Abstract
Aims/hypothesis
MicroRNAs regulate a broad range of biological mechanisms. To investigate the relationship between microRNA expression and type 2 diabetes, we compared global microRNA expression in insulin target tissues from three inbred rat strains that differ in diabetes susceptibility.
Methods
Using microarrays, we measured the expression of 283 microRNAs in adipose, liver and muscle tissue from hyperglycaemic (Goto–Kakizaki), intermediate glycaemic (Wistar Kyoto) and normoglycaemic (Brown Norway) rats (n = 5 for each strain). Expression was compared across strains and validated using quantitative RT-PCR. Furthermore, microRNA expression variation in adipose tissue was investigated in 3T3-L1 adipocytes exposed to hyperglycaemic conditions.
Results
We found 29 significantly differentiated microRNAs (padjusted < 0.05): nine in adipose tissue, 18 in liver and two in muscle. Of these, five microRNAs had expression patterns that correlated with the strain-specific glycaemic phenotype. MiR-222 (padjusted = 0.0005) and miR-27a (padjusted = 0.006) were upregulated in adipose tissue; miR-195 (padjusted = 0.006) and miR-103 (padjusted = 0.04) were upregulated in liver; and miR-10b (padjusted = 0.004) was downregulated in muscle. Exposure of 3T3-L1 adipocytes to increased glucose concentration upregulated the expression of miR-222 (p = 0.008), miR-27a (p = 0.02) and the previously reported miR-29a (p = 0.02). Predicted target genes of these differentially expressed microRNAs are involved in pathways relevant to type 2 diabetes.
Conclusion
The expression patterns of miR-222, miR-27a, miR-195, miR-103 and miR-10b varied with hyperglycaemia, suggesting a role for these microRNAs in the pathophysiology of type 2 diabetes, as modelled by the Gyoto–Kakizaki rat. We observed similar patterns of expression of miR-222, miR-27a and miR-29a in adipocytes as a response to increased glucose levels, which supports our hypothesis that altered expression of microRNAs accompanies primary events related to the pathogenesis of type 2 diabetes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-010-1667-2) contains supplementary material, which is available to authorised users.
Keywords: Expression, MicroRNA, Murine diabetes model
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that bind to regulatory sites of target mRNA and modify their expression, either by translational repression or target mRNA degradation, resulting in decreased protein production. MiRNA–mRNA targeting is achieved through both perfect and imperfect sequence alignment mechanisms. A single miRNA may potentially regulate the expression of several hundred genes. MiRNAs are increasingly recognised as being involved in a broad range of biological functions [1] and are known to be expressed in a tissue-specific manner [2]. A growing body of evidence implicates miRNAs in type 2 diabetes and pathophysiological elements of the cardiometabolic syndrome characterised by hyperglycaemia (reviewed by Krutzfeldt and Stoffel [3]). MiRNAs are required for pancreatic development [4], the regulation of glucose-stimulated insulin secretion [5, 6] and amino acid catabolism [7]. They are also involved in many functional aspects of insulin target tissues, such as adipocyte differentiation [8], myogenesis and myoblast differentiation [9] and fatty acid synthesis in hepatocytes [10].
MiRNA profiling using microarray technology has recently been developed and applied to the study of a variety of conditions [11, 12]. To date, only one study has investigated miRNA expression in the context of type 2 diabetes [13], using the Goto–Kakizaki (GK) rat, a model of spontaneous, lean type 2 diabetes [14]. Fifteen miRNAs were found to differ in expression by more than 1.5-fold between GK and Wistar control rats in skeletal muscle. As global miRNA expression was investigated in only one tissue and only in small samples (two rats per group), further research in this area is needed.
Animal models offer great advantages for expression studies, as their homogeneous genetic background and controlled environment decrease the variability of gene expression levels among colony members. A number of polygenic animal models are available for type 2 diabetes studies [15], including the widely studied GK strain. The GK strain was derived by selectively breeding outbred Wistar rats that spontaneously developed glucose intolerance [14] and is a particularly good model for type 2 diabetes. The defects of these rats include glucose-stimulated insulin secretion, peripheral insulin resistance, stable fasting hyperglycaemia and hyperinsulinaemia, which can be detected as early as 2–4 weeks of age [14, 16]. Additionally, the GK strain is non-obese and consequently not affected by the confounding risk factors for type 2 diabetes introduced by obesity.
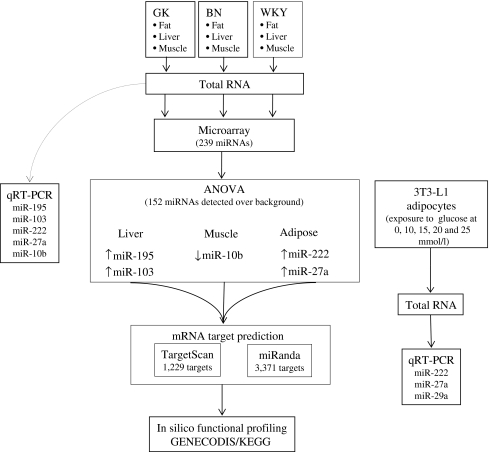
Given the critical roles miRNAs are suggested to play in several aspects of glucose homeostasis, we hypothesised that miRNA expression profiles of insulin target tissues differ between hyperglycaemic and normoglycaemic status. We employed microarray technology to generate miRNA expression profiles in adipose tissue, liver and skeletal muscle from GK rats, and compared these with expression profiles from normoglycaemic, genetically related Wistar Kyoto (WKY) rats, and normoglycaemic, genetically distant Brown Norway (BN) rats, which are extensively used to study genetic determinants of type 2 diabetes phenotypes in F2 hybrids and congenic strains [17]. Phenotypic data for these strains, including fasting glucose levels, show that the WKY strain has an intermediate phenotype relative to GK and BN strains. Therefore, we aimed to identify miRNAs showing differential expression across strains, and specifically those with intermediate expression levels in WKY rats. Predicted target genes of differentially expressed miRNAs were identified and their biological functions assessed using bioinformatics tools (see Fig. 1 for an outline of the study design).
Fig. 1.
