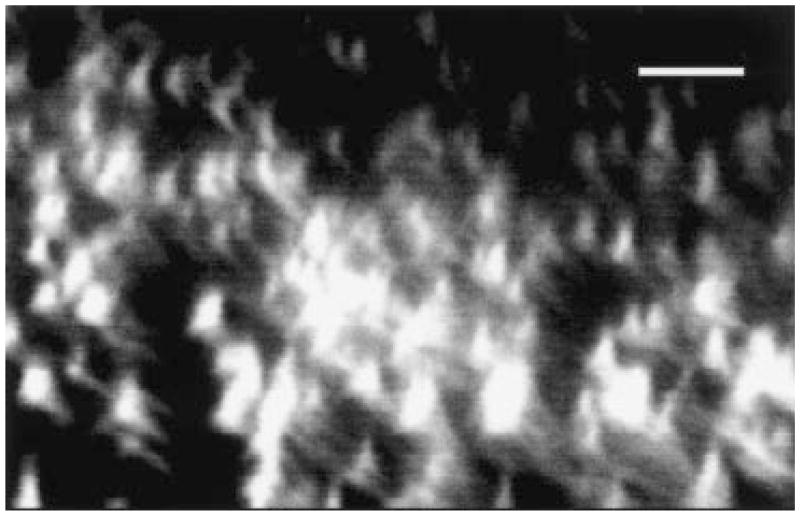
Flow cells have generally been used for the growth and study of three-dimensional biofilms (Lawrence et al., 1991). Such systems, however, require a specialized apparatus, maintenance of sterility over weeks and a large amount of liquid medium. In the experiments reported here, we placed a glass coverslip in a conical tube partially filled with medium, inoculated the medium with the strain of interest and incubated the bacteria with the coverslip surface without agitation. The coverslip was then removed from the medium for observation. For V. cholerae, such an experimental design results in the development of a three-dimensional biofilm with pillars of bacteria and water channels (Fig. 1). Such biofilms reach a thickness of ≈ 25 μM after 5 days of incubation at room temperature. Although the flow cell is an excellent means of assessing biofilm formation, the systems described here greatly simplify the processing of a large number of strains or mutants.
Fig. 1.
Confocal scanning laser micrograph through the xz-plane of a 5-day-old biofilm of V. cholerae. Biofilms were formed with V. cholerae that constitutively express GFP from a plasmid. Thus, bright areas represent biofilm-associated bacteria. The substratum is located at the bottom of the micrograph. Bar = 5 μm.
Genetic screen for biofilm-deficient mutants
A screen for biofilm-deficient mutants of V. cholerae El Tor was undertaken. Briefly, transposon mutagenesis was used to create a library of mutants that was then screened for the ability to form biofilms in the wells of a polyvinylchloride (PVC) microtitre dish as described in Experimental procedures. Of the 10 000 mutants screened, 80 were operationally defined as defective in biofilm formation because of their inability to produce a crystal violet ring when stained in the microtitre dish assay. All of these mutants were screened for defects in flagellar-mediated motility. Nine of the 80 mutants were non-motile using the motility agar assay. The sequence of the transposon junctions of five of these mutants was determined. All five were shown to have insertions in known flagellar structural or motility genes. Sequence analysis of arbitrary polymerase chain reaction (PCR) products showed that transposon insertions in 45 mutants mapped to various genes involved in the production of a functional MSHA pilus. These results were confirmed by haemagglutination assays. Twenty-six additional mutants were both motile and able to agglutinate red blood cells. Sequence analysis of arbitrary PCR products demonstrated that five of these mutants had transposon insertions in the vps locus, which is responsible for exopolysaccharide production (Yildiz and Schoolnik, 1999). Six mutants had transposon insertions in genes of unknown function, and we were unable to obtain transposon insertion sequences from 15 additional mutants.
Description of mutant groups
Table 1 shows representative mutants of the three groups isolated, namely, mutants in the MSHA biogenesis and secretion pathways (MSHA), mutants in synthesis of exopolysaccharide (EPS) and flagellar mutants (MOT).
Table 1.
