Summary
In plants, type I and II S-adenosyl-L-methionine-dependent O-methyltransferases (OMTs) catalyze most hydroxyl group methylations of small molecules. A homology-based RT-PCR strategy using Catharanthus roseus (Madagascar periwinkle) RNA previously identified six new type I plant OMT family members. We now describe the molecular and biochemical characterization of a seventh protein. It shares 56–58% identity with caffeic acid OMTs (COMTs), but it failed to methylate COMT substrates, and had no activity with flavonoids. However, the in vitro incubations revealed unusually high background levels without added substrates. A search for the responsible component revealed that the enzyme methylated dithiothreitol (DTT), the reducing agent added for enzyme stabilization. Unexpectedly, product analysis revealed that the methylation occurred on a sulfhydryl moiety, not on a hydroxyl group. Analysis of 34 compounds indicated a broad substrate range, with a preference for small hydrophobic molecules. Benzene thiol (Km 220 μM) and furfuryl thiol (Km 60 μM) were the best substrates (6–7-fold better than DTT). Small isosteric hydrophobic substrates with hydroxyl groups, like phenol and guaiacol, were also methylated, but the activities were at least 5-fold lower than with thiols. The enzyme was named C. roseus S-methyltransferase 1 (CrSMT1). Models based on the COMT crystal structure suggest that S-methylation is mechanistically identical to O-methylation. CrSMT1 so far is the only recognized example of an S-methyltransferase in this protein family. Its properties indicate that a few changes in key residues are sufficient to convert an OMT into a S-methyltransferase (SMT). Future functional investigations of plant methyltransferases should consider the possibility that the enzymes may direct methylation at sulfhydryl groups.
Keywords: Catharanthus roseus, S-methyltransferase, O-methyltransferase, evolution, protein modeling, homology-based cDNA cloning
Introduction
S-adenosyl-L-methionine (SAM)-dependent hydroxyl methyltransferases (OMTs) are involved in the biosynthesis of lignin and many plant secondary products, and often the methylations are essential in determining specific physiological functions of the resultant molecules. Two distinct protein families with less than 25% identity are responsible for these reactions in plants. Prototypes of the so-called (Noel et al., 2003) class II family are the proteins methylating caffeoyl-CoA (biosynthesis of lignin), but the family also contains examples that methylate flavonoids and phenylpropanoid conjugates (Ibdah et al., 2003; Lukacin et al., 2004; Vogt, 2004). These proteins exist as homodimers with subunit molecular weights of 23–28 kDa, and they typically require a divalent cation (e.g. Mg2+) to mediate the deprotonation of the acceptor hydroxyl group prior to the methyl group transfer. One of these proteins has been recently crystallized (Ferrer et al., 2005) and shows high structural similarity to the animal catechol OMT that also requires Mg2+ (Vidgren et al., 1994).
The class I OMTs are also homodimers, but are distinguished by larger subunit sizes (38–43 kDa) and mechanistic independence from divalent cations. Instead, the type I OMTs appear to use general acid/base catalysis to prepare the methyl-accepting hydroxyl moiety for SAM-dependent transmethylation. Typical examples of this family are the caffeic acid OMTs (COMTs) involved in lignin biosynthesis. The class I family is much more functionally diverse than the class II family, containing enzymes that methylate other phenylpropanoid derivatives such as eugenol and chavicol, as well as those that methylate flavonoids, isoflavonoids, alkaloids, coumarins, orcinols, and polyalcohols (e.g. inositol). The range of possible substrates may be much larger than we presently realize as many of the family members have only been identified based upon sequence similarity to previously characterized type I plant OMTs, which does not necessarily translate to functional similarity. Often, changes in one or a few amino acids are sufficient for pronounced alterations in substrate specificity (Frick and Kutchan, 1999; Gang et al., 2002). The recent crystallization of three type I plant OMTs provided a structural basis for understanding OMT architecture, selectivity and catalytic mechanisms (Zubieta et al., 2001, 2002). Nevertheless, it is still difficult to predict the physiological substrates of newly identified family members based upon structural models alone. Moreover, several enzymes possess broad substrate specificities in vitro that do not permit easy conclusions about in vivo substrates (Chiron et al., 2000; Gauthier et al., 1998; Schröder et al., 2002; Wein et al., 2002).