Outline of study design and results
Methods
Research design and methods Four-month-old male rats from GK (n = 5), WKY (n = 5) and BN (n = 5) colonies were fed with standard laboratory chow pellets (B&K Universal, Hull, UK) and kept on a 12 h light/12 h dark cycle. Animal procedures were carried out in accordance with UK Home Office and institutional guidelines. GK and BN strains were obtained from Oxford colonies (GK/Ox and BN/Ox) and WKY rats from a commercial supplier (Harlan, Bicester, UK). Fasting glycaemia was determined in representative animals from the GK (n = 8), BN (n = 4) and WKY (n = 5) colonies after a 16–18 h fast, using an intraperitoneal glucose tolerance test (IPGTT) [18]. Plasma concentrations of glucose were determined using diagnostic kits (ABX, Shefford, UK) on a Cobas Mira Plus automatic analyser (ABX).Liver, white adipose tissue (retroperitoneal fat pad) and skeletal muscle (soleus) samples were obtained from each of the 15 rats. Total RNA was then extracted from homogenised tissue samples using TRI Reagent (Sigma, Gillingham, UK) in accordance with the manufacturer’s instructions. RNA quality was assessed using a spectrophotometer (NanoDrop; Labtech International, Ringmer, UK) and a Bioanalyzer 2100 (Agilent, South Queensferry, UK). RNA was generally of high quality (average 260/280 ratio 2.13 and average 260/230 ratio 1.83). Following RNA quality control steps, 15 muscle samples (GK = 5, BN = 5 and WKY = 5), 14 fat samples (GK = 4, BN = 5 and WKY = 5) and 13 liver samples (GK = 4, BN = 5 and WKY = 4) were used. Pancreatic tissue was not analysed in this study because of the difficulty of extracting total RNA from pancreatic beta cells, the number of which is significantly reduced in GK animals.
LNA-based miRNA microarray MiRNA profiling of liver, fat and skeletal muscle samples was performed using the locked nucleic acid (LNA mercury [19]) microarray from Exiqon (Vedbaek, Denmark), which contains probes for 239 rat-specific miRNAs (based on mirBASE v8.1). Briefly, 2 μg of total RNA from experimental samples and from a common reference rat sample were labelled in separate reactions with Hy3 and Hy5 (Exiqon) fluorescent labels respectively. Each pair of labelled experimental and reference samples was combined, denatured and hybridised to the miRCURY LNA Array v.8.1 (Exiqon). This array contains four replicates of each of the 239 rat miRNA probes as well as spike-ins, negative controls and proprietary Exiqon miRNA probes. Hybridisation was performed according to the miRCURY LNA array manual using a Tecan HS4800 hybridisation station (Tecan, Männedorf, Germany). Low- and high-stringency washes were carried out to minimise non-specific hybridisation and the microarrays were then dried. Images were acquired using an Axon GenePix 4000B scanner (Axon Instruments-MDS, Sunnyvale, CA, USA) and GenePix software (Sunnyvale, CA, USA).
Microarray data processing Raw intensity data were read into the R statistical package [20] for further processing using the ‘marray’ and ‘limma’ [21] libraries from BioConductor [22]. Data were background-corrected using negative control probes on the array to estimate background signal. Probes were considered expressed in a given sample when the intensity exceeded a threshold equal to the median background (across all samples) + 2 SD units. These data were then used to identify probes detected in at least one of the three tissues; a probe was considered expressed in a tissue if the median intensity exceeded the background threshold. The data were normalised using the locally weighted scatterplot smoothing (LOESS) regression algorithm within each array, to adjust for any intensity-dependent dye bias. Because of the expected differences in miRNA expression between tissues, between-array normalisation was not performed. Inspection of box plots of the M-ratios (log [experimental/reference]) showed that all arrays for a given tissue had comparable distributions. Rat-specific miRNAs that were expressed above background in at least one tissue were included in the subsequent analyses (n = 152). Differential expression between tissues was calculated using functions in the limma package, using data from all three strains (adipose tissue, n = 14; liver, n = 13; muscle, n = 14).Then, data for each tissue were analysed for differential expression between strains, again using limma. All results of expression analysis were corrected for multiple testing using the false discovery rate (FDR) method of Hochberg and Benjamini [23] and FDR-adjusted p values <0.05, corresponding to an FDR of 5%, were considered significant.
Direct miRNA quantification Validation of miRNA expression data for five miRNAs (miR-222, miR-103, miR-27a, miR-195, miR-10b) by quantitative RT-PCR was performed using total RNA from the 42 rat samples studied and microRNA specific primers (Applied Biosystems [ABI], Warrington, UK). Briefly, reverse transcription was carried out in a total reaction volume of 15 µl containing 5 µl total RNA (concentration 10 ng/µl), 3 µl of reverse transcription primer, 1.50 µl of 10× RT buffer, 1.00 µl MultiScribe Reverse Transcriptase (50 U/µl), 0.15 µl of 100 mmol/l dNTPs (with dTTP), 0.19 µl of RNase Inhibitor, 20 U/µl and 4.16 µl of nuclease-free water (all reagents supplied by ABI). Reactions were incubated according to the manufacturer’s recommendations. Quantitative RT-PCRs were performed in triplicate; the 10 µl PCR reaction contained 1.33 µL reverse transcription product, 10 µl 2× PCR Master Mix, 1 µl microRNA primer (ABI) and 7.67 µl of nuclease-free water. The reactions were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 35 s. The highly conserved and universally expressed snoRNA and 4.5 S RNA (H-rat) were used as normalising endogenous controls in the qRT-PCR. Fold changes (FC) in expression were calculated using the 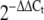 method [24].
method [24].
Exposure of 3T3-L1 adipocyte cells to altered glucose concentration We cultured 3T3-L1 fibroblasts (obtained from ECACC (European Collection of Cell Cultures, Salisbury, UK) in DMEM (containing 25 mmol/l glucose) supplemented with 4 mmol/l glutamine and 10% calf serum in addition to penicillin (100 U/ml) and streptomycin (100 μg/ml) in humidified air at 37°C and 5% CO2. Two days after confluence, cells were differentiated by replacing calf serum with 10% FBS and addition of 250 nmol/l dexamethasone, 0.5 mmol/l 3-isobutyl-1-methylxanthine (IBMX) and 100 nmol/l insulin to the growing medium. After 2 days, cells were placed in DMEM containing 4 mmol/l glutamine, 10% FBS and 100 nmol/l insulin for two more days. Following differentiation, cells were maintained in DMEM containing 5 mmol/l glucose plus 4 mmol/l glutamine and 10% FBS for an additional 2 days to recover at a low glucose concentration and subsequently incubated for 24 h with 5, 10, 15, 20 or 25 mmol/l glucose. Four biological replications of this glucose stimulation experiment were carried out. Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Crawley, UK). Expression levels of miR-222, miR-27a and the previously reported miR-29a (used here as a positive control) were measured by qRT-PCR as above, using microRNA specific primers (ABI) on the Bio-Rad (Bio-Rad, Hemel Hempstead, UK) platform (PCR reactions were run in triplicate). MiRNA expression was normalised to the expression of the mouse controls snoRNA202 and snoRNA234. Fold changes in expression were calculated using the 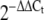 method [24] by comparing miRNA expression at 10, 15, 20 and 25 mmol/l with expression at baseline (5 mmol/l). Analysis of variance was used to compare differences in the mean values across all glucose exposures.
method [24] by comparing miRNA expression at 10, 15, 20 and 25 mmol/l with expression at baseline (5 mmol/l). Analysis of variance was used to compare differences in the mean values across all glucose exposures.