Representative biofilm mutants.
| Mutant group | Identity/similarity (organism) | Function |
|---|---|---|
| Mannose-sensitive haemagglutination | MshA (V. cholerae) | Attachment to abiotic surfaces (Watnick et al., 1999) |
| PilT (P. aeruginosa) | Twitching motility (Whitchurch et al., 1991) | |
| Exopolysaccharide production | WcaJ (E. coli ) | Colanic acid biosynthesis (Stevenson et al., 1996) |
| EpsD (Ralstonia solanacearum) | Exopolysaccharide biosynthesis (Huang and Schell, 1995) | |
| Cps19fF (Streptococcus pneumoniae) | Putative UDP-N-acetyl-D-mannosamine transferase (Morona et al., 1997) | |
| Motility | FlrC (V. cholerae) | σ54 transcriptional activator (Klose and Mekalanos, 1998) |
| MotY (V. alginolyticus) | Sodium-driven flagellar motor (Okunishi et al., 1996) | |
| FlgF (E. coli ) | Flagellar basal body rod protein (Homma et al., 1990; Blattner et al., 1997) |
The first group of mutants included transposon insertions in many open reading frames (ORFs) of the operons controlling both biosynthesis and secretion of the MSHA pilus (Marsh and Taylor, 1999). All these mutants failed to agglutinate red blood cells. The MSHA group also included two mutants with independent transposon insertions in a gene with 66% identity in protein sequence to the pilT gene of P. aeruginosa. PilT, which has also been identified in Myxococcus xanthus and Neisseria gonorrhoeae, is believed to be responsible for the retraction of type IV pili (Whitchurch et al., 1991; Wu et al., 1997; Wolfgang et al., 1998). In these three bacteria, although pilT mutants are hyperpiliated and produce pili that are indistinguishable from wild-type pili when visualized by electron microscopy, they do not exhibit twitching motility. Using electron microscopy of negatively stained V. cholerae to study multiple specimens, we were able to detect single pili on occasional wild-type cells and multiple pili on occasional pilT mutant cells (data not shown). The frequency of piliated MSHA mutant cells was ≈ 20-fold less, possibly suggesting that another pilus may be present infrequently under these conditions. In V. cholerae El Tor, however, we have thus far been unable to demonstrate MSHA-based twitching motility by standard experimental techniques. We hypothesize, however, by analogy with P. aeruginosa, N. gonorrhoeae and M. xanthus, that MSHA pili do impart twitching motility to V. cholerae and that pilT mutants of V. cholerae possess MSHA pili that are non-functional. MSHA pili, however, may not be able to generate sufficient force to effect twitching under the conditions we have tested.
The second group of mutants was defective in flagellar motility (MOT). Based on similarity to previously studied flagellar genes, this group included both mutants lacking flagella and mutants with paralyzed flagella. The predicted presence or absence of flagella was confirmed by electron microscopy. These mutants were also unable to agglutinate red blood cells. The inability of non-motile mutants of classical V. cholerae to haemagglutinate has been reported previously (Gardel and Mekalanos, 1996). Using electron microscopy of negatively stained V. cholerae, no pili were observed on non-motile cells, suggesting that motility affects the production of MSHA pili.
The third group of biofilm-deficient mutants had transposon insertions in genes involved in exopolysaccharide biosynthesis (EPS). Two of these genes have been identified previously as essential for exopolysaccharide production in the rugose form of V. cholerae El Tor (Yildiz and Schoolnik, 1999). All these mutants were able to agglutinate red blood cells in a mannose-sensitive fashion, indicating the presence of functional MSHA pili.
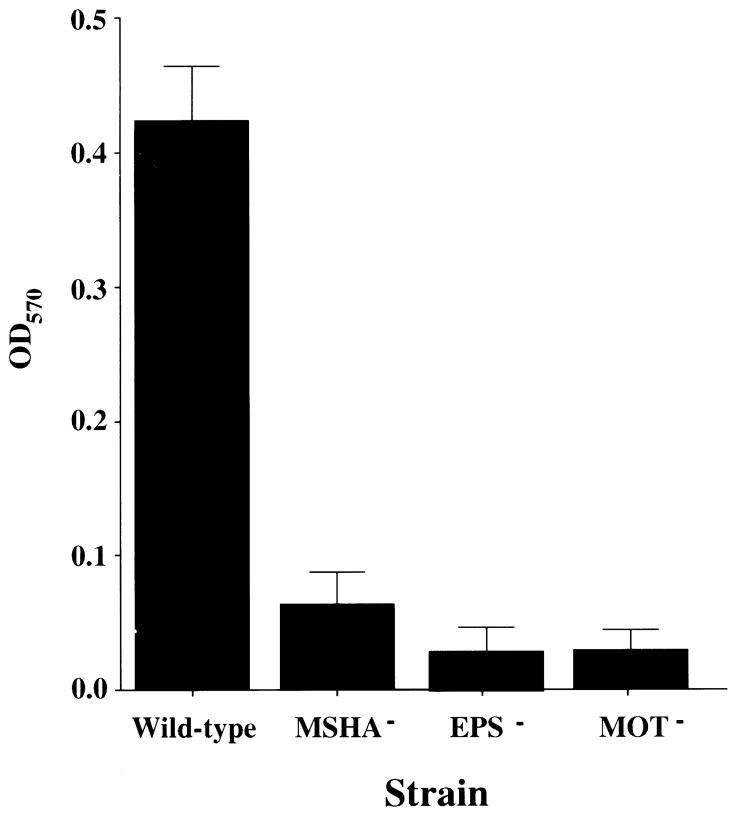
A crystal violet quantification of the biofilms formed by representative mutants on borosilicate glass after a 24 h incubation is shown in Fig. 2. This demonstrates (i) that biofilm formation defects of the mutants from all these classes are virtually indistinguishable by crystal violet staining at 24 h; and (ii) that mutants defective in biofilm formation on PVC are also defective on glass.