We have a long-standing interest in the biosynthesis of flavonoids and indole alkaloids in Catharanthus roseus (Madagascar periwinkle). To search for class I OMTs in these two secondary metabolic pathways, we used biochemical approaches (enzyme purification) and homology-based RT-PCR strategies. Six type I plant OMT family members have been isolated and characterized thus far. One such enzyme turned out to be a COMT having broad substrate specificity (Schröder et al., 2002), while the other five proteins formed a group of closely related proteins with >60% identity. We succeeded in functionally identifying two of these proteins as OMTs involved in flavonoid biosynthesis (Cacace et al., 2003; Schröder et al., 2004), but we were unable to elucidate the functions of the three other family members, either because the recombinant protein was insoluble or because all attempts to detect SAM-dependent activity with available substrates failed.
As part of these studies we isolated the cDNA for a seventh class I OMT that upon initial biochemical characterization also exhibited no activity with any of the substrates tested. Curiously, the control reactions (i.e. incubations lacking substrate to correct for background radioactivity) revealed unexpectedly high background rates when compared with previously examined type I OMTs. Closer experimental analysis of this unexpected background activity, described here, uncovered a methyltransferase with a strong preference for sulfhydryl groups, in contrast to the expected preference for hydroxyl moieties.
Results
Cloning and initial characterization
We used RT-PCR with RNA from Catharanthus roseus cell cultures and degenerate primers designed for cDNAs of the OMT family (Cacace et al., 2003) to amplify partial EST sequences for putative class I OMTs. The complete cDNA for a newly identified sequence was obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) techniques with specific primers. The highest BLAST scores (Altschul et al., 1997) with the deduced protein revealed 56–58% identity with many COMTs, including the C. roseus COMT previously isolated and characterized (Schröder et al., 2002). Much lower values (30–35%) were scored with the other five OMTs described from C. roseus (Cacace et al., 2003; Schröder et al., 2004).
The moderate sequence similarities with functionally identified OMTs suggested that the protein could be a candidate either for the 7-OMT expected for flavonoid biosynthesis in Madagascar periwinkle (Carew and Krueger, 1976; Milo et al., 1985; Piovan et al., 1998) or for the hydroxytabersonine OMT that had been a focus of our previous work (Cacace et al., 2003; Schröder et al., 2004). The cDNA was subcloned for expression of a His-tagged recombinant protein in Escherichia coli. SDS-PAGE analysis of whole cells lysed in the presence of SDS indicated that the protein was expressed at high levels, and analysis of extracts prepared with non-denaturing buffers revealed that about 30–40% of the expressed protein was in the soluble fraction. The resultant recombinant protein was purified to near homogeneity by Ni-chelation affinity chromatography, based on the His-tag appended to the N-terminus of the expressed protein.
Biochemical tests using a large collection of flavonoids (flavanones, flavones, dihydroflavonols, and flavonols) investigated during the course of previous studies (Cacace et al., 2003; Schröder et al., 2004), and using substrates typical for caffeic acid OMTs (Schröder et al., 2002) failed to reveal activity above background with any of these substances. The protein was also inactive with 16-hydroxytabersonine, the substrate of the hydroxyl-directed OMT in the biosynthetic pathway to vindoline (Cacace et al., 2003; De Luca et al., 1986; Fahn et al., 1985).
Identification of the new protein as an S-methyltransferase (SMT)
Although no activity could be detected with hydroxyl group-containing compounds, a closer inspection of the assay results revealed an unusually high background level of transmethylation activity. In contrast to all other OMTs examined thus far, the control incubations containing all assay components except the hydroxyl-containing substrates showed exceptionally high radioactive values after the standard extraction with ethyl acetate (EtOAc). The extracted counts per minute (cpm) sometimes accounted for up to 3–4% of the total input S-adenosyl-L-[methyl-14C]methionine (14C-SAM). Moreover, thin layer chromatography analysis revealed distinct peaks of radioactivity that did not correspond to 14C-SAM or its known degradation products. This was not observed in incubations with the other OMTs cloned from C. roseus, i.e. with the recombinant proteins identified as caffeic acid OMT (CrCOMT) (Schröder et al., 2002), the two flavonoid OMTs (flavonoid 3′,5′-OMT and 4′-OMT) and with the other C. roseus OMTs for which no substrate could be identified (Cacace et al., 2003; Schröder et al., 2004). The observations suggested that the new putative OMT possessed a unique catalytic property linked either directly or indirectly to the transfer of the radioactive methyl group of SAM to a component of the incubation mixture. Experiments to identify the responsible assay component demonstrated that the amount of radioactive product extracted by EtOAc treatment was greatly reduced although not eliminated if dithiothreitol (DTT) (usually 2 mM) was omitted from reactions. We suspected that the residual activity was caused by DTT contained in the protein storage buffer (4 mM, added routinely for enzyme stabilization), which amounted to 0.15 to 0.3 mM DTT in the incubations, depending on the protein preparation used.