In silico functional profiling of target genes Potential miRNA target genes were identified and retrieved using the algorithms implemented by the Sanger microRNA database [25] and by TargetScan 4.2 [26]. Biological gene annotation was generated using GENECODIS [27]. The GENECODIS algorithm uses the hypergeometric distribution to determine whether individual pathways or combinations of pathways are significantly overrepresented among the genes of interest. The p values computed for each pathway were adjusted using the FDR method of Hochberg and Benjamini [23] to control the false discovery rate, and corrected p values <0.01 were considered significant.
Results
Characteristics of GK, BN and WKY rats To avoid possible long-term modification of gene expression (including miRNAs), which might be caused by intervention during glucose tolerance testing (in particular the use of anaesthesia), phenotyping was not performed in the same rats used in this experiment, but in 4-month-old male rats representative of the GK (n = 8), BN (n = 4) and WKY (n = 5) colonies (see Electronic supplementary material [ESM] Fig. 1). Cumulative glycaemia was significantly increased in GK vs BN (p < 0.001), GK vs WKY (p < 0.001) and WKY vs BN (p < 0.05; ESM Fig. 1). An IPGTT showed a significant increase in glycaemia in response to glucose stimulus measured over 120 min, again with significant differences between the strains (ESM Fig. 1). Although WKY animals are normoglycaemic, they have plasma glucose levels and cumulative glycaemia higher than those of BN rats but lower than those of GK rats, placing WKY animals at an intermediate phenotypic level (ESM Fig. 1). These phenotypic data are a reasonable assessment of the glycaemic status of the rats used in this study, as they were of the same age and from the same colonies. In addition, several studies have reported increases in postprandial plasma glucose, insulin levels and fasting plasma glucose in GK rats starting a few weeks after birth [14, 28, 29].
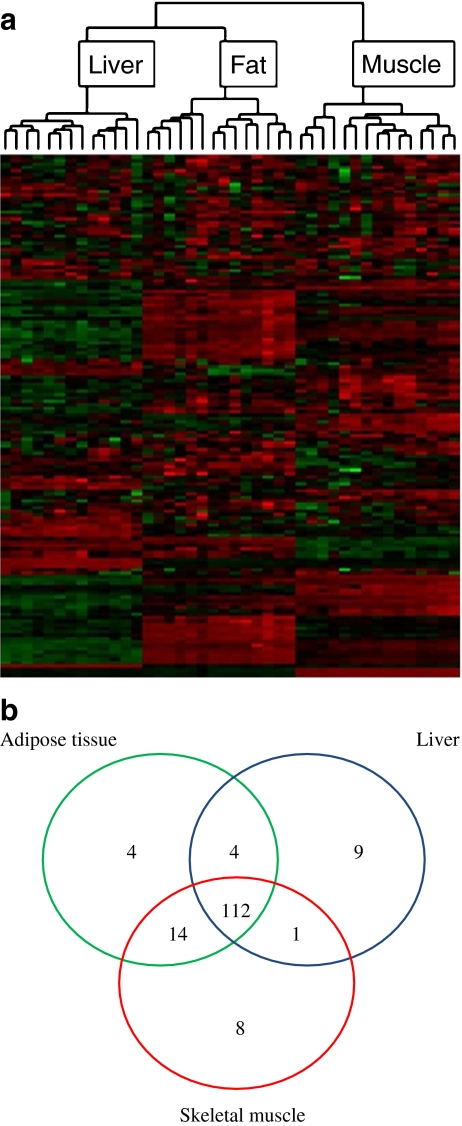
MiRNA expression differs between insulin target tissues MiRNA expression was characterised using the Exiqon microarray platform in 13 liver, 15 muscle and 14 adipose tissue samples that passed quality control tests (from a total of 15 samples per tissue). Unsupervised hierarchical clustering showed that each tissue had a distinct miRNA expression profile (Fig. 2a). Of the 239 miRNA probes evaluated, 152 were expressed over background in at least one of the tissues, with 112 expressed in all three (Fig. 2b). Among the miRNAs detected in only one of three tissues studied were several with known tissue specificity, such as miR-1, miR-206 and miR-133 in muscle and miR-122 in liver (ESM Table 1). The vast majority of miRNAs (123/152, 81%) showed statistically significant differences in expression (adjusted p < 0.05) between at least one pair of tissues, all with greater than 1.5-fold change (ESM Table 1). These results highlight the tissue-dependent pattern of miRNA expression.
Fig. 2.
miRNA expression was specific in insulin target tissues from GK, WKY and BN rats. a Unsupervised hierarchical clustering analysis for expression of 152 miRNAs differentiated samples by tissue. Each row represents an miRNA and each column a tissue sample. Colours represent relative intensity of the detected signal in each sample, red representing high expression and green representing low expression. b Summary of miRNA expression across three insulin target tissues (adipose tissue, liver and skeletal muscle) for 152 miRNAs detected above background levels
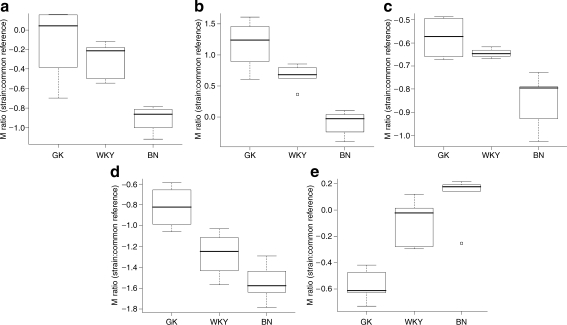
MiRNA expression differs between diabetic and normoglycaemic rat strains Analysis of differential expression between the three rat strains using ANOVA revealed 29 significant miRNAs (adjusted p < 0.05). Of these, nine were observed in adipose tissue, 18 in liver and two in muscle (Table 1; ESM Fig. 2). Pairwise comparisons between strains (GK/BN, BN/WKY and GK/WKY) were also carried out to confirm the origin of the differences detected in the ANOVA analysis (Table 1). The expression levels of five miRNAs corresponded approximately to a linear relationship between the glycaemic phenotypes of the three strains. These were: miR-222 and miR-27a in adipose tissue (both with BN<WKY<GK); miR-195 and miR-103 in liver (both with BN<WKY<GK); and 10b in skeletal muscle (BN>WKY>GK; Fig. 3). In liver, miR-191 was more highly expressed in the GK rats than in either of the normoglycaemic strains (GK>BN∼WKY), while miR-200a had higher expression in the normoglycaemic strains (BN∼WKY>GK; ESM Fig. 2). While these patterns may be of biological interest (ESM Fig. 2), possibly reflecting threshold effects of miRNA expression in relation to the phenotype, we decided to focus on the five miRNAs with an approximately linear relationship in expression levels between strains.