Fig. 2.
Quantification of bacteria in biofilms formed by wild-type V. cholerae, an MSHA mutant (MSHA), an exopolysaccharide mutant (EPS) and a motility mutant (MOT). The biofilms were stained with crystal violet. Crystal violet was solubilized, and the optical density of the resultant solution was measured.
Rates of biofilm formation by biofilm-deficient mutants
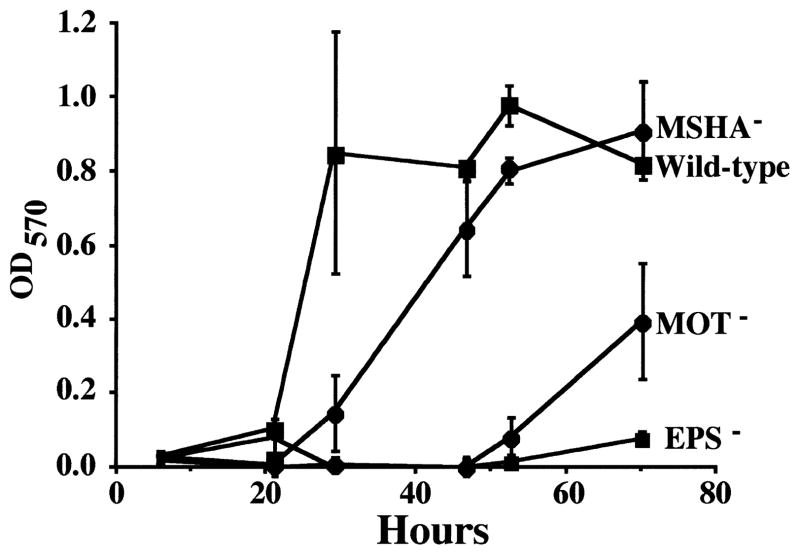
We hypothesized that some of the biofilm mutants might have a lower rate of biofilm formation, and that, if given sufficient time, they would be able to form biofilms indistinguishable from the wild-type strain of V. cholerae El Tor. To begin to test this hypothesis, biofilm development by both wild-type and mutant V. cholerae was studied over the course of 72 h. Figure 3 quantifies biofilm formation over 72 h for the V. cholerae wild-type strain and representative biofilm-deficient mutants. All mutants in each group were tested and found to follow the pattern of the representative mutant illustrated in Fig. 3. In addition, time course assays were repeated on borosilicate with similar results.
Fig. 3.
Biofilm development over 72 h for V. cholerae El Tor and representative derivative mutants. OD570 quantifies the amount of crystal violet associated with the biofilm after staining.
As illustrated in Fig. 3, the wild-type strain reached a plateau in crystal violet staining after 29 h of incubation, while the MSHA mutant reached an equivalent plateau in crystal violet staining after ≈ 48 h. Furthermore, the rate of biofilm formation by the pilT mutant was indistinguishable from that of other MSHA mutants. This suggests that the presence of a non-functional MSHA pilus does not measurably enhance biofilm-forming ability beyond that of an MSHA structural mutant.
EPS mutants did not form a biofilm detectable by crystal violet staining during the course of the experiment. This suggests that EPS mutants are not simply slower but unable to form a biofilm under the conditions of this experiment.
Flagellar mutants were able to form a biofilm measurable by crystal violet staining after ≈ 72 h. No difference in the rate of biofilm formation was observed between mutants with a paralyzed flagellum and mutants with no flagellum at all. As was found for MSHA, therefore, the presence of a non-motile flagellum does not improve biofilm-forming ability beyond that of a flagellar structural mutant.
Microscopic analysis of wild-type and mutant biofilm structure
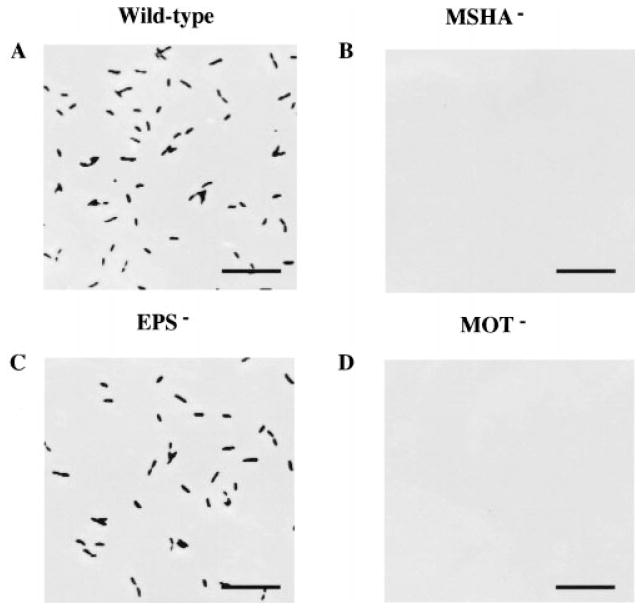
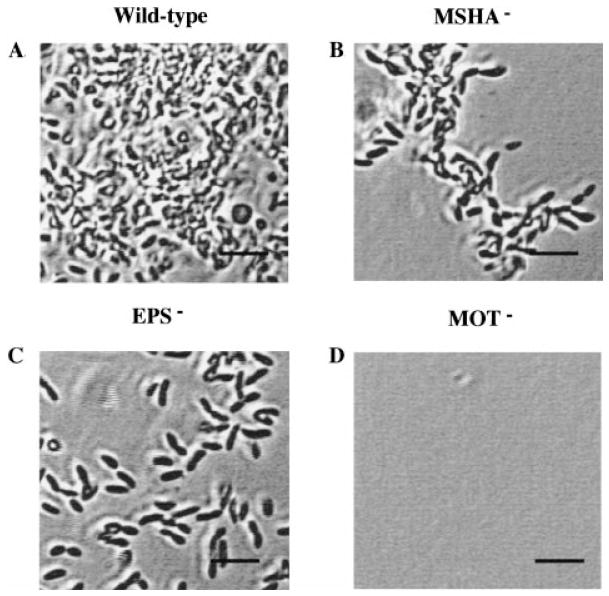
Although measurement of the crystal violet incorporated into a biofilm allows easy estimation of the bacterial biomass adherent to a surface, microscopy is required to determine the spatial distribution of the adherent bacteria. To study the development of the wild-type and mutant biofilm architectures further, we observed the biofilms microscopically at 15 min, 28 h and 72 h after inoculation. These experiments were repeated multiple times with similar results. Furthermore, the total surface area examined in each of these studies was at least 0.06 cm2, and images shown are representative of what was observed in multiple fields. Figure 4 shows attachment of the wild-type and mutant cells to the bottom of a microtitre well at 15 min. This demonstrates clearly that, while wild-type and EPS mutants were able to attach readily in 15 min, no surface-associated MSHA and flagellar mutants were observed. This pattern was also observed on borosilicate (results not shown).
Fig. 4.
Phase-contrast micrographs comparing the attachment of wild-type V. cholerae and representative mutants after 15 min of incubation with a polystyrene surface. Bar = 5 μm.
After 24 h of incubation with a borosilicate glass coverslip, the pattern of attachment we observed for wild-type and representative V. cholerae mutants was quite different (Fig. 5). The wild-type V. cholerae formed a confluent biofilm on the coverslip, the MSHA mutant was just beginning to form the microcolonies that are characteristic of incipient biofilms, the EPS mutant remained a monolayer of isolated, attached cells, and the flagellar mutant was not associated with the surface.
Fig. 5.
Phase-contrast micrographs comparing the attachment of wild-type V. cholerae and representative mutants after 24 h of incubation with a borosilicate coverslip. Bar = 2 μm.
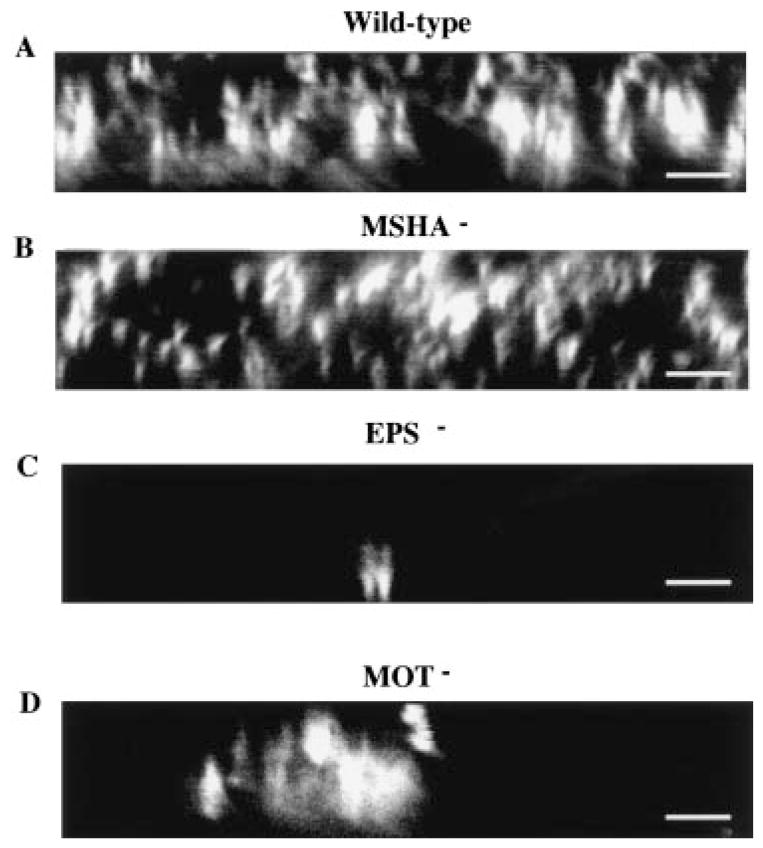
After 72 h of incubation on the borosilicate surface, the wild-type biofilm reached a thickness of 10–15 μm, and confocal microscopy was necessary to visualize the biofilm structure. In Fig. 6, representative vertical cross-sections are shown for 72 h biofilms formed by the various mutants. In this simple system, wild-type V. cholerae forms a biofilm with the characteristic architecture of pillars of bacteria and water channels that is observed in continuous flow systems (Costerton et al., 1995). Furthermore, the MSHA mutant has formed a biofilm that is indistinguishable from the wild type. In contrast, the flagellar mutant has a few localized areas of attachment. Within these areas, however, the biofilm is approximately as thick as the wild type and contains water channels that characterize the wild-type biofilm. Finally, even after 3 days, the EPS mutant remains attached to the surface as a monolayer. Taken together, these results indicate (i) that pili and flagella facilitate but are not absolutely required for attachment to the surface; (ii) that flagella are required for spread along the surface; and (iii) that EPS is necessary for formation of the three-dimensional biofilm structure.
Fig. 6.
Confocal scanning laser micrographs comparing the attachment of wild-type V. cholerae and representative mutants after 72 h of incubation with a borosilicate coverslip. Micrographs represent optical sections in the xz-plane with the substratum located at the bottom of the micrograph. Bar = 5 μm.
Discussion
We are interested in the role of biofilm formation in the emergence of new pathogens from aquatic environments. In order to understand the genetic basis of biofilm formation in our prototype, V. cholerae, we have isolated and characterized transposon insertion mutants that are defective in biofilm development. The mutants identified can be divided into three groups: (i) those with insertions in genes involved in type IV pili biogenesis and function; (ii) those with insertions in genes involved in flagellar motility; and (iii) those with insertions in genes involved in EPS synthesis.
Two external structures, the flagellum and MSHA type IV pili, are involved in attachment to the abiotic surface. These two structures may promote attachment to a surface either by acting as specific adhesins or through the generation of force. Time course studies of biofilm formation by the relevant mutants demonstrate that flagella and MSHA pili accelerate, but are not required for, attachment to abiotic surfaces. Furthermore, there is no difference in the rate of biofilm formation by mutants that have no flagellum and mutants with a paralyzed flagellum. We also see no difference in the rate of biofilm formation between mshA mutants and pilT mutants, suggesting, by analogy with the pilT gene of P. aeruginosa, that the presence of a non-functional pili does not enhance the ability of the bacterium to attach to a surface. These two observations (i) that both pili and flagellar mutants eventually attach to abiotic surfaces; and (ii) that the rates of biofilm formation by mutants in which the flagellum and the MSHA type IV pili are either paralyzed or absent are indistinguishable, suggest that the generation of force is the essential property of these structures in accelerating attachment to an abiotic surface.
The requirement for the generation of force in attachment of V. cholerae to an abiotic surface is consistent with the existence of a repulsion between the bacterium and the abiotic surface. Thus, if the flagellum and pili are absent, there is still a finite probability that the bacterium will collide with the surface with enough force to overcome the repulsion, and attachment to the surface will occur, albeit more slowly. Electrostatic interactions have been hypothesized to be responsible for a repulsion between the bacterium and the surface that is sensed over a distance of 10–20 nm (Fletcher, 1996). Once the bacterium overcomes this repulsion, it may reach a distance of < 1 nm from the abiotic surface, where specific interactions are thought to be responsible for the strong attraction between the bacterium and the surface that results in attachment (Gristina, 1987; Fletcher, 1996). Experimental measurements of these types of forces have only been undertaken recently (Razatos et al., 1998). More detailed studies are required to understand the nature and magnitude of these forces and their effect on attachment to a surface.
We have also isolated biofilm-defective mutants that initially attach to abiotic surfaces in a manner that is indistinguishable from the wild type but are then unable to develop into a three-dimensional biofilm. These are the mutants in EPS production. EPS has been implicated in the distinctive colony morphology of the rugose variant of V. cholerae El Tor (Yildiz and Schoolnik, 1999). Interestingly, the work of Yildiz and Schoolnik (1999) has shown that EPS was absent from colonies of wild-type V. cholerae El Tor. Our studies imply, however, that EPS is expressed in biofilm-associated cells of wild-type V. cholerae El Tor. This suggests that EPS may be expressed under different conditions in the wild type and rugose variant and, perhaps, that a part of the regulatory machinery that controls EPS production in the wild type is bypassed in the rugose variant. Exopolysaccharide production has been implicated in biofilm formation previously (Davies et al., 1993; Hoyle et al., 1993; Yildiz and Schoolnik, 1999). Our finding supports a role for exopolysaccharide in stabilizing the three-dimensional biofilm structure. Possible mechanisms for this stabilization include physical constraint and minimization of intercellular repulsions both by maintenance of an intercellular distance and by shielding of the electrostatic charges on the bacterial surface.
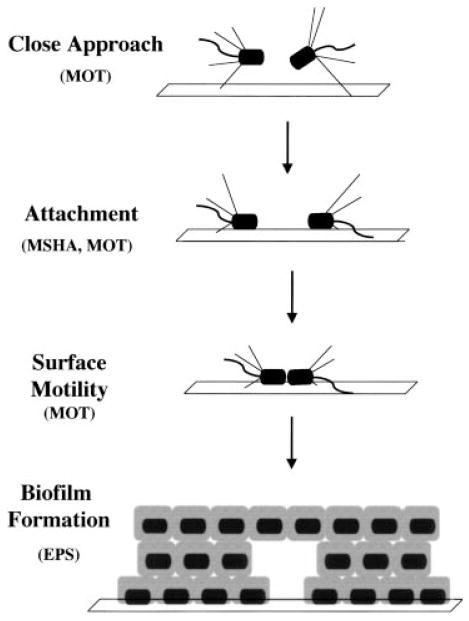
We have isolated and characterized V. cholerae mutants that are defective in attachment, surface motility and formation of a three-dimensional structure. These mutants define a developmental pathway for biofilm formation by V. cholerae El Tor. Figure 7 depicts our current model of the role that these gene products play in the development of a three-dimensional biofilm. Flagellar motility allows the bacterium to swim through a repulsive potential towards the abiotic surface. As the bacterium nears the surface, the MSHA pilus is able to tether the bacterium and pull it onto the surface, where attractive interactions with the surface lead to attachment. Movement along the surface again involves the generation of force by flagella, because the attraction between the bacterium and the surface must be overcome in order to move. Finally, EPS is required to stabilize cell–cell interactions that are integral to the formation of a three-dimensional biofilm.
Fig. 7.
Proposed steps in the formation of a three-dimensional V. cholerae biofilm on an abiotic surface. Rods are V. cholerae with polar type IV MSHA pili and one polar flagellum. The bacteria swim towards the surface using flagellar motility. The bacteria attach to the surface using type IV pili, which then retract to draw the bacterium onto the surface. Flagella allow movement along the surface and microcolony formation, and EPS is important for the three-dimensional arrangement of bacteria.
V. cholerae El Tor is the predominant cause of cholera in the world today. This suggests that it has taken over the niche of classical V. cholerae in the environment. The biofilms produced by V. cholerae El Tor are thicker and more densely packed than those produced by classical V. cholerae (P. I. Watnick and R. Kolter, unpublished results). Because biofilm formation is such an important part of survival in the aquatic environment, we hypothesize that its superior ability to form biofilms may impart a survival advantage to V. cholerae El Tor. This has perhaps contributed to its worldwide predominance. We plan to address this hypothesis in future studies.
Experimental procedures
Bacterial strains and plasmids
N16961Sm, a streptomycin-resistant mutant of a V. cholerae El Tor strain isolated from the seventh pandemic, was used for all biofilm studies. KFV11 (Watnick et al., 1999), a mutant of N16961Sm containing a large, in frame deletion in the mshA gene, was used as the prototype for studies of biofilm formation by all mutants blocked in MSHA secretion and biogenesis. Transposon mutagenesis was carried out by mating with the E. coli strain β-2155, which is a diaminopimelate (DAP) auxotroph and λpir lysogen (Kolter et al., 1978), containing pBSL180, a suicide plasmid carrying a mini-Tn10 transposon derivative marked with a kanamycin (Km) cassette (Pridmore, 1987; Alexeyev and Shokolenko, 1995). For fluorescent labelling of biofilms, V. cholerae was transformed with pSMC2 (Bloemberg et al., 1997), a plasmid containing a constitutively expressed gene for the green fluorescent protein (GFP). Approximately 60% of the cells lose plasmid-mediated ampicillin resistance during 24 h of growth unless antibiotic selection is provided. Thus, GFP-labelled V. cholerae biofilms were formed in the presence of antibiotic selection.
Transposon mutagenesis and screen for defects in biofilm formation
Matings of V. cholerae (strain N16961Sm) with E. coli strain β-2155 (pBSL180) were carried out for 2 h at 37°C on Luria–Bertani agar plates containing 0.3 mM DAP. Transposon insertion mutants of V. cholerae were then selected by plating on LB agar containing 50 μg ml−1 added Km. The screen for biofilm-defective mutants was performed in a fashion similar to that described previously for P. aeruginosa and E. coli (O’Toole and Kolter, 1998b; Pratt and Kolter, 1998). Briefly, Km-resistant colonies were transferred to polyvinylchloride (PVC) microtitre dish wells filled with 100 μl of LB broth. The dishes were allowed to incubate at room temperature for 24 h. At this point, the wells were rinsed and stained with crystal violet as described previously (O’Toole and Kolter, 1998b). Mutants that were unable to produce a biofilm in the microtitre dish assay were retrieved from the master plate and retested in triplicate. Mutants that failed to make biofilms on the secondary screen were stored in glycerol at −80°C for further study.
Arbitrary PCR
For sequence analysis of transposon junctions, an arbitrary PCR protocol was used as described previously (O’Toole and Kolter, 1998b). Arbitrary PCR involves two rounds of PCR. The first round includes a primer unique to the transposon and, in this case, two arbitrary primers that are designed to hybridize to an arbitrary sequence on the chromosome. The second round includes a nested primer unique to the transposon as well as a primer that is identical to the 5′ end of the arbitrary primer. This round is designed specifically to amplify PCR products obtained in the first round. Arbitrary PCR products were obtained using the following two sets of primers: (i) P12 with ARB1 and ARB6 (round 1), P11 with ARB2 (round 2); or (ii) P20 with ARB1 and ARB6 (round 1) followed by P2 with ARB2 (round 2). The sequences of these primers are listed below. Both these sets of primers were intended for sequence analysis upstream of the left transposon junction.
Transposon primers
P12: 5′-CAGCGCATCGCCTTCTAT CGC-3′; P11: 5′-CTTGACGAGTTCTGAGCGGG-3′; P20: 5′-CCGCGGTGGAGCTCC-3′; P2: 5′-ATGACAAG ATGTG-TATCCACC-3′.
Arbitrary primers
ARB1: 5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT-3′; ARB6: 5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC-3′; ARB2: 5′-GGCCACGCGTCGACTAGTAC-3′.
Motility and haemagglutination assays
Motility and haemagglutination assays with human red blood cells were performed as described previously (Gardel and Mekalanos, 1996; Pratt and Kolter, 1998).
Time course assays
Sterile, covered polystyrene microtitre dish wells were filled with 100 μl of LB broth with 100 μg ml−1 added streptomycin. Mutants were inoculated in triplicate and allowed to incubate for times varying from 8 h to 72 h. Before biofilm quantification, the growth rate of mutant strains was assessed by measuring the OD630 of all wells using a microtitre plate reader. Growth defects were not noted for any of the mutants selected for further study. Wells were subsequently rinsed, adherent bacteria were stained with crystal violet, and 300 μl of dimethylsulphoxide (DMSO) was added to each well to resuspend attached crystal violet. Biofilm formation was then quantified by measuring an OD570 directly for each well using a microtitre plate reader.
Phase microscopy
For observation of initial attachment to a surface, biofilms were formed on the bottoms of sterile, 24-well polystyrene microtitre dishes. This was done by growing cultures to mid-log phase, adding 400 μl of the culture to a well and observing attachment to the bottom of the well over time using an inverted-phase microscope equipped with a CCD video camera system (Optronics Engineering) and computer interface (magnification 400×). For incubations of 24 h or more, biofilms of the V. cholerae strain of interest were formed at room temperature on borosilicate coverslips placed in 50 ml Falcon tubes that had been filled with 6 ml of LB broth with appropriate antibiotics added and a 1:100 dilution of an overnight V. cholerae culture. At the desired end-point, the coverslip was rinsed with LB broth or distilled water to remove non-adherent bacteria and placed over a concave microscope slide filled with LB broth. Bacteria associated with the biofilm were then observed at 600× magnification using an Optiphot-2 microscope (Nikon) equipped with a CCD video camera system (Optronics Engineering) and computer interface.
Confocal scanning laser microscopy
Biofilms of the GFP-expressing V. cholerae strain of interest were formed on borosilicate coverslips as described above. Confocal microscopy was performed on 72 h biofilms. A MRC-1024 confocal microscope (Bio-Rad) was used to collect z-sections through the biofilm of interest using a 488 nm excitation wavelength for visualization of the fluorescent bacteria (magnification 600×).
Acknowledgments
We thank George O’Toole, Leslie Pratt, Dianne Newman, Patrick Stragier and other members of the Kolter laboratory for many helpful discussions and careful review of the manuscript. We especially thank Laura Croal for sequence analysis of several of the transposon insertion mutants. This work was supported by NIH grant K08AI01588 to P.I.W. and NIH grant GM58213 to R.K.
References
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- Anwar H, Strap JL, Costerton JW. Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother. 1992;36:1347–1351. doi: 10.1128/aac.36.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett GI, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bloemberg GV, O’Toole GA, Lugtenberg BJ, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- Colwell RR, Spira WM. The ecology of Vibrio cholerae. In: Barua D, Greenough WBI, editors. Cholera. New York: Plenum; 1992. pp. 107–127. [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: Substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Fletcher M. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies. In: Fletcher M, editor. Bacterial Adhesion: Molecular and Ecological Diversity. New York: Wiley-Liss; 1996. pp. 1–24. [Google Scholar]
- Gardel CL, Mekalanos JJ. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristina AG. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- Homma M, Kutsukake K, Hasebe M, Iino T, Mac-Nab RM. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- Hoyle BD, Williams LJ, Costerton JW. Production of mucoid exopolysaccharide during development of Pseudomonas aeruginosa biofilms. Infect Immun. 1993;61:777–780. doi: 10.1128/iai.61.2.777-780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- Huq A, Huq SA, Grimes DJ, O’Brien M, Chu KH, Capuzzo JM, et al. Colonization of the gut of the blue crab (Callinectes sapidus) by Vibrio cholerae. Appl Environ Microbiol. 1986;52:586–588. doi: 10.1128/aem.52.3.586-588.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose KE, Mekalanos JJ. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173:6558–6567. doi: 10.1128/jb.173.20.6558-6567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JW, Taylor RK. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J Bacteriol. 1999;181:1110–1117. doi: 10.1128/jb.181.4.1110-1117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AL, Powelson DK. Bacterial interactions with surfaces in soils. In: Fletcher M, editor. Bacterial Adhesion: Molecular and Ecological Diversity. New York: Wiley-Liss; 1996. pp. 25–57. [Google Scholar]
- Morona JK, Morona R, Paton JC. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998a;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998b;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Pridmore RD. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- Razatos A, Ong Y, Sharma MM, Georgiou G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc Natl Acad Sci USA. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla BN, Singh DV, Sanyal SC. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol Med Microbiol. 1995;12:113–120. doi: 10.1111/j.1574-695X.1995.tb00182.x. [DOI] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vess RW, Anderson RL, Carr JH, Bond WW, Favero MS. The colonization of solid PVC surfaces and the acquisition of resistance to germicides by water micro-organisms. J Appl Bacteriol. 1993;74:215–221. doi: 10.1111/j.1365-2672.1993.tb03018.x. [DOI] [PubMed] [Google Scholar]
- Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Hobbs M, Livingston SP, Krishapillai V, Mattick JS. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, Lauer P, Park H, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- Wu SS, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]