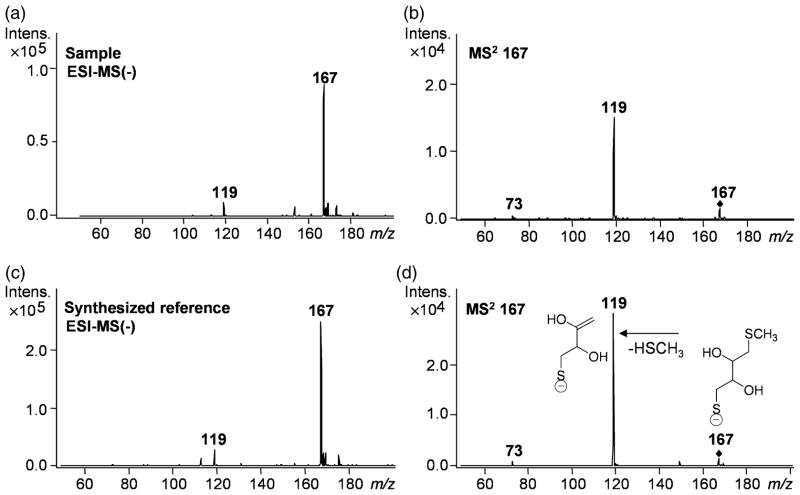
We next prepared recombinant enzyme purified in the absence of DTT. When this DTT-free sample was tested, enzyme assays revealed the expected low background levels of extractable radioactivity commonly observed with other type I OMTs. Finally, addition of DTT restored the previously observed high background levels. These series of experimental observations strongly suggested that DTT itself was the assay component undergoing SAM-dependent methylation by the recombinant protein. As methylation of DTT had not been described before in plants, we investigated the product of scaled-up incubations. Using liquid chromatography–electrospray ionization–UV–tandem mass spectrometry (LC-ESI-UV-MSn) analysis, we obtained a pseudo-molecular ion [M-H]− at m/z 167 for the mono-methylated product of DTT (Figure 1a), which yielded a fragment ion at m/z 119 (Figure 1b) as HSCH3 was eliminated as the parent ion underwent fragmentation (MS2 167). The molecular weight of the product and the elimination of methyl thiol pointed to methylation of one of the sulfur atoms of DTT. We then synthesized the monomethyl thioether of DTT [confirmed by 1H nuclear magnetic resonance (NMR) spectroscopy] and characterized it by LC-ESI-UV-MSn. The reference compound displayed the same retention time, mass spectrum (Figure 1c) and product ion spectrum (Figure 1d) as the SAM-dependent product of the enzymatic reaction with DTT. The results conclusively demonstrate that DTT is methylated, but not as expected at one of the hydroxyl groups, but at one of the sulfhydryl moieties. The enzyme was therefore named C. roseus S-methyltransferase 1 (CrSMT1).
Figure 1.
Liquid chromatography–electrospray ionization–UV–tandem mass spectrometry analysis of the product formed from dithiothreitol (DTT) by Catharanthus roseus S-methyltransferase 1 (CrSMT1), and of the synthesized reference compound DTT monomethyl thioether. Mass spectra in the negative mode of the sample (a) and the reference (c) as well as the product ion spectrum of m/z 167 of the sample (b) and the reference (d) are shown. The chemical structures in (d) show the deduced fragmentation pattern.