Table 1.
miRNAs that showed different expression among strains GK, WKY and BN in three insulin target tissues
| Tissue | miRNA | Rat chromosome location | Accession number | Location | Strand | p value | Comparison | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GK/BN | BN/WKY | GK/WKY | ||||||||||||
| Start | End | Fold change | p value | Fold change | p value | Fold change | p value | |||||||
| Adipose | rno-miR-140* | 19 | MI0000611 | 37422704 | 37422802 | + | 1.4 × 10−2 | 1.20 | 2.6 × 10−1 | −1.45 | 3.9 × 10−3 | −1.22 | 4.2 × 10−1 | |
| Adipose | rno-miR-207 | 5 | MI0003479 | 58101186 | 58101263 | + | 2.1 × 10−2 | −1.39 | 9.3 × 10−2 | 1.55 | 8.1 × 10−3 | 1.11 | 8.6 × 10−1 | |
| Adipose | rno-miR-21 | 10 | MI0000850 | 74864500 | 74864591 | − | 4.7 × 10−4 | 1.11 | 5.7 × 10−1 | −1.76 | 3.3 × 10−4 | −1.59 | 8.6 × 10−3 | |
| Adipose | rno-miR-222 | X | MI0000962 | 14935056 | 14935158 | + | 4.7 × 10−4 | 2.43 | 1.4 × 10−4 | −1.70 | 3.9 × 10−3 | 1.42 | 2.1 × 10−1 | † |
| Adipose | rno-miR-27a | 19 | MI0000860 | 25638578 | 25638664 | − | 5.5 × 10−3 | 1.74 | 5.1 × 10−3 | −1.52 | 1.4 × 10−2 | 1.15 | 8.4 × 10−1 | † |
| Adipose | rno-miR-29a | 4 | MI0000863 | 58107760 | 58107847 | − | 7.9 × 10−4 | 1.18 | 2.8 × 10−1 | −1.68 | 3.3 × 10−4 | −1.42 | 4.0 × 10−2 | |
| Adipose | rno-miR-335 | § | MI0000612 | – | – | n/a | 5.5 × 10−3 | 1.96 | 3.0 × 10−2 | −2.24 | 3.6 × 10−3 | −1.14 | 9.0 × 10−1 | |
| Adipose | rno-miR-347 | § | MI0000635 | – | – | n/a | 6.2 × 10−3 | −1.66 | 3.0 × 10−2 | 1.85 | 3.8 × 10−3 | 1.11 | 9.0 × 10−1 | |
| Adipose | rno-miR-487b | 6 | MI0003547 | 1.34 × 108 | 1.34 × 108 | + | 1.2 × 10−3 | −1.40 | 5.1 × 10−3 | 1.43 | 1.1 × 10−3 | 1.02 | 9.5 × 10−1 | |
| Liver | rno-miR-100 | 8 | MI0000885 | 44518670 | 44518749 | + | 3.7 × 10−3 | −1.00 | 9.8 × 10−1 | −1.41 | 2.7 × 10−3 | −1.41 | 4.2 × 10−3 | |
| Liver | rno-miR-103 | 10 | MI0000888 | 20695027 | 20695112 | + | 4.1 × 10−2 | 1.21 | 6.0 × 10−2 | −1.16 | 9.2 × 10−2 | 1.05 | 9.6 × 10−1 | † |
| Liver | rno-miR-125b | 8 | MI0000896 | 44570155 | 44570241 | + | 3.7 × 10−3 | −1.09 | 5.3 × 10−1 | −1.33 | 5.7 × 10−3 | −1.44 | 3.7 × 10−3 | |
| Liver | rno-miR-126 | 3 | MI0000898 | 4768117 | 4768189 | + | 3.3 × 10−2 | −1.40 | 6.0 × 10−2 | 1.43 | 2.4 × 10−2 | 1.02 | 9.9 × 10−1 | |
| Liver | rno-miR-140* | 19 | MI0000611 | 37422704 | 37422802 | + | 3.5 × 10−2 | 1.14 | 3.9 × 10−1 | −1.32 | 1.5 × 10−2 | −1.16 | 3.6 × 10−1 | |
| Liver | rno-miR-146 | 10 | MI0000919 | 28476368 | 28476462 | − | 7.0 × 10−3 | 1.24 | 6.0 × 10−2 | −1.36 | 4.2 × 10−3 | −1.10 | 6.4 × 10−1 | |
| Liver | rno-miR-15b | 2 | MI0000843 | 1.59 × 108 | 1.59 × 108 | + | 7.0 × 10−3 | −1.20 | 6.0 × 10−2 | 1.32 | 4.2 × 10−3 | 1.10 | 6.1 × 10−1 | |
| Liver | rno-miR-191 | 8 | MI0000934 | 1.14 × 108 | 1.14 × 108 | + | 3.3 × 10−2 | 1.27 | 6.0 × 10−2 | −1.02 | 8.7 × 10−1 | 1.25 | 6.9 × 10−2 | |
| Liver | rno-miR-193 | 10 | MI0000936 | 65901048 | 65901133 | + | 3.5 × 10−2 | 1.32 | 6.0 × 10−2 | 1.00 | 1.0 | 1.32 | 6.9 × 10−2 | |
| Liver | rno−miR-195 | 10 | MI0000939 | 57074170 | 57074256 | + | 6.2 × 10−3 | 1.65 | 6.1 × 10−3 | −1.21 | 2.0 × 10−1 | 1.37 | 6.9 × 10−2 | † |
| Liver | rno-miR-200a | 5 | MI0000943 | 1.73 × 108 | 1.73 × 108 | − | 7.0 × 10−3 | −1.55 | 6.0 × 10−2 | −1.17 | 4.4 × 10−1 | −1.81 | 6.0 × 10−3 | |
| Liver | rno-miR-222 | X | MI0000962 | 14935056 | 14935158 | + | 2.3 × 10−2 | 1.17 | 1.9 × 10−1 | −1.35 | 8.0 × 10−3 | −1.15 | 3.6 × 10−1 | |
| Liver | rno-miR-27b | 17 | MI0000859 | 7351449 | 7351545 | − | 3.5 × 10−2 | −1.24 | 1.1 × 10−1 | 1.31 | 2.3 × 10−2 | 1.06 | 9.6 × 10−1 | |
| Liver | rno-miR-290 | 1 | MI0000964 | 64274926 | 64275006 | − | 3.7 × 10−3 | 1.13 | 5.7 × 10−1 | −1.87 | 2.7 × 10−3 | −1.66 | 1.4 × 10−2 | |
| Liver | rno-miR-292-5p | 1 | MI0000966 | 64274340 | 64274421 | − | 3.2 × 10−2 | 1.05 | 9.1 × 10−1 | −2.17 | 1.8 × 10−2 | −2.07 | 6.9 × 10−2 | |
| Liver | rno-miR-296 | 3 | MI0000967 | 1.65 × 108 | 1.65 × 108 | − | 2.3 × 10−2 | 1.11 | 5.3 × 10−1 | −1.39 | 8.0 × 10−3 | −1.25 | 1.6 × 10−1 | |
| Liver | rno-miR-335 | § | MI0000612 | – | – | n/a | 1.1 × 10−2 | −1.05 | 7.0 × 10−1 | 1.38 | 5.7 × 10−3 | 1.31 | 4.7 × 10−2 | |
| Liver | rno-miR-422b | 18 | MI0003719 | 57377805 | 57377869 | − | 3.2 × 10−2 | −1.22 | 1.4 × 10−1 | 1.36 | 1.3 × 10−2 | 1.11 | 7.0 × 10−1 | |
| Skeletal muscle | rno−let-7e | 1 | MI0000832 | 56487134 | 56487226 | + | 3.5 × 10−3 | 1.04 | 8.5 × 10−1 | −1.36 | 4.0 × 10−3 | −1.30 | 2.0 × 10−2 | |
| Skeletal muscle | rno-miR-10b | 3 | MI0000842 | 57340853 | 57340961 | + | 3.5 × 10−3 | −1.59 | 1.5 × 10−3 | 1.14 | 4.0 × 10−1 | −1.40 | 2.6 × 10−2 | † |
Rno-miR-422b is now called rno-mir-378 (accession number MI0003719; located on rat chromosome 18).
Strain differences in expression were detected by ANOVA, All p value s are adjusted for FDR = 5%
§Indicates that genomic microRNA location is unknown (rat chromosome location column)
†miRNAs for which quantitative RT-PCR was attempted
n/a, strand unknown
Fig. 3.
Box plots of expression in strains GK, WKY and BN (relative to a common reference) for five miRNAs whose expression was consistent with glycaemic status. The pattern GK>WKY>BN was observed for miR-27a (a) and miR-222 (b) in adipose tissue, and for mir-103 (c) and miR-195 (d) in liver, whereas GK<WKY<BN was observed for miR-10b in muscle (e). Circles represent outliers
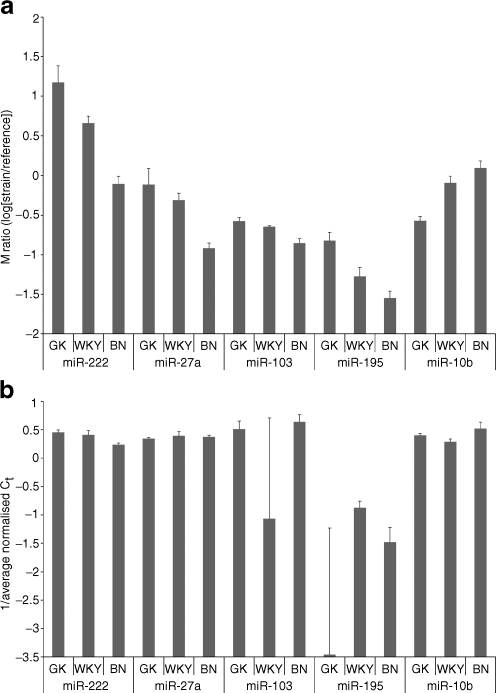
Validation of differential expression between hyperglycaemic and normoglycaemic rats The five miRNAs (miR-222, miR-27a, miR-195, miR-103, miR-10b) with expression patterns consistent with glycaemic status (GK<WKY<BN or GK>WKY>BN) were selected for validation using the same samples as in the microarray study. Relative levels of expression across all strain comparisons for miR-222 in adipose tissue and miR-103 in liver were validated by qRT-PCR (Fig. 4). However, inconsistent results between array and quantitative RT-PCR data were obtained for at least one of the three possible strain comparisons for mir-27a in adipose tissue, miR-195 in liver and miR-10b in muscle (Fig. 4). It was notable that miR-222 showed the greatest expression differences between strains, and was the most successful validation. Thus, the modest fold changes could be one explanation for the relatively poor validation rate, which we have estimated at 60% (nine replicated fold changes out of 15 comparisons). However, our results with respect to validation are in agreement with other studies that have shown the success of qRT-PCR, as a validation tool for miRNA expression, to range between 50 and 100% [30, 31]. The fold change detected in the qRT-PCR experiment was much greater than that in the microarray experiment; for miR-222 (4.8-fold increase [GK/BN] in adipose tissue compared with a 2.4-fold change in the microarray experiment), this may reflect the higher sensitivity and larger dynamic range of quantitative RT-PCR, and has been observed in other studies [32].
Fig. 4.
Signal intensity ratios between strains GK, WKY and BN for array data (a) and quantitative reverse transcriptase PCR (b) for miR-222 and miR-27a in adipose tissue, miR103 and miR-195 in liver and miR-10b in muscle. a Microarray log-ratio intensities for array data for each sample relative to the common reference. Positive values indicate higher expression in the experimental sample relative to the common reference, and vice versa for negative values. Error bars show the SE. The larger the mean log-ratio for a given strain, the more highly expressed the corresponding miRNA. b Expression of miRNAs detected using qRT-PCR. Data are presented as the inverse of the Ct score. Columns indicate expression relative to the control genes (snoRNA and 4.5S RNA) and error bars show the SE
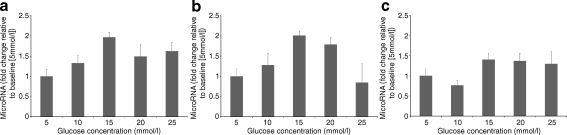
MicroRNA expression increases in response to hyperglycaemia in adipocytes Exposure of 3T3-L1 adipocytes to increasing concentrations of glucose (10, 15, 20 and 25 mmol/l) resulted in an overall overexpression of miR-222, miR-27a and miR-29a when compared with baseline (5 mmol/l). MiR-27a expression declined initially (FC = 0.76) at 10 mmol/l and then increased to values ranging between 1.29 and 1.4 (ANOVA, p = 0.023). MiR-222 was initially overexpressed at 10 (FC = 1.27), 15 (FC = 2.01) and 20 (FC = 1.79) mmol/l of glucose. At the highest concentration of glucose (25 mmol/l), the expression of miR-222 fell below expression at the baseline (FC = 0.8; ANOVA, p = 0.0084). The expression of miR-29a (our positive control) followed a pattern similar to that presented by miR-222 but with a more gradual decline towards higher glucose levels and never reaching expression below the baseline level (FC = 1.32 at 10 mmol/l, FC = 1.96 at 15 mmol/l, FC = 1.49 at 20 mmol/l and FC = 1.69 at 25 mmol/l; ANOVA, p = 0.023; Fig. 5).
Fig. 5.
Relative changes in expression of miRNAs in 3T3-L1 adipocytes following stimulation with glucose. Fold changes in expression of a miR-29a, b miR-222 and c miR-27a in response to exposure of 3t3-L1 adipocytes to glucose 5, 10, 15, 20 and 25 mmol/l for 24 h. MicroRNA expression was assayed by qRT-PCR. Fold changes were calculated using the 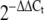 method and are relative to baseline expression at 5 mmol/l. Error bars represent the standard deviation
method and are relative to baseline expression at 5 mmol/l. Error bars represent the standard deviation
In silico functional profiling To gain insight into the potential roles of the miRNAs that followed the observed glycaemic phenotype between the three strains, a list of potential gene-target transcripts was generated for miR-222, miR-27a, miR-195, miR-103 and miR-10b. Predicted target genes for these five miRNAs were identified using two algorithms: miRanda and TargetScan, which produced 3,371 and 1,229 unique targets, respectively. Surprisingly, there was no overlap between these two lists, highlighting the current difficulty in inferring likely downstream effects of altered miRNA expression. Each list was then profiled by querying the annotated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using GENECODIS [27] and pathways significant in both analyses are reported. The genes predicted to be targets of these miRNAs were significantly enriched (FDR, adjusted p < 0.01) for a number of signalling pathways related to glucose homeostasis. Among the top-ranking pathways in each analysis were the mitogen-activated protein kinase (MAPK) signalling pathway, the insulin signalling pathway and the gonadotropin-releasing hormone signalling pathway (ESM Table 2). As miR-222 was validated both biologically and technically, a similar approach was used to query the miRanda and TargetScan lists of predicted target genes of this specific miRNA. Among the significant pathways overlapping between the two predictions, we observed the MAPK signalling pathway (ESM Table 3).
Discussion
There is increasing evidence that miRNAs play a role in many aspects of metabolism and glucose homeostasis, and hence may be involved in the pathogenesis of disorders such as type 2 diabetes. In this study we used microarrays to obtain global expression profiles of miRNAs in three insulin target tissues from hyperglycaemic rats (GK) and compared them with expression profiles from controls derived from genetically related (WKY) and genetically distant (BN) strains (for study design and data breakdown, see Fig. 1).
Phenotypic data indicated that the WKY rats represent an intermediate glycaemic phenotype between hyperglycaemic GK rats and normoglycaemic BN rats (ESM Fig. 1). We therefore focused on five miRNAs with relative expression levels correlating with glycaemic status: miR-222 and miR-27a are upregulated with hyperglycaemic status in adipose tissue, miR-195 and miR-103 are upregulated in liver, and miR-10b is downregulated in skeletal muscle. Thus, four of the miRNAs showed highest expression in GK and lowest in BN, while one showed the reverse pattern. These five miRNAs showed a wide range of expression levels in different tissues (averaged across strains) and relative to each other (ESM Table 1). With the exception of miR-27a, these miRNAs were expressed at their highest levels in adipose tissue and lowest levels in liver. Indeed, miR-222, miR-27a and miR-10b were only expressed close to background levels in liver. Interestingly, each of the five miRNAs showed the significant pattern in only one of the three tissues studied, i.e. relative expression between strains differed between tissues for the same miRNA. This may indicate that the changes in miRNA expression in a given tissue relate to particular processes occurring in that tissue, rather than to a general dysregulation of miRNA expression. The inclusion of a strain with an intermediate phenotype offered increased sensitivity in detecting miRNAs that vary coordinately with the hyperglycaemic phenotype, as different sets of miRNAs were identified as differentially expressed when the GK strain was compared with BN or WKY separately (see Comparison columns in Table 1). The most dramatic changes were observed for miR-222 in adipose tissue (BN<WKY<GK), and were confirmed both by qRT-PCR and exposure of adipocytes to increased glucose levels in vivo to be significantly correlated with the hyperglycaemic phenotype. Upregulation of miR-222 has been shown to be involved in cell cycle regulation through its control of cyclin-dependent kinase inhibitor (p27Kip1) expression [33, 34]. MiR-222 expression was highest in the hyperglycaemic GK strain, and in view of microRNA’s regulatory role we would expect the expression of its target genes to be affected in GK adipose tissue as a result. Although not included in the design or the scope of this study, direct control of target gene expression exerted by miRNAs may be established through functional studies that evaluate correlations with mRNA and protein levels [35, 36].
Comparison of miRNA expression between the two normoglycaemic strains (BN vs WKY) in the present study showed significant differences in the expression levels of 30 out of 152 (∼20%) detected miRNAs, with modest fold changes. These changes could reflect the strong genetic divergence between WKY and BN strains [37] as well as signatures of physiological features that characterise these strains; for example, the WKY strain shows resistance to mammary cancers, exhibits exaggerated neuroendocrine and behavioural responses to stress and is used as a model of depression [38, 39]. Further evidence of possible strain effects that might interfere with the interpretation of microRNA expression data arises from a previous study comparing the expression of 89 miRNAs in GK and outbred Wistar rats, in which overexpression of miR-29a, miR-29b and miR-29c was detected in muscle, liver and adipose tissue from 3-month-old GK rats compared with Wistar rats [13]. Our microarray findings indicate that miR-29a had significantly different expression between the three strains (adjusted p = 0.0008) only in adipose tissue, and the expression of this miRNA across these strains followed a BN<GK<WKY pattern. The disparity of these observations could indicate that variation in the expression of miR-29a is not due to the hyperglycaemic phenotype of the GK rat, but represents a strain/genetic difference in miRNA expression. However, exposure of 3T3-L1 adipocytes to experimental hyperglycaemic conditions showed that increased glucose stimulated the expression of miR-29a, suggesting that this increase in expression is a genuine observation and not a strain effect.
Because hyperglycaemia is already present in GK animals and, to some extent (compared with BN levels), in the WKY strains, it is not possible through a microarray/qRT-PCR approach to determine whether these differences in miRNA expression are primary or secondary to the type 2 diabetes phenotype. The effect of glucose levels on the expression of these miRNAs was evaluated using cultured adipocytes exposed to hyperglycaemia. We observed significantly increased expression of miR-222, miR-27a and the previously observed miR-29a [13], suggesting that, at least in adipose tissue, increased expression of these miRNAs may be involved in the initial cellular responses to hyperglycaemia and be part of the early cellular events related to the pathogenesis of type 2 diabetes.
The in silico prediction of miRNA target genes and subsequent functional analysis can provide clues as to what biological processes may be disrupted by altered miRNA expression. The MAPK and insulin signalling pathways were suggested by analysis of the combined list of predicted target genes of miR-222, miR-27a, miR-10b, miR-103 and miR-197. Defects in these pathways have been implicated in the pathogenesis of type 2 diabetes [40–42]. Inferring functional effects caused by differential miRNA expression is complex because of the poor overlap between predicted target-gene lists. In an effort to account for this, we report only the pathways significant in the analysis of two lists of predicted target genes generated by the TargetScan and MiRanda algorithms (ESM Table 2).
In conclusion, these findings provide novel information on the molecular mechanisms that may account for genetic loci linked to glucose homeostasis, adiposity [43], lipid metabolism [44] and metabolite abundance [45] in the GK strain. This study comprehensively assesses miRNA expression levels in three insulin target tissues in the GK rat model of type 2 diabetes. As well as confirming that miRNA expression is highly tissue-dependent, we found significant differences between the hyperglycaemic GK rat and intermediate/normoglycaemic controls in muscle, liver and adipose tissue. Furthermore, our data suggest that overexpression of miR-222 and miR-27a and the confirmation of miR29a overexpression in adipocytes represent a response to hyperglycaemia, further implicating altered microRNA expression in the pathophysiology of type 2 diabetes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Median intensity of expression for 152 miRNAS detected above background in at least one insulin target tissue, ranked by adjusted p value for differential expression between tissues (FDR < 0.05) (PDF 127 kb)
Pathway analysis of predicted miR-222, miR-487b, miR-10b, miR-125b and miR-103 target genes generated by theTargetScan and the MiRanda algorithm (PDF 54.4 kb)
Pathway analysis of predicted miR-222 target genes generated by TargetScan and the MiRanda algorithm (PDF 32 kb)
Phenotypingof GK, BN and WKY strains. a Differences in plasma glucose after IPGTT in GK, BN and WKY strains measured over a period of 120 min. a Cumulative glycaemia in GK, BN and WKY rats. b Differences in body weight. c Differences in total cholesterol, HDL-cholesterol and plasma triacylglycerol. *p < 0.05, **p < 0.01, ***p < 0.001, significantly different from BN. †p < 0.05, ††p < 0.01, †††p < 0.001, significant difference between GK and WKY (PDF 27 kb)
Box plots of expression in GK, WKY and BN rats (relative to a common reference) for 29 microRNAs with significant differential expression across strains. Expression patterns of miR-222 and miR-27a in adipose tissue (a), miR-195 and miR-103 in liver (b) and miR-10b in skeletal muscle (c) correlated to fasting glucose phenotypes (GK>WKY>BN or GK<WKY<BN; see ESM Fig. 1). Measured expression levels for: miR-29a, miR-21, miR-347, miR-140*, miR-207 in adipose tissue. MiR-let-7e in skeletal muscle, and miR-290, miR-146, miR15b, miR-296, miR-422b, miR-140* in liver levels which correlated with HDL cholesterol and total cholesterol phenotypes (the order BN<GK<WKY or BN>GK>WKY). In contrast, miR-100, miR-335 and miR-292-5p displayed the WKY>(GK∼BN) or WKY<(GK∼BN) pattern in liver, suggesting that strain effects and not phenotypic effects control both HDL and total cholesterol levels (see ESM Fig. 1). MiR-125b and miR-193 expression in the liver corresponded approximately to a linear relationship between plasma triacylglycerol levels across the three strains (GK>BN>WKY or GK<BN<WKY), while the expression of miR-487b and miR-335 (in adipose tissue) and of miR-27b and miR-126 (in the liver) followed a BN>(GK∼WKY) or a BN<(GK∼WKY) expression pattern, suggesting a strain effect regulating plasma triacylglycerol levels (PDF 85 kb)
Acknowledgements
Funding was provided by Diabetes UK (grant number BDA:RD06/0003287), the Throne-Holst Foundation and the Nuffield Department of Medicine. M. Ria is supported by a postdoctoral fellowship from the Swedish Research Council (524-2007-7839) and a Marie Curie fellowship (PIEF-GA-2008-221255) of the European Commission. D. Gauguier holds a Wellcome Senior Fellowship in Basic Biomedical Science (057733). A. L. Gloyn is an MRC New Investigator (81696). C. M. Lindgren is a Wellcome Trust Career Development Fellow.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Abbreviations
- BN
Brown Norway
- GK
Goto–Kakizaki
- FC
Fold change
- FDR
False discovery rate
- IPGTT
Intraperitoneal glucose tolerance test
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-activated protein kinase
- WKY
Wistar Kyoto
Footnotes
B. M Herrera and H. E. Lockstone contributed equally to the preparation of this manuscript.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 5.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 6.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 7.Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum Mol Genet. 2005;14:3371–3377. doi: 10.1093/hmg/ddi368. [DOI] [PubMed] [Google Scholar]
- 8.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 9.Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219–225. doi: 10.1089/dna.2006.0556. [DOI] [PubMed] [Google Scholar]
- 10.Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2007;9(Suppl 2):67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 14.Goto Y, Kakizaki M, Masaki N. Production of spontaneous diabetic rats by repetition of selective breeding. Tohoku J Exp Med. 1976;119:85–90. doi: 10.1620/tjem.119.85. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan K, Ramarao P. Animal models in type 2 diabetes research: an overview. Indian J Med Res. 2007;125:451–472. [PubMed] [Google Scholar]
- 16.Portha B, Giroix MH, Serradas P, et al. Beta-cell function and viability in the spontaneously diabetic GK rat: information from the GK/Par colony. Diabetes. 2001;50(Suppl 1):S89–S93. doi: 10.2337/diabetes.50.2007.S89. [DOI] [PubMed] [Google Scholar]
- 17.Gauguier D. The rat as a model physiological system. London: Wiley; 2006. [Google Scholar]
- 18.Solberg LC, Valdar W, Gauguier D, et al. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome. 2006;17:129–146. doi: 10.1007/s00335-005-0112-1. [DOI] [PubMed] [Google Scholar]
- 19.Castoldi M, Schmidt S, Benes V, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R_project (2008) R: A language and environment for statistical computing. Available from www.R-project.org, accessed 28 December 2009
- 21.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed]
- 22.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begum N, Ragolia L. Altered regulation of insulin signaling components in adipocytes of insulin-resistant type II diabetic Goto–Kakizaki rats. Metabolism. 1998;47:54–62. doi: 10.1016/S0026-0495(98)90193-7. [DOI] [PubMed] [Google Scholar]
- 29.Portha B, Serradas P, Bailbe D, Suzuki K, Goto Y, Giroix MH. Beta-cell insensitivity to glucose in the GK rat, a spontaneous nonobese model for type II diabetes. Diabetes. 1991;40:486–491. doi: 10.2337/diabetes.40.4.486. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg I, Eran A, Nishino I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 32.Loscher CJ, Hokamp K, Kenna PF, et al. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 34.Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saar K, Beck A, Bihoreau MT, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dugovic C, Solberg LC, Redei E, Reeth O, Turek FW. Sleep in the Wistar–Kyoto rat, a putative genetic animal model for depression. Neuroreport. 2000;11:627–631. doi: 10.1097/00001756-200002280-00038. [DOI] [PubMed] [Google Scholar]
- 39.Will CC, Aird F, Redei EE. Selectively bred Wistar–Kyoto rats: an animal model of depression and hyper-responsiveness to antidepressants. Mol Psychiatry. 2003;8:925–932. doi: 10.1038/sj.mp.4001345. [DOI] [PubMed] [Google Scholar]
- 40.Bouzakri K, Roques M, Gual P, et al. Reduced activation of phosphatidylinositol-3 kinase and increased serine 636 phosphorylation of insulin receptor substrate-1 in primary culture of skeletal muscle cells from patients with type 2 diabetes. Diabetes. 2003;52:1319–1325. doi: 10.2337/diabetes.52.6.1319. [DOI] [PubMed] [Google Scholar]
- 41.Koistinen HA, Chibalin AV, Zierath JR. Aberrant p38 mitogen-activated protein kinase signalling in skeletal muscle from type 2 diabetic patients. Diabetologia. 2003;46:1324–1328. doi: 10.1007/s00125-003-1196-3. [DOI] [PubMed] [Google Scholar]
- 42.Leng Y, Karlsson HK, Zierath JR. Insulin signaling defects in type 2 diabetes. Rev Endocr Metab Disord. 2004;5:111–117. doi: 10.1023/B:REMD.0000021432.84588.f6. [DOI] [PubMed] [Google Scholar]
- 43.Gauguier D, Froguel P, Parent V, et al. Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet. 1996;12:38–43. doi: 10.1038/ng0196-38. [DOI] [PubMed] [Google Scholar]
- 44.Argoud K, Wilder SP, McAteer MA, et al. Genetic control of plasma lipid levels in a cross derived from normoglycaemic Brown Norway and spontaneously diabetic Goto–Kakizaki rats. Diabetologia. 2006;49:2679–2688. doi: 10.1007/s00125-006-0396-z. [DOI] [PubMed] [Google Scholar]
- 45.Dumas ME, Wilder SP, Bihoreau MT, et al. Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nat Genet. 2007;39:666–672. doi: 10.1038/ng2026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Median intensity of expression for 152 miRNAS detected above background in at least one insulin target tissue, ranked by adjusted p value for differential expression between tissues (FDR < 0.05) (PDF 127 kb)
Pathway analysis of predicted miR-222, miR-487b, miR-10b, miR-125b and miR-103 target genes generated by theTargetScan and the MiRanda algorithm (PDF 54.4 kb)
Pathway analysis of predicted miR-222 target genes generated by TargetScan and the MiRanda algorithm (PDF 32 kb)
Phenotypingof GK, BN and WKY strains. a Differences in plasma glucose after IPGTT in GK, BN and WKY strains measured over a period of 120 min. a Cumulative glycaemia in GK, BN and WKY rats. b Differences in body weight. c Differences in total cholesterol, HDL-cholesterol and plasma triacylglycerol. *p < 0.05, **p < 0.01, ***p < 0.001, significantly different from BN. †p < 0.05, ††p < 0.01, †††p < 0.001, significant difference between GK and WKY (PDF 27 kb)
Box plots of expression in GK, WKY and BN rats (relative to a common reference) for 29 microRNAs with significant differential expression across strains. Expression patterns of miR-222 and miR-27a in adipose tissue (a), miR-195 and miR-103 in liver (b) and miR-10b in skeletal muscle (c) correlated to fasting glucose phenotypes (GK>WKY>BN or GK<WKY<BN; see ESM Fig. 1). Measured expression levels for: miR-29a, miR-21, miR-347, miR-140*, miR-207 in adipose tissue. MiR-let-7e in skeletal muscle, and miR-290, miR-146, miR15b, miR-296, miR-422b, miR-140* in liver levels which correlated with HDL cholesterol and total cholesterol phenotypes (the order BN<GK<WKY or BN>GK>WKY). In contrast, miR-100, miR-335 and miR-292-5p displayed the WKY>(GK∼BN) or WKY<(GK∼BN) pattern in liver, suggesting that strain effects and not phenotypic effects control both HDL and total cholesterol levels (see ESM Fig. 1). MiR-125b and miR-193 expression in the liver corresponded approximately to a linear relationship between plasma triacylglycerol levels across the three strains (GK>BN>WKY or GK<BN<WKY), while the expression of miR-487b and miR-335 (in adipose tissue) and of miR-27b and miR-126 (in the liver) followed a BN>(GK∼WKY) or a BN<(GK∼WKY) expression pattern, suggesting a strain effect regulating plasma triacylglycerol levels (PDF 85 kb)