Abstract
In Saccharomyces cerevisiae, Snf1 kinase, the ortholog of the mammalian AMP-activated protein kinase, is activated by an increase in the phosphorylation of the conserved threonine residue in its activation loop. The phosphorylation status of this key site is determined by changes in the rate of dephosphorylation catalyzed by the yeast PP1 phosphatase Glc7 in a complex with the Reg1 protein. Reg1 and many PP1 phosphatase regulatory subunits utilize some variation of the conserved RVxF motif for interaction with PP1. In the Snf1 pathway, the exact role of the Reg1 protein is uncertain since it binds to both the Glc7 phosphatase and to Snf1, the Glc7 substrate. In this study we sought to clarify the role of Reg1 by separating the Snf1- and Glc7-binding functions. We generated a series of Reg1 proteins, some with deletions of conserved domains and one with two amino acid changes in the RVxF motif. The ability of Reg1 to bind Snf1 and Glc7 required the same domains of Reg1. Further, the RVxF motif that is essential for Reg1 binding to Glc7 is also required for binding to Snf1. Our data suggest that the regulation of Snf1 dephosphorylation is imparted through a dynamic competition between the Glc7 phosphatase and the Snf1 kinase for binding to the PP1 regulatory subunit Reg1.
Keywords: Snf1 kinase, AMP-activated protein kinase, Glc7, PP1 phosphatase, Reg1
1. Introduction
Yeast cells respond to glucose limitation by activating the Snf1 kinase signaling pathway. A key step in the activation of Snf1 is the phosphorylation of the Snf1 activation loop threonine residue [1] by one of three Snf1-activating kinases [2, 3]. An analogous pathway is operative in mammalian cells where the Snf1 ortholog, the AMP-activated protein kinase (AMPK) is activated by phosphorylation of the homologous threonine residue in response to energy stress [4]. For many years it was thought that identifying the upstream activating kinase(s) in this pathway would lead to an understanding of regulation. In recent years, the upstream activating kinases have been identified in both mammalian and yeast systems [2, 3, 5–7] but they do not appear to be the site of regulation. Instead, it is the dephosphorylation reaction that is the regulated step. In mammalian cells, energy stress causes an increase in AMP which binds to the gamma subunit of the AMPK complex making the enzyme resistant to dephosphorylation [8, 9]. In yeast, the role of AMP is uncertain but the dephosphorylation reaction is the step that is responsive to changes in glucose levels. The rate of dephosphorylation in low glucose is over 10-fold slower than the rate in high glucose [10]. Understanding how the dephosphorylation reaction is regulated appears to be the key to understanding how this pathway is controlled by glucose and energy stress.
The dephosphorylation of the Snf1 kinase is catalyzed by the PP1 phosphatase Glc7 in a complex with the Reg1 protein [11]. Members of the PP1 family of protein phosphatases are commonly found in association with one or more regulatory subunits that direct the phosphatase to distinct subcellular localizations and specify substrate selectivity [12]. Evidence that the Glc7/Reg1 complex is responsible for the dephosphorylation of Snf1 is substantial. Deletion of Reg1 leads to hyperphosphorylated and constitutively active Snf1 [1]. Similarly, the T152K point mutation in Glc7 which interferes with the ability of Reg1 to associate with Glc7 also leads to hyperactivated Snf1 [13]. A simple model for this pathway would propose that the association of Reg1 with Glc7 targets the phosphatase to Snf1. However, the situation is more complex. First, Reg1 without its partner Glc7 is found as a major component of the Snf1 complex [14]. Second, the association of Reg1 with Snf1 is greater in low glucose conditions [15] when Snf1 is protected from dephosphorylation. The activity of the Glc7/Reg1 is not directly regulated by glucose, since the phosphatase is active toward other substrates in low glucose [10]. Thus, a more complicated picture emerges in which glucose-mediated regulation of this pathway appears to lie within the Snf1 kinase complex where binding to Reg1 and the access of the activation loop to the phosphatase are somehow responsive to glucose levels.
PP1 phosphatases are directed to specific subcellular localizations and to specific substrate proteins by association with regulatory proteins [12]. Many PP1 regulatory proteins bind to PP1 using a sequence motif with the consensus sequence of RVxF. Structural studies of the rabbit PP1 phosphatase in complex with the myosin phosphatase targeting subunit show the valine and phenyalanine side chains of this motif are buried in a hydrophobic groove present on the surface of the PP1 enzyme and distinct from the active site [16]. In Reg1, this motif has the sequence RHIHF with the isoleucine and phenylalanine side chains predicted to be critical to interaction with PP1. Indeed, mutation of these residues has been shown to disrupt interaction with the yeast PP1 phosphatase Glc7 and to lead to constitutively active Snf1 kinase [17, 18]. The interaction of Reg1 and Glc7 is not limited to the RHIHF sequence fitting into the hydrophobic grove. The Glc7 protein bearing the T152K mutation also shows reduced binding to Reg1 and increased activity of the Snf1 kinase [13]. Examination of the structure of homologous PP1 phosphatases predicts that the threonine residue at position 152 is well separated from the hydrophobic groove on Glc7 that interacts with the RHIHF sequence [16]. Thus the interaction of Reg1 with Glc7 is likely to involve other structural domains distinct from the RHIHF sequence.
In this study we engineered mutations in the Reg1 protein in an attempt to separate the Glc7 and Snf1 binding determinants. Our results indicate that the same domains and even the same amino acids in the Reg1 protein that are needed for Glc7 binding are also required for interaction with Snf1. Thus, Reg1, the PP1 regulatory subunit, binds both the phosphatase and the substrate of the phosphatase. This finding leads to a model in which the dynamic competition between Glc7 and Snf1 for binding to Reg1 may contribute to the regulation of the Snf1 kinase.
2. Materials and methods
2.1 Yeast strains, media and genetics methods
The Saccharomyces cerevisiae strains used in this study are all derived from S288C and are described in Table 1. Strains with multiple gene deletions were produced by genetic crosses and sporulation. The yeast strains MSY990 and MSY987 (Table 1) were created by introducing the reg1Δ::HIS allele into strains MSY182 and FY1193 by homologous recombination using a linear DNA fragment containing the HIS3 gene flanked with DNA from the REG1 locus. The reg1Δ::HIS3 allele contained a complete replacement of the REG1 open reading frame. The REG1 flanking DNA used to direct homologous transformation included 819 bp upstream of the REG1 ORF (nucleotides −948 to −130 relative to the ATG codon) and 239 bp downstream from the REG1 ORF (nucleotides 3188 to 3436 relative to the ATG codon). Accurate and complete replacement of the REG1 gene was confirmed by PCR. The heterozygous diploid containing the glc7Δ::KAN allele was purchased from Open Biosystems (Huntsville, Alabama, USA). The deletion of the GLC7 gene was confirmed by PCR. The glc7Δ::KAN strain was transformed with a plasmid containing the GLC7 gene expressed from its own cognate promoter and tagged with 3 copies of the HA epitope at the C-terminus. This strain was subjected to sporulation and viable, haploid strains resistant to kanamycin were isolated. Growth of cells utilized standard media at 30°C [19]. Raffinose media contained 2% raffinose, 0.01% glucose (g/100 ml) and 1μg/ml antimycin A. Sucrose media contained 2% sucrose (g/100 ml) and where indicated 0.02% 2-deoxy-glucose (g/100 ml). Transformation of yeast strains used the lithium acetate method [20].
Table 1.
S. cerevisiae strains.
| Strain | Genotype |
|---|---|
| FY1193 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 snf1Δ10 |
| MSY182 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 |
| MSY990 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200reg1Δ::HIS3 |
| MSY987 | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 snf1Δ10reg1Δ::HIS3 |
| MSY1028 | MATa ura3 leu2 his3 trp1Δ63 reg1Δ::HIS3 glc7Δ::KAN [ pGLC7-3HA CEN LEU2 ] |
| MSY1030 | MATa ura3 leu2 his3 trp1Δ63 reg1Δ::HIS3 glc7Δ::KAN [ pGLC7 CEN LEU2 ] |
2.2 Epitope tagging and plasmid constructions
The Reg1 protein was epitope tagged with five tandem copies of the V5 epitope [21] placed at the C-terminus of the REG1 open reading frame. This construct was introduced to yeast cells on a low-copy number CEN plasmid derived from pRS316 [22] and was expressed from the native REG1 promoter. Deletions (Reg1-C1, Reg1-C2, Reg1-C3 and Reg1-N1) and point mutations (I466A, F468M) were created with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by DNA sequencing. The Glc7 protein was tagged with 3 copies of the HA epitope placed at the C-terminus of the GLC7 reading frame. This construct was introduced to yeast cells on a low-copy number CEN plasmid derived from pRS315 [22] and was expressed from the native GLC7 promoter. The Snf1 protein tagged with 3 copies of the HA epitope has been described [1].
2.3 Invertase assays
Invertase activity of log-phase cells grown in high or low glucose medium was quantitatively assayed using a colorimetric assay coupled to glucose oxidase [23]. Three independent cultures were assayed, and the mean values are shown with the error bars representing one standard error [24].
2.4 Western analysis
Protein extracts were prepared from cells grown to mid-log phase in high or low glucose conditions. Cells were lysed by vortexing in the presence of glass beads in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1 % (w/v) SDS, 1 % Nonidet P40, 0.5 % sodium deoxycholate) supplemented with protease and phosphatase inhibitors. Twenty micrograms of total protein were separated on SDS polyacrylamide gels and transferred to membranes. Proteins tagged with the HA epitope were detected with HA-probe HRP mouse monoclonal antibody (Santa Cruz). The Reg1 protein tagged with V5 epitope was detected with Anti-V5-HRP mouse monoclonal antibody (Invitrogen). Chemiluminescent detection was performed with ECL Plus Western Blotting Detection System (GE Healthcare). Quantitative western blots were processed using the Snap i.d. system (Millipore) and blocked with Odyssey Blocking Buffer (Li-Cor). HA epitope-tagged proteins were detected with HA probe (Santa Cruz) diluted 1:2000. V5 epitope-tagged proteins were detected with Anti-V5 (Invitrogen) diluted 1:1000. The secondary antibody for both HA and V5 blots was Anti-mouse IRDye 800CW (Li-Cor) diluted 1:5000. Blots were visualized using an Odyssey Infrared Imager (Li-Cor) and band quantification was performed using Odyssey software.
2.5 Immunoprecipitation
Proteins tagged with the HA epitope (Snf1 or Glc7) were collected from protein extracts (500 μg) using 20 μl HA-coupled agarose beads (Santa Cruz). Beads were incubated in the extract at 4°C for 2 hours. Unbound proteins were removed by washing the beads 3 times in 1 ml of RIPA buffer. Bound proteins were eluted from the beads by incubation in SDS sample buffer at 95°C for 5 minutes, resolved on SDS polyacrylamide gels and detected by western blotting.
2.6 Statistical analysis
Statistical analysis was done using Student’s t test. p < 0.05 was considered significant.
3. Results
3.1 Structure of the Reg1 protein and design of Reg1 mutations
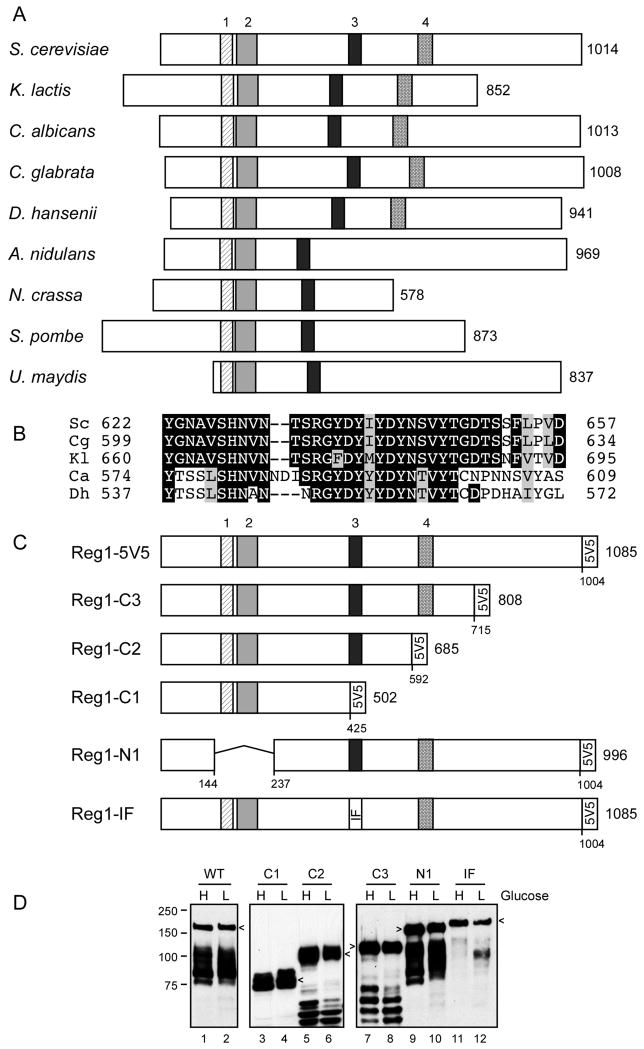
The Reg1 protein of S. cerevisiae is a PP1 phosphatase regulatory protein. The yeast PP1 phosphatase Glc7 interacts with numerous regulatory proteins that direct Glc7 to different localizations and to different substrates [25]. While some of the Glc7 regulatory subunits have orthologs in other species including mammals, orthologs of the Reg1 protein are restricted to the dikarya subkingdom of fungi that includes the ascomycete and basidiomycete species. Comparison of the Reg1 primary sequence with orthologs from four other species revealed the presence of three conserved blocks of sequence [26]. Blocks one and two are needed for interaction with the yeast 14-3-3 proteins but are not essential for Reg1 mediated repression of SUC2 [26]. Block three contains the conserved RVxF motif that is known to mediate interaction with PP1 phosphatases [27]. In Reg1, the RVxF motif has the sequence RHIHF. Mutations in this motif eliminate Reg1 function and disrupt its ability to bind to Glc7 [17, 18]. We have extended the sequence analysis of Reg1 by comparing it to sequences now available from additional species and have uncovered a fourth conserved sequence block (Fig. 1A and 1B). In order to further characterize the structure-function relationships of the Reg1 protein, we generated plasmids that express Reg1 proteins with five copies of the V5 epitope at the C-terminus. Three C-terminal truncations were generated (C1, C2 and C3); each retained the V5 tag but removed C-terminal sequences that included blocks 3 and 4, block 4 alone or the C-terminal 270 amino acids that lack any conserved sequence elements (Fig. 1C). In addition, a Reg1 with precise deletion of blocks 1 and 2 (Reg1-N1) and another (Reg1-IF) with two point mutations in the PP1 interacting sequence were also generated. Cells lacking the entire REG1 gene were transformed with low-copy number plasmids that expressed these REG1 variants using the native transcriptional promoter and terminator sequences. Extracts were prepared from cells grown in high glucose or after shifting to low glucose and were examined by western blotting using anti-V5 epitope monoclonal antibodies (Fig. 1D). The wild type Reg1 protein and all 5 variants are readily detected in both high and low glucose samples.
Fig. 1.
Conserved domains of the Reg1 protein. (A) Schematic representation of Reg1 orthologs from nine fungal species. The number of amino acids in each protein is shown on the right. The size and position of four conserved blocks of sequences are drawn to scale. Blocks 1 through 3 were previously noted [17]. (B) Multiple sequence alignment of conserved block 4. (C) Reg1 deletion constructs and the double point mutation (IF: I466M, F468A) are shown with the 5 tandem copies of the V5 epitope on the C-termini. The total length of each protein and deletion junctions are shown. (D) Western blot of Reg1-V5 proteins isolated from cells grown in high (H) or low (L) glucose. Arrows indicate mobility of the full-length protein. Faster migrating species represent proteolytic fragments.
3.2 Growth phenotypes of cells expressing mutant Reg1 proteins
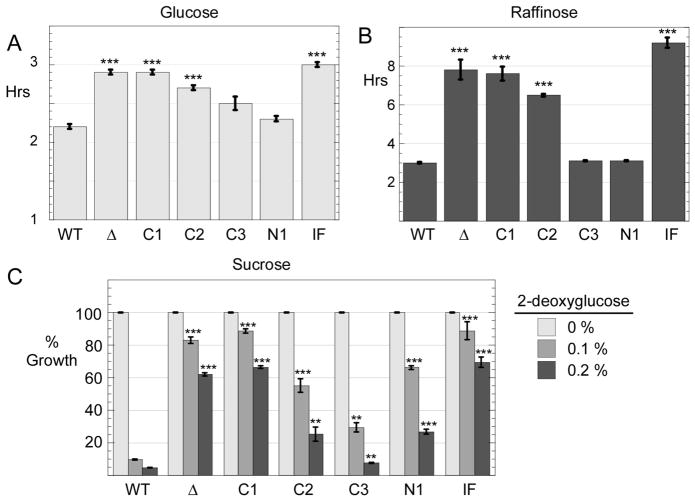
Cells lacking the Reg1 protein display a number of phenotypes including slow growth on glucose as the carbon source [17]. We engineered a complete knockout of the REG1 gene and introduced various REG1 alleles on low copy number plasmids using the URA3 gene for selection. Cells were grown in synthetic complete media lacking uracil with glucose as the carbon source. When the wild type REG1 gene was present on a plasmid, the doubling time was 2.2 hours (Fig. 2A). Cells lacking REG1 (transformed with empty plasmid vector) had a doubling time of 3.0 hours and showed reduced growth on agar plates in spot dilution assays (Fig. 2B). Cells expressing Reg1-C1 and Reg1-IF proteins also had doubling times over 3.0 hours while the Reg1-N1 and Reg1-C3 grew at rates comparable to the cells provided with wild type Reg1. Cells expressing Reg1-C2 had an intermediate doubling time on glucose of 2.6 hours.
Fig. 2.
Growth phenotypes of Reg1 mutants. Cells lacking the REG1 gene (MSY990) were transformed with a low copy number plasmid vector (Δ) or plasmids expressing either wild type Reg1 protein or a Reg1 mutant as indicated. Cells were grown in media with either glucose (A) or raffinose (B) as the carbon source. Absorbance at 600 nm was measured and the doubling time was calculated. The values plotted are the mean of three independent transformants with error bars representing one standard error. ***, p<0.001 versus wild type cells (WT) was calculated by Student’s t test. (C) Yield of cells after 18 hours of growth in sucrose media with increasing concentrations of 2-deoxyglucose is plotted after normalization to the yield of cells in the absence of 2-deoxyglucose. The values plotted are the mean of three independent transformants with error bars representing one standard error. Statistical significance was calculated in comparison to the wild type data in the presence of 2-deoxyglucose. *, p < 0.05; **, p < 0.01; *** p < 0.001
We also determined the doubling times of these cells when grown on raffinose, a less favorable carbon source. Cells expressing wild type Reg1, Reg1-C3 or the Reg1-N1 protein all had doubling times of approximately 3.3 hours. Cells without any Reg1 protein or those expressing the Reg1-C1, Reg1-C2 or Reg1 IF proteins all had greatly reduced growth rates with doubling times of greater than 6 hours.
A second phenotype observed in cells lacking REG1 function is the increased ability to grow in the presence of 2-deoxyglucose. This compound is taken up by cells and phosphorylated to 2-deoxyglucose-6-phosphate, however the next step in glycolysis is blocked by the 2-deoxy substitution. Cells exposed to 2-deoxyglucose undergo glucose repression which interferes with growth on alternative carbon sources like sucrose. Cells lacking REG1 function escape from glucose repression and are resistant to 2-deoxyglucose. To measure resistance to 2-deoxyglucose, we inoculated liquid cultures at an A600 of 0.1 in medium containing sucrose and increasing concentrations of 2-deoxyglucose. Cultures are then grown for 18 hours and the A600 value recorded. Cells with or without wild type Reg1 had an A600 of between 3 and 5. Addition of 2-deoxyglucose at 0.1% or 0.2% (g/100 ml) to cells with wild type Reg1 resulted in a reduction in growth of over 90% (Fig. 2B). Cells expressing no Reg1 protein or expressing the Reg1-C1, or Reg1-IF proteins showed resistance to 2-deoxyglucose, a finding consistent with a defect on glucose repression. Cells expressing the Reg1-C2, Reg1-C3 or the Reg1-N1 proteins displayed an intermediate level of resistance to 2-deoxyglucose. Finally, it is worth noting that the reduced growth rates observed in glucose and raffinose with some Reg1 variants does not strictly correlate with resistance to 2-deoxyglucose. The Reg1-N1 protein provided seemingly normal growth rates on glucose and raffinose but also displayed significant resistance to 2-deoxyglucose. We conclude that reg1 mutations are pleiotropic and that mutations do not affect all reg1 phenotypes equally.
3.3 Effect of Reg1 mutations on SUC2 expression
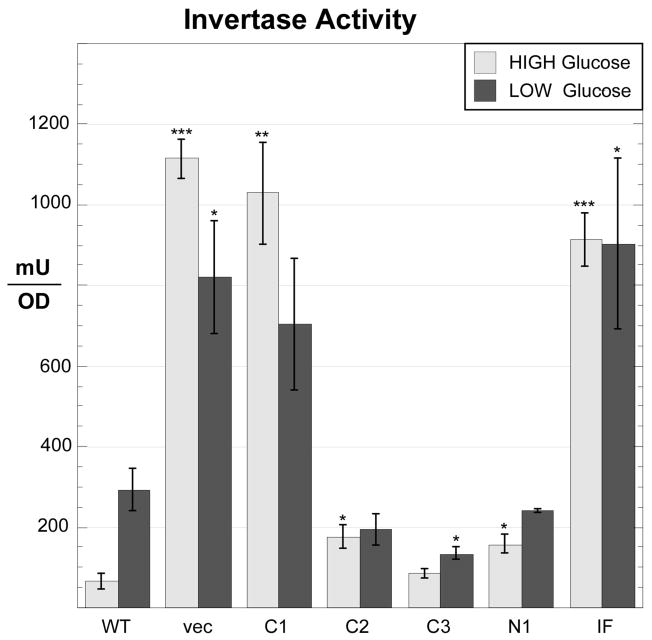
Cells lacking the Reg1 protein show numerous changes in gene expression that are mediated by hyperactive Snf1 kinase [11]. Here we analyzed the function of our panel of Reg1 mutants with regard to the expression of the SUC2 gene. SUC2 expression is normally repressed in high glucose due to the action of the Mig1 transcriptional repressor protein. In low glucose, activated Snf1 kinase phosphorylates Mig1, leading to its export from the nucleus [28] and the induction of SUC2 expression. In the absence of any Reg1 protein, invertase expression is completely derepressed showing extremely high levels of enzyme activity in cells grown in both high and low glucose media (Fig. 3). A similar pattern of invertase overexpression was observed when cells were transformed with plasmids expressing either the Reg1-C1 deletion or the Reg1-IF point mutation. We conclude that these two Reg1 constructs lack any ability to mediate SUC2 repression. The other mutants, Reg1-C2, Reg1-C3 and Reg1-N1 showed some level of SUC2 repression, yet none behaved like wild type Reg1 protein. For instance, for the Reg1-C2 deletion which lacks the conserved block 4 (Fig. 1), the expression of invertase was largely unaffected by glucose. Still, cells expressing Reg1-C2 were not able to fully induce SUC2 expression, suggesting that the Reg1-C2 protein was functional at some level by interfering with SUC2 induction but was not responsive to changes in glucose levels.
Fig. 3.
Invertase expression in cells expressing Reg1 deletions. Invertase expression was assayed in cells grown in 2% glucose or after shifting to 0.05% glucose for three hours. The yeast strain (MSY990) containing a complete deletion of the REG1 gene was transformed with low copy number plasmids expressing REG1 mutants as shown. The mean invertase expression from three independent cultures is plotted with errors bars representing one standard error. Statistical significance was calculated in comparison to the wild type data in either high or low glucose. *, p < 0.05; **, p < 0.01; *** p < 0.001
3.4 Effect of Reg1 mutations on binding to Glc7
In order to measure the association of Reg1 with Glc7, a coimmunoprecipitation assay was used. The yeast strain MSY1028 with complete deletion of the REG1 and GLC7 genes was transformed with plasmids that express these proteins with or without epitope tags. Extracts were then prepared from cells grown in high and low glucose. Glc7 protein tagged with three copies of the HA epitope was isolated by incubation of the extracts with agarose beads conjugated to monoclonal antibodies directed against the HA epitope. Bound proteins were eluted with SDS sample buffer and analyzed by western blotting. This analysis was performed a minimum of three times with similar results. A representative experiment is shown in Figure 4. The association of wild type Reg1 with Glc7 is evident and only detectable when the Glc7 protein is expressed with the HA tag at its C-terminus (Fig. 4a; lanes 3 and 4). The amount of Reg1 associated with Glc7 appears to be greater when cells are grown in low glucose, however the total amount of Reg1 present in extracts from cells grown in low glucose is greater than that observed in extracts from cells grown in high glucose.
Fig. 4.
Effect of Reg1 deletions on interaction with Glc7. (A) Reg1 forms a complex with Glc7. Protein extracts were prepared from cells expressing either untagged (MSY1030) or HA-tagged Glc7 (MSY1028) and untagged or V5-tagged Reg1 as indicated. Proteins were pulled down with HA-beads, eluted in SDS sample buffer, and probed by western blotting with V5 antibodies (top panel). (B) Protein extracts were prepared from cells expressing either wild type V5-tagged Reg1 (lanes 1 and 2) or the C2 and C3 deletion mutants. (C) Protein extracts were prepared from cells expressing either wild type V5-tagged Reg1 (lanes 1 and 2), the C1 or N1 deletion mutants or the Reg1-IF point mutation.
When our panel of Reg1 mutants was analyzed for the ability to bind Glc7, we found that only the Reg1-N1 and Reg1-C3 proteins were able to associate with Glc7. Thus the conserved blocks 1 and 2 deleted in Reg1-N1 and the C-terminal 270 residues deleted in Reg1-C3 are not required for association with Glc7. All of the mutants were readily detectable in the cell extracts (middle panel; V5 western) although the relative amounts was somewhat variable. Nonetheless, we fail to detect any association with Glc7 by the Reg1-C1, Reg1-C2 or Reg1-IF mutants. The failure of the Reg1-IF protein to associate with Glc7 was expected since this double point mutation has been previously shown to disrupt Glc7 binding [17]. However, the Reg1-C2 construct from which the conserved block 4 has been deleted also fails to bind to Glc7 despite the presence of the PP1-binding motif. Thus, the PP1-binding motif in Reg1 is necessary but not sufficient for stable interaction with Glc7.
3.5 Effect of Reg1 mutations on binding to Snf1
An abundance of biochemical and genetic evidence demonstrate a direct interaction between the Reg1 protein and the Snf1 kinase [6, 14, 15, 18, 29]. We investigated the ability of our panel of Reg1 mutants to interact with Snf1 kinase using the same coimmunoprecipitation assay. Protein extracts were prepared from cells expressing the Snf1 kinase tagged with the HA epitope and Reg1 tagged with the V5 epitope. Snf1 protein was isolated by incubation with agarose beads conjugated with monoclonal antibodies to HA. Bound proteins were eluted in SDS sample buffer and any bound Reg1 protein was detected by western blotting. This analysis was performed a minimum of three times with similar results. A representative experiment is shown in Figure 5. As a control for the specificity of this pull down assay, extracts were also made from cells in which the Snf1 and Reg1 proteins were present but untagged. When Snf1-HA and Reg1-V5 were expressed in the same cells, the presence of Reg1 protein in a complex with Snf1 is readily detected (Fig. 5a). We consistently detect more Reg1 in the complex when extracts are prepared from cells grown in low glucose. No Reg1 is detected in the immune complex when extracts were prepared from cells expressing untagged Snf1 (lanes 3 and 4). Therefore, this assay specifically detects the Snf1-Reg1 complex.
Fig. 5.
Effect of Reg1 deletions on interaction with Snf1. (A) Reg1 forms a complex with Snf1. Protein extracts were prepared from cells (MSY987) expressing either untagged or HA-tagged Snf1 and untagged or V5-tagged Reg1 as indicated. Proteins were pulled down with HA-beads, eluted in SDS sample buffer, and probed by western blotting with V5 antibodies (top panel). (B) Protein extracts were prepared from cells expressing either wild type V5-tagged Reg1 (lanes 1 and 2) or the N1, C1 and C2 deletion mutants. (C) Protein extracts were prepared from cells expressing either wild type V5-tagged Reg1 (lanes 1 and 2), the C3 deletion mutant or the Reg1-IF point mutation.
When we tested the ability of our Reg1 mutants to form a stable complex with Snf1, we found that only the Reg1-N1, Reg1-C3 mutants were able to bind Snf1 (Fig. 5b and c). Reg1-C1, Reg1-C2 and Reg1-IF were never detected in the Snf1 pull down complexes. Since Reg1-N1 can bind Snf1, the conserved sequence blocks 1 and 2 are not required for this interaction. Further, deletion of C-terminal 270 amino acids of Reg1 did not interfere with Snf1 association. Additional deletions in the C-terminus that removed conserved block 4 (Reg1-C2) or blocks 3 and 4 (Reg1-C1) completely abrogate association with Snf1. These findings are consistent with earlier studies using the two-hybrid assay that mapped the Snf1 interaction domain to this region of Reg1 [18]. The Reg1-IF protein harbors two point mutations in the PP1 phosphatase interaction motif. Changes in these residues have been shown to interfere with the ability of Reg1 to bind to the PP1 phosphatase Glc7 [17, 18]. In this experiment, we show that Reg1-IF protein is no longer able to interact with Snf1 (Fig. 4c; lanes 7 and 8). Thus the domains of Reg1 that mediate binding to the Snf1 kinase are the same as those that are needed for association with the PP1 phosphatase Glc7.
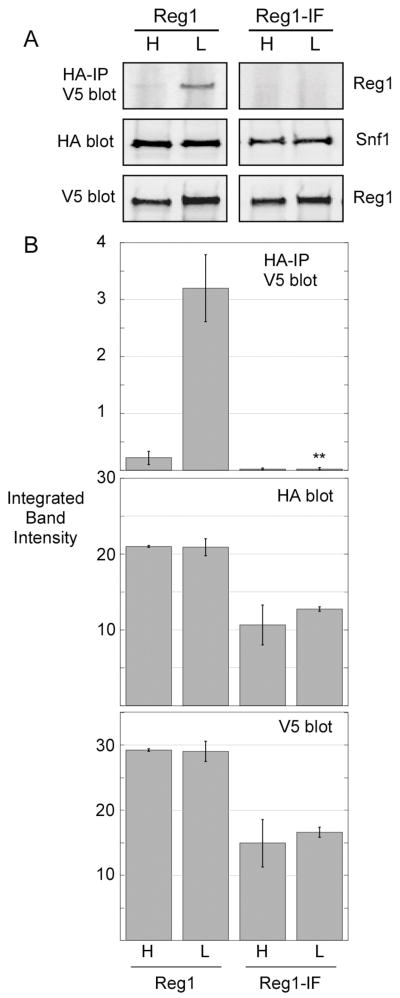
The involvement of the PP1-binding motif of Reg1 in the association with Snf1 was an unexpected finding. Earlier studies which used the two-hybrid system to detect Reg1-Snf1 interaction showed that mutation of the phenylalanine residue in this motif to arginine did not block binding to Snf1 [18]. Our studies used a double point mutation (I466M, F468A) previously shown to abrogate interaction with Glc7 [17]. To confirm that the Reg1-IF protein is unable to bind Snf1, we performed the coimmunoprecipitation assay using an infra-red detection system that allows quantification of the western signal. Extracts were prepared in triplicate and representative blots are shown (Fig. 6A). Cells expressing the Reg1-IF protein grow more slowly and the absolute level of the Reg1 protein and Snf1 proteins are reduced by about 2-fold. However, the amount of associated Reg1 is reduced by greater than 30-fold. Even if the association were reduced 10-fold, it would still have been detectable in this assay. Therefore, we conclude that I466M F468A mutations in the Reg1 RHIHF motif interfere with its ability to associate with the Snf1 protein.
Fig. 6.
Effect of Reg1-IF point mutations on Snf1 interaction. HA-tagged Snf1 and associated proteins were collected by immunoprecipitation from extracts expressing wild type V5-tagged Reg1 or V5-tagged Reg1-IF. Cells were grown in either high glucose (H) or low glucose (L) prior to protein extraction. Bound Reg1 proteins and the levels of Snf1 and Reg1 proteins in the protein extracts were determined by quantitative western blotting. Extracts were prepared and analyzed in triplicate. (A) Representative blots are shown. (B) Mean integrated band intensity values from triplicate samples are plotted with the error bars representing one standard error. Statistical analysis of the data for associated Reg1 in low glucose using Student’s t test. **, p < 0.001.
3.6 Phosphorylation of Snf1 but not activity is required for Reg1 association
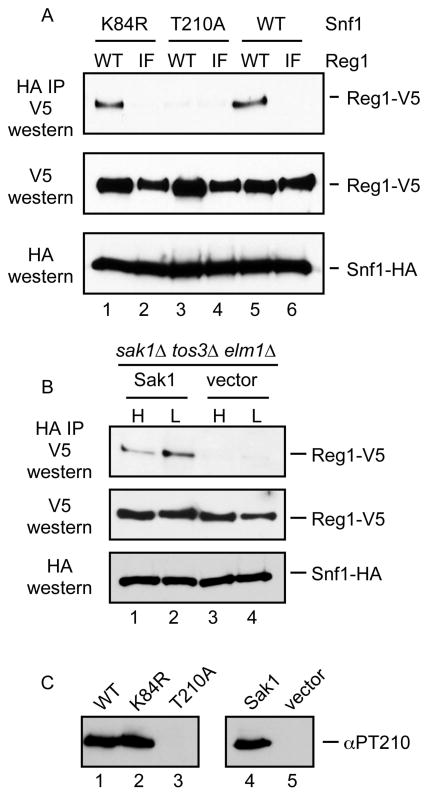
The failure of the Reg1-IF protein to associate with Snf1 can be interpreted in two ways. First it is possible that the RHIHF motif of Reg1 makes direct contacts with Snf1 and mutation of these residues destabilizes the interaction. An alternative interpretation is that the Reg1-IF mutant is unable to interact with Glc7 which disrupts the ability of the Glc7/Reg1 complex to dephosphorylate and inactivate Snf1. Thus the Reg1-IF mutation leads to the hyperactivity of Snf1 which may be the underlying cause that disrupts the interaction of Snf1 and Reg1. To distinguish between these two possibilities, we analyzed the ability of Reg1-IF to associate with Snf1 mutants that were incapable of becoming hyperactive. We used the SNF1-K84R gene which bears a mutation in the ATP binding site and is phenotypically similar to the complete deletion of the SNF1 gene [30] The Snf1-K84R protein is still phosphorylated on its activation loop threonine in response to glucose limitation [1]. In addition we used an activation-impaired allele, SNF1-T210A, which expresses a Snf1 protein that cannot be phosphorylated or activated [1]. When these Snf1 proteins are used in the coimmunoprecipitation assays in conjunction with wild type Reg1 and Reg1-IF, we found that the Snf1-K84R protein could associate with Reg1 but not with Reg1-IF. Representative blots are shown in Figure 7. Therefore, the defect in Snf1 association with Reg1-IF is not due to Snf1 hyperactivation. The Snf1-T210A protein was not able to associate with Reg1 or with Reg1-IF. Earlier studies with the two-hybrid assay have also reported that this mutation blocked association with Reg1 [15]. We sought to determine whether the failure of the Snf1-T210A protein to associate with Reg1 was due to change of the activation loop threonine to alanine or to the loss of phosphorylation at this site. We expressed wild type Snf1 protein in cells that lack the genes for all three of the Snf1-activating kinase genes, SAK1, TOS3 and ELM1. These cells were transformed with a low copy plasmid with and without the SAK1 gene (Fig. 7B). In this way, the wild type Snf1 protein would either be phosphorylated or unphosphorylated on its activation loop threonine. We found that Snf1 association with Reg1 requires at least one of the Snf1-activating kinases. Therefore, phosphorylation of the Snf1 activation loop is necessary for association with Reg1. We confirmed the phosphorylation status of the activation loop threonine by western blotting (Fig. 7C) the extracts used in these experiments with a phosphopeptide antibody specific for Snf1 phosphorylated on threonine 210 [1].
Fig. 7.
Requirements for Snf1-Reg1 association. (A) Association of Snf1 and Reg1 was measured by coimmunoprecipitation. Reg1 plasmids expressed with wild type (WT) or Reg1-IF proteins. Snf1 plasmids expressed either wild type or Snf1 with the K84R or T210A point mutations. (B) The requirement for Snf1 phosphorylation was examined by coimmunoprecipitation from cells lacking all three Snf1-activating kinases. Extracts were prepared from cells transformed with a plasmid that expresses Sak1 or empty vector. (C) The phosphorylation status of the Snf1 activation loop in the cells used in panels A and B was measured using a phosphopeptide specific antibody (αPT210).
4. Discussion
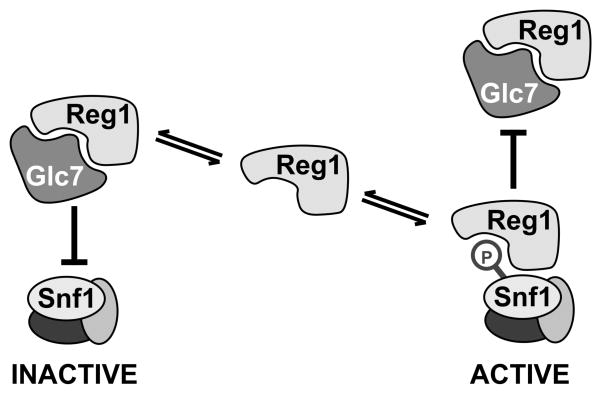
The dephosphorylation of Snf1 is catalyzed by the PP1 phosphatase Glc7 in a complex with the regulatory subunit Reg1. However, the exact role played by Reg1 is uncertain because it binds to Snf1 independently of the Glc7 protein [14] and its association is greater under conditions when Snf1 is protected from dephosphorylation [15]. We began this study with the goal of identifying and separating the Glc7-binding and Snf1-binding determinants of Reg1. In this way, we sought to determine whether the association of Reg1 with Snf1 was required for promotion of or protection from dephosphorylation. We were surprised to find that the domains of Reg1 that are needed to associate with Glc7 are the same domains that are needed to associate with Snf1. Indeed, mutation of the RHIHF sequence in Reg1 eliminated binding of Reg1 to both Glc7 and Snf1. It is uncertain why our experiments found that the RHIHF sequence of Reg1 was required for binding Snf1 (Fig. 6) when an earlier study reported that mutation of phenyalanine 468 did not affect interaction with Snf1 [18]. It is possible that using the two-hybrid system in which a catalytically active protein kinase is recruited to a transcriptional promoter can result in transcriptional activation by mechanisms other than reconstitution of two-hybrid activator [31]. We feel that the coimmunoprecipitation assay is a more direct measure of Reg1-Snf1 association. Mutation of the RHIHF motif in Reg1 completely abrogates association with Snf1. The failure of Reg1-IF to bind to Snf1 is not due to hyperactivation of the Snf1 kinase since the Reg1-IF protein is also unable to bind the catalytically impaired Snf1-K84R protein (Fig. 7A). The simplest interpretation is that the RHIHF residues make direct contact with the proteins which associate with Reg1. It is difficult to imagine that the Glc7 and Snf1 proteins make simultaneous contact with the same residues on Reg1. A more likely scenario would have both Glc7 and Snf1 competing for the same site on Reg1. Thus, the regulation of Snf1 kinase may be mediated in part through a competition between Glc7 and Snf1 for the binding of Reg1 (Fig. 8).
Fig. 8.
Competitive Binding Model. Snf1 and Glc7 compete for binding to the same surface of Reg1. When bound to Glc7, Reg1 directs the phosphatase to the Snf1 activation loop thereby promoting the inactivation of Snf1 kinase activity. When bound to Snf1, Reg1 protects the Snf1 activation loop from dephosphorylation thereby stabilizing the active state of the Snf1 kinase.
Yeast encode numerous PP1 regulatory subunits that can bind to Glc7 and direct the phosphatase to distinct localizations and targets. Evidence has accumulated that the PP1 regulatory subunits compete with one another for limiting Glc7. Overexpression of the PP1 subunit Gac1 redirected Glc7 to glycogen synthase and interfered with glucose repression mediated by the Glc7/Reg1 complex [32]. Gac1 possesses its own RVxF motif that is required for association with Glc7. Overexpression of Gip3 protein caused the redistribution of Glc7 localization [33]. Gip3 contains a sequence that fits the RVxF consensus but its requirement for Glc7 association has not yet been demonstrated. Thus the regulation of Glc7 localization and function may be controlled by a dynamic competition between regulatory subunits for limiting Glc7.
The Reg1 protein is a PP1 regulatory subunit that binds to both PP1 and to a PP1 substrate. We embarked on this project to assess the functional importance of several domains in Reg1 which are conserved among species, and use this information to build a model for Snf1 regulation in yeast. We found that a newly identified block of conserved amino acids designated as block 4 (Fig. 1) is important for binding to both Glc7 and Snf1. Reg1-C3 has a deletion of 270 C-terminal residues but retains all four conserved blocks of sequence. Reg1-C3 associates with both Glc7 and Snf1. Reg1-C2 contains a larger C-terminal deletion that removes block 4 and is no longer able to associate with either Glc7 or Snf1. Even more intriguing is the finding that the Reg1-IF mutant (I466M, F468A) is unable to bind Glc7 and Snf1. Failure to bind Snf1 is not a consequence of hyperactivation of Snf1 kinase activity since the Reg1-IF protein is unable to associate with a catalytically impaired Snf1 protein (Fig. 7A). Our data suggest that the PP1 regulatory subunit uses the RVxF motif to mediate interaction with both the PP1 phosphatase as well as a substrate of the PP1 phosphatase.
Evidence for competition between PP1 phosphatase and another molecule for the RVxF site has been seen in mammals. The protein kinase A associated protein AKAP149 contains an RVxF motif in its C-terminal KH domain. Mutation of the valine residue to glutamate abrogates binding to both PP1 as well as to RNA [34]. Phosphorylation of a nearby serine residue affects the competition between PP1 and RNA for binding AKAP149 by promoting association with RNA and destabilizing association with PP1. The Reg1 protein is known to be phosphorylated in a Snf1-dependent manner [18]. It remains to be seen whether the phosphorylation of Reg1 is catalyzed directly by Snf1 and whether phosphorylation of Reg1 affects the competition between PP1 and Snf1 for Reg1 binding.
5. Conclusion
The ability of the Reg1 protein to associate with the PP1 phosphatase Glc7 and with the PP1 substrate Snf1 was measured by coimmunoprecipitation. Mutation of the RVxF motif or deletion of the conserved amino acid sequence C-terminal to the RVxF motif eliminated Reg1 binding to both Glc7 and Snf1. Since the same point mutations blocked binding to both proteins, it is reasonable to think that these proteins bind exclusively of the other and that they compete for the same site. We propose a model in which the balance between active and inactive Snf1 is controlled by the competition between Glc7 and Snf1 for binding to the Reg1 protein.
Acknowledgments
This work was supported by grants GM46443 (MS) and DK007729 (ST) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCartney RR, Schmidt MC. Journal of Biological Chemistry. 2001;276(39):36460. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 2.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Proc Natl Acad Sci U S A. 2003;100(15):8839. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. Curr Biol. 2003;13(15):1299. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 4.Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Journal of Biological Chemistry. 1996;271(44):27879. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 5.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. J Biol. 2003;2(4):28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nath N, McCartney RR, Schmidt MC. Mol Cell Biol. 2003;23(11):3909. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. Curr Biol. 2003;13(22):2004. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. J Biol Chem. 2006;281(43):32207. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- 9.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Biochem J. 2007;403(1):139. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubenstein EM, McCartney RR, Zhang C, Shokat KM, Shirra MK, Arndt KM, Schmidt MC. J Biol Chem. 2008;283(1):222. doi: 10.1074/jbc.M707957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedbacker K, Carlson M. Front Biosci. 2008;13:2408. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen PT. J Cell Sci. 2002;115(Pt 2):241. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 13.Tu J, Carlson M. Embo J. 1995;14(23):5939. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbing K, McCartney RR, Schmidt MC. Biochem J. 2006;393(Pt 3):797. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludin K, Jiang R, Carlson M. Proc Natl Acad Sci U S A. 1998;95(11):6245. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Nature. 2004;429(6993):780. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 17.Dombek KM, Voronkova V, Raney A, Young ET. Mol Cell Biol. 1999;19(9):6029. doi: 10.1128/mcb.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanz P, Alms GR, Haystead TA, Carlson M. Mol Cell Biol. 2000;20(4):1321. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 20.Gietz RD, Schiestl RH, Willems AR, Woods RA. Yeast. 1995;11:355. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 21.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Journal of General Virology. 1991;72( Pt 7):1551. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski RS, Hieter P. Genetics. 1989;122(1):19. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein A, Lampen JO. Methods Enzymol. 1975;42C:504. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC. Mol Cell Biol. 1999;19:4561. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedelini L, Marquina M, Arino J, Casamayor A, Sanz L, Bollen M, Sanz P, Garcia-Gimeno MA. J Biol Chem. 2007;282(5):3282. doi: 10.1074/jbc.M607171200. [DOI] [PubMed] [Google Scholar]
- 26.Dombek KM, Kacherovsky N, Young ET. J Biol Chem. 2004;279(37):39165. doi: 10.1074/jbc.M400433200. [DOI] [PubMed] [Google Scholar]
- 27.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. EMBO J. 1997;16(8):1876. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Vit MJ, Waddle JA, Johnston M. Molecular Biology of the Cell. 1997;8(8):1603. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Nature. 2002;415(6868):141. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 30.Celenza JL, Carlson M. Mol Cell Biol. 1989;9(11):5034. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuchin S, Treich I, Carlson M. Proc Natl Acad Sci U S A. 2000;97(14):7916. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Hart H, Cheng C, Roach PJ, Tatchell K. Mol Genet Genomics. 2001;265(4):622. doi: 10.1007/s004380100455. [DOI] [PubMed] [Google Scholar]
- 33.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Mol Cell Biol. 2006;26(7):2648. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogne M, Stokka AJ, Tasken K, Collas P, Kuntziger T. Hum Mol Genet. 2009;18(5):978. doi: 10.1093/hmg/ddn425. [DOI] [PubMed] [Google Scholar]