Substrates and biochemical properties of CrSMT1
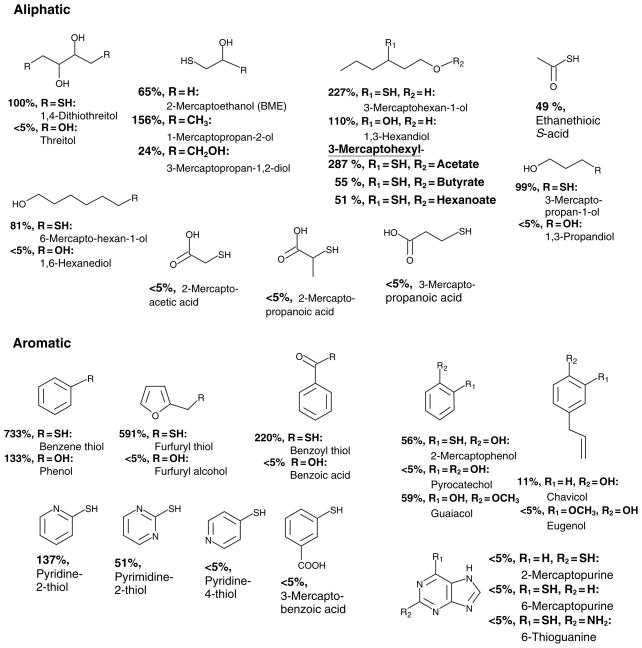
The observed transmethylation activity of CrSMT1 directed against DTT led us to investigate other sulfhydryl-containing compounds. The data are summarized in Figure 2, with the relative activities normalized to DTT as the first substrate identified (=100%). Substances with structural similarity to DTT, that is, aliphatic compounds with sulfydryl and hydroxyl groups, all served as substrates for CrSMT1. The aliphatic substrates exhibiting the highest levels of CrSMT1-mediated transmethylation were 3-mercaptohexan-1-ol (227%) and its acetate ester (287%). Esterification with larger acids strongly reduced the activity, suggesting that 3-mercaptohexylacetate represented an upper size limit on substrates acceptable to CrSMT1. Of the three smallest substrates tested, 2-mercaptoethanol (BME) was the most active, with nearly two-thirds of the activity of DTT, but this activity increased to 156% with the addition of a terminal methyl group (1-mercaptopropan-2-ol), while the addition of a second hydroxyl group (3-mercaptopropan-1,2-diol) reduced the activity again to a quarter of that of DTT. The removal of the hydroxyl on position 2 (3-mercaptopropan-1-ol) increased the activity to DTT levels, and doubling the length of the carbon chain reduced the activity only slightly (6-mercaptohexan-1-ol = 81%). It is worth noting that the simple repositioning of the sulfhydryl group of 6-mercaptohexan-1-ol to the 3 position (3-mercaptohexan-1-ol) transformed it into one of the best substrates tested thus far, with 227% of the activity of DTT. Of the other two smallest substrates tested, ethanethioic S-acid had half of the activity of DTT, and Na2S was inactive. The presence of carboxyl groups abolished CrSMT1 activity. The three aliphatic acids (2-mercaptoacetic acid, 2-mercaptopropanoic acid, 3-mercaptopropanoic acid) had activities just above the detection limit (<5%, normalized to DTT, and <1% when normalized to the best substrate, benzene thiol).
Figure 2.
Activity of Catharanthus roseus S-methyltransferase 1 (CrSMT1) with sulfhydryl compounds and structurally related substances with hydroxyl groups. The per cent values were normalized to the dithiothreitol (DTT) activity, where 100% = 223 μkat kg−1. <5% = close to detection limit; these values correspond to <1% when normalized to the best substrate (benzene thiol).
In addition to aliphatic substrates, we also tested aromatic compounds possessing sulfhydryl groups (Figure 2). Of these aromatic sulfhydryl-containing compounds, benzene thiol and furfuryl thiol exhibited the highest levels of methylation compared with other substrates examined, with about 7× and 6× that of DTT, respectively. All deviations from these core structures or derivatizations reduced the acceptability of the substrates. For example, altering the side chain of benzene thiol to include a carbonyl (benzoyl thiol) reduced the activity to a level comparable to that of the best aliphatic compounds assayed, while adding a nearby hydroxyl group (2-mercaptophenol) reduced the activity even further to half of that of DTT. The activity of benzoyl thiol (220%) suggests that ethanethioic acid, which has low activity (49%) despite having a similar configuration, could simply be too small to be a good substrate. The presence of one or even two nitrogens in the aromatic rings was tolerated (pyridine-2-thiol = 137%; pyrimidine-2-thiol = 51%), but the positioning was critical (almost no activity with pyridine-4-thiol). Increasing the substrate size to a purine backbone abolished activities to near background levels, as observed with 2- and 6-mercaptopurine as well as with 6-thioguanine. Like the carboxylated aliphatic compounds, the one carboxylated aromatic compound tested, 3-mercaptobenzoic acid, was not accepted as a substrate for CrSMT1, suggesting that the carboxyl group interfered with CrSMT1 activity.
These results defined the substrate preferences of CrSMT1 in vitro: small hydrophobic molecules, with a preference for simple aromatic rings. The best aliphatic substrates (3-mercaptohexan-1-ol and its esters) fit this definition because they are flexible enough to assume shapes similar to those in benzene thiol and furfuryl thiol, suggesting that the aromaticity of the rings is not a defining feature of CrSMT1 substrates. The substrate specificity explained the initial finding that neither phenylpropanoids nor flavonoids were methylated (sizes are too large), but it left open the question of whether small hydrophobic hydroxylated molecules might be acceptable. We therefore tested several possible substrate pairs that were only distinguished from each other by the presence of a sulfhydryl or hydroxyl group. In most cases the hydroxylated compounds exhibited much lower activities than their sulfydryl-containing counterparts. For example, threitol, the hydroxyl-containing analogue of DTT, had barely detectable activity. Similarly, furfuryl alcohol (an analog to furfuryl thiol, 591%) possessed negligible activity. The same was true for benzoic acid (an analog to benzoyl thiol, 220%), 1,3-propandiol (an analog to 3-mercaptopropan-1-ol, 99%), 1,6-hexanediol (an analog to 6-mercaptohexan-1-ol, 81%), and pyrocatechol (an analog to 2-mercaptophenol, 56%). Pyrocatechol has often been used as a prototypical standard substrate for COMTs (Maury et al., 1999; Pellegrini et al., 1993). The low activity of CrSMT1 using pyrocatechol therefore stresses the functional distinction of CrSMT1 relative to typical COMTs, even though they possess upwards of 50–60% sequence similarity.
Two hydroxyl-containing compounds, however, proved to be quite good substrates. Phenol, an analog to the best substrate, benzene thiol (733%), was also the best hydroxyl-containing substrate examined, with 133% of the activity of DTT, while 1,3-hexanediol, an analog to 3-mercaptohexan-1-ol (one of the best CrSMT1 substrates), was also slightly more active than DTT (110%). In total, the results indicate a definite preference for sulfhydryl groups as the ultimate methyl acceptor, but also point to significant activities with hydroxylated compounds if these substrates maintain acceptable size and hydrophobicity characteristics. This hypothesis was confirmed in assays using guaiacol, which revealed 59% activity when compared with DTT. Increasing the size of such hydrophobic molecules by adding the side chain characteristic for phenylpropanoids led to marked reduction in methylation activity (Figure 2; chavicol 11% and eugenol <5%). In summary, these results of a plethora of biochemical assays confirm the strict size limitation of the active site pocket of CrSMT1.
With some of these substrates it was not obvious, in the absence of further chemical characterization, whether the methylation was directed at a sulfhydryl or a hydroxyl group. We performed capillary gas chromatography–mass spectrometry (GC-MS) product analyses for four of the bestCrSMT1 substrates found, including benzene thiol, furfuryl thiol, 3-mercaptohexan-1-ol, and 1-mercaptopropan-2-ol, two of which contain both sulfhydryl and hydroxyl groups available for transmethylation. In the case of all four substrates, the retention times and mass spectra of the extracted products of scaled-up incubations matched those of the synthesized thioether-containing reference compounds(data not shown). In the case of the 3-mercaptohexanol series, S-methylation was also evident from the fact that the esters containing hydroxyl blocking groups were nonetheless active as CrSMT1 substrates, and sometimes exhibited even more transmethylation activity relative to the unprotected free alcohols (3-mercaptohexan-1-ol versus its acetyl ester).
The pH dependence (testing from pH 6.5 to 11, with furfuryl thiol, the best substrate at that point) revealed highest values at pH 9–9.3, with 75% of the activity at pH 8.1. This value was chosen for the standard incubations because it was closer to physiological values. Assays in the range from 10 to 30°C showed a broad optimum centered at 20°C, with 70% activity at 30°C.
The apparent Michaelis constant, Km, values determined for the best substrates, benzene thiol and furfuryl thiol, were 218 ± 28 μM and 56 ± 11 μM, respectively, and according to the catalytic constant, kcat/Km criterion the catalytic efficiency with furfuryl thiol was about 60% higher than that with benzene thiol. The Km for SAM was 96 ± 19 μM when assayed with both substrates. The apparent maximum velocity, Vmax was 236 ± 9 μkat kg−1 for benzene thiol, 100 × 5 μkat kg−1 for furfuryl thiol, and 266 × 11 μkat kg−1 for SAM. The effect of divalent cations was investigated with benzene thiol, furfuryl thiol, and DTT as substrates. Mg2+ had no significant effect when tested at 0.1 and 1 mM. Zn2+ at 1 mM inhibited all activities by 90% or more, but at low concentrations the effect was substrate-dependent. With benzene thiol and furfuryl thiol just 50 μM was sufficient for almost complete inhibition, but the activity with DTT was inhibited only 60% at this low Zn2+ concentration. DTT is a zinc chelator (Krezel et al., 2001), and thus it is likely that sequestration of the metal ion by this particular substrate was responsible for the relief from inhibition. A stimulating effect by divalent cations was not detected.
In silico bioinformatics analysis
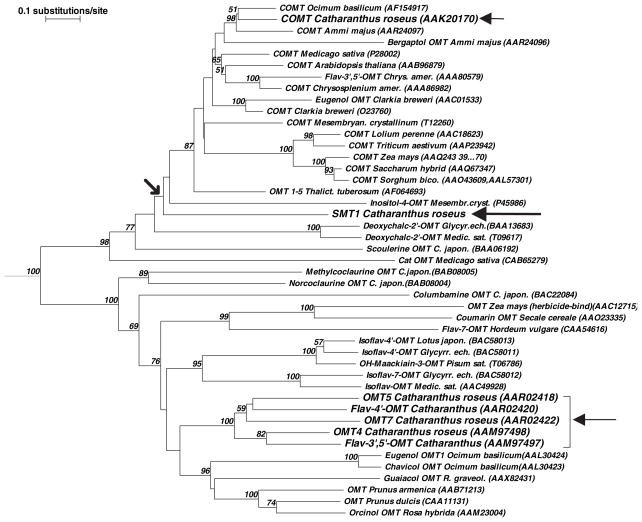
The unexpected identification of a new class I plant methyltransferase capable of efficient sulfhydryl-directed SAM-dependent methylation, CrSMT1, prompted a more detailed bioinformatic analysis. First, a relationship tree based upon primary sequence identity and similarity was constructed using COMTs and other type I plant methyltransferases (MTs) with known substrate specificities (Figure 3). The resultant tree showed that CrSMT1 clearly belonged in the class I OMT protein family, while its position on an obvious divergent subbranch off the main tree confirmed that it was not closely related to any of the other type I family members included in the analysis. In particular, CrSMT1 exhibited a distinctly distant relationship with COMT from its host organism C. roseus and with the cluster formed by the five other OMTs known from C. roseus (arrows in Figure 3).
Figure 3.
Relationship tree of selected plant O-methyltransferases (OMTs). The tree was developed with the program TREECON for Windows (Van de Peer and De Wachter, 1994), using the inbuilt matrix for amino acid sequences, and the neighbor-joining method for distance calculations (Saitou and Nei, 1987). The outgroup (root) was the O-demethylpuromycin-O-methyltransferase from Streptomyces anulatus (DMPM_STRLP). The length of the branches is a measure of the number of substitutions per site (top left). The numbers at the forks are bootstrap values indicating the per cent values for obtaining this particular branching in 1000 repetitions of the analysis; only the values above 50% are shown. Chrys.amer., Chrysosplenium americanum; Sorghum bico., Sorghum bicolor; Thalict. tuberosum, Thalictrum tuberosum; Medic.sat., Medicago sativa; Glyc.ech., Glycyrrhiza echinata; R. graveol., Ruta graveolens; C. japon., Coptis japonica.
The crystal structures of three MT enzymes of the class I OMT family have revealed key amino acids for catalytic activity and residues important for the recognition and stable binding of hydroxyl-containing substrates (Zubieta et al., 2001, 2002). Because of the close structural similarity amongst the experimentally determined type I OMT enzymes, we constructed homology models of CrSMT1 based upon sequence conservation with type I OMTs and compared CrSMT1 with Medicago sativa COMT (Zubieta et al., 2002) to look for unusual residues that might be responsible for the properties of CrSMT1 as a sulfhydryl-directed MT. Figure 4 shows the primary sequence alignment of CrSMT1 and M. sativa COMT, highlighting residues that, with reference to the CrSMT1 three-dimensional model, are discussed in more detail below.
Figure 4.
Alignment of Catharanthus roseus S-methyltransferase 1 (CrSMT1) with the crystallized caffeic acid O-methyltransferase (COMT) from Medicago sativa. Marked positions; residues and numbering from the M. sativa COMT (Zubieta et al., 2002): a, catalysis (transmethylation): W266, H269, D270, E297 and E330; b, SAM binding: D206–G212, D231, L232, D251–F253, K265 and W271; c, methyl acceptor binding: A162, A322, H323 and N324; d, binding of propanoid tail (e.g. of caffeic acid) in COMT: M130, M180, I316 and M320.
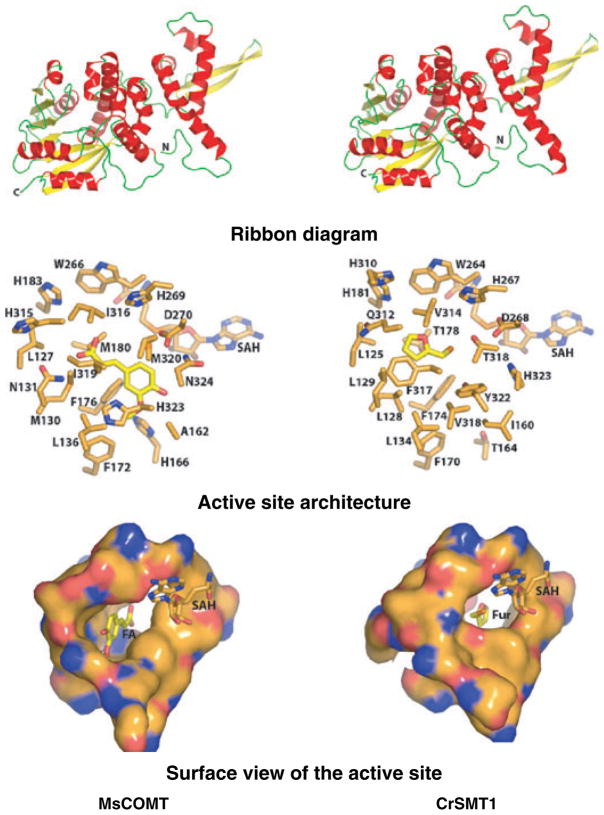
The overall architecture of the experimentally determined structure of COMT and the homology model of CrSMT1 are nearly identical (Figure 5, top). As expected from the primary sequence alignment and the placement of CrSMT1 in the type I family of plant small molecule MTs, both enzymes contain a small N-terminal domain and a larger C-terminal domain. The N-terminal domain, consisting of four alpha helices and two beta strands, has been shown to facilitate dimerization in type I plant MTs such as COMT. The C-terminal domain contains a core Rossmann fold commonly found in SAM-dependent MTs (Zubieta et al., 2001, 2002). SAM co-substrate binding, and catalysis occur mainly in the C-terminal domain. Slight differences between COMT and the modeled CrSMT1 appear in the last helix that participates in the structure of the substrate binding pocket.
Figure 5.
Comparison of the crystallized Medicago sativa caffeic acid O-methyltransferase (COMT) (left) and the Catharanthus roseus S-methyltransferase 1 (CrSMT1) model (right). The figures were produced with the program PYMOL (DeLano, 2002). Top: ribbon diagrams. Alpha helices are shown in red, connecting loops in green and beta strands in yellow. The N- and C-termini are marked. Middle: close-up view of the COMT–S-adenosyl-L-homocysteine (SAH)–ferulate complex and model of the CrSMT1 active site containing SAH and a docked molecule of furfuryl thiol. Bottom: close-up molecular surface view of the active site. FA, ferulate; Fur, furfuryl thiol.
Figure 5 (middle) shows a close-up view of the active site for the experimentally determined COMT–S-adenosyl-L-homocysteine (SAH)–ferulate complex (protein data bank code 1KYZ) and the comparison with the CrSMT1 model containing a docked SAH and furfuryl thiol (one of the most efficiently turned over substrates of CrSMT1; the model with benzene thiol showed close structural congruity with furfuryl thiol). Moreover, as suggested from the primary sequence alignment, the proposed catalytic residues for general base catalysis and SAM binding are conserved in both COMT and CrSMT1 (Figure 4, in COMT: W266, H269, D270, E297, E330; D206–G212, D231, L232, D251–F253, K265 and W271). Given this high level of sequence identity in key catalytic residues, it is likely that CrSMT1 uses an analogous catalytic mechanism to that of other described type I plant MTs such as COMT, i.e. deprotonation of the thiol group using a histidine general base prior to transfer of the reactive methyl group of SAM to the thiolate anion.
In contrast to the highly conserved SAM-binding pocket described above, large portions of the methyl acceptor binding site contained amino acid substitutions that were likely to modulate the specificity for the methyl-accepting chemical compound. The substitution of A162 in COMT for I160 in CrSMT1, A322 for V320, H323 for Y322, and N324 for H323 leads to a substantial contraction of the available space in the phenolic ring binding pocket that is described for COMT with bound ferulate. In particular, the side chain of Y322 of CrSMT1 protrudes into one end of the active site pocket, making it impossible for CrSMT1 to accommodate the large phenylpropanoid structure characteristic of COMT substrates (e.g. caffeic acid).
However, the replacement of M130, M180, I316 and M320 in COMT by L128, T178, V314 and T318, respectively, in CrSMT1 (see also Figure 4) effectively enlarged the available volume at the other end of the pocket, i.e. corresponding to the region in COMT that sequestered the propanoid tail of COMT substrates. The more spacious architecture of the CrSMT1 in this area could be one of the reasons for the broad range of acceptable substrates in CrSMT1. Finally, I319 of COMT was replaced by F317 in CrSMT1, possibly providing added van der Waals interactions, at the expense of a small restriction of available volume. The bottom part of Figure 5 illustrates the differences between COMT and CrSMT1 by showing a molecular surface view of the active site close-ups.
Discussion
CrSMT1 was discovered as part of a RT-PCR based screen aimed at finding ESTs and eventually full-length clones for small molecule OMTs in C. roseus. CrSMT1 so far is the only SMT identified in this plant MT family. This finding is important particularly because most plant MTs have been and continue to be identified and annotated based only on the similarity of their primary amino acid sequences with known OMTs, in absence of confirming experimental evidence. Sequence alignments and three-dimensional modeling show that CrSMT1 contains all of the key residues previously identified in OMTs as being necessary for SAM binding and catalysis (Zubieta et al., 2001, 2002). Moreover, the almost absolute conservation of key catalytic and SAM-binding residues suggests that O- and S-methylation are probably mechanistically very similar. This hypothesis is supported by the methylation of some hydroxylated compounds by CrSMT1, in particular in those cases where hydroxyl and sulfhydryl substrates were otherwise identical (e.g. in the most striking example phenol versus benzene thiol). The mechanistic reasons for the catalytic preference of CrSMT1 for thiols has thus far resisted explanation even in light of the informative structural models. In addition to differences in pKa values between –SH and –OH moieties, subtle changes in the catalytic machinery associated with the histidine general base may play roles that are not apparent from the three-dimensional models, and explanations for the unusual thiol-directed specificity of CrSMT1 await more definitive structural elucidations. Nevertheless, the overall analysis suggests that a fairly small number of key residue differences are sufficient to convert an OMT into a SMT. Certainly, experimental structures of CrSMT1 combined with site-directed mutagenesis should uncover the residues responsible for the activity of CrSMT1. These studies are underway. In the meantime, our findings suggest that future attempts to identify the function of newly identified and putative OMTs should in general consider the possibility that they too might be directed at sulfhydryl moieties rather than at the more widely accepted hydroxyl groups of plant small molecules.
The other six type I OMT family members from C. roseus had no significant activity with the prototype substrate DTT initially used to identify CrSMT1. It therefore appears unlikely that S-methylation is a common property of all MTs, but it seems possible that CrSMT1 is the only recognized member of a subset of SMTs in this family. DTT methylation has also been found in crude extracts from strawberry fruits Fragaria ananassa (Wein et al., 2002), but the product was not identified, and similar findings in other plants might easily have been ignored as artifacts. It should also be noted that experiments with DTT can result in pseudosubstrate activities of MTs under artificial in vitro conditions, as reported recently (Burga et al., 2005). Based on our initial finding with CrSMT1 from C. roseus, the possibility of SAM-dependent DTT methylation was investigated with a Ruta graveolens type I OMT with identified hydroxyl-containing substrates (methoxylated phenols, e.g. guaiacol). Methylation activity with DTT was barely detectable under standard assay conditions, and probably would have gone unnoticed without specifically searching for DTT methylation. However, a high rate of DTT-directed S-methylation was detected in the presence of Zn2+, which was otherwise a strong inhibitor of the OMT activity with hydroxyl-containing substrates. Kinetic data showed that high enzyme activity with DTT required a zinc/DTT ratio of 1:1. DTT is a zinc chelator (Krezel et al., 2001), and the stoichiometry indicated that a soluble zinc–DTT complex on the one hand protected the enzyme against the metal-dependent inhibition, and on the other hand shifted the thiol–thiolate equilibrium of DTT towards the thiolate anion that presumably serves as the active substrate. Although this represented in vitro pseudosubstrate specificity (Burga et al., 2005), it is noteworthy that the kinetic analysis suggested a competition of DTT and the hydroxylated substrate for the same active site, because this supports the notion that O- and S-methylation are mechanistically very similar. No such stimulating effect of zinc on methylation of DTT or other substrates was observed with CrSMT1, and all of the evidence supports a conclusion that CrSMT1 is a bona fide S-methyltransferase.
The in planta physiological role of CrSMT1 remains to be elucidated. The activity with benzene thiol was also identified in crude extracts from young shoots, indicating that it is not restricted to cell suspension cultures, but a detailed analysis of organ-specific expression remains to be carried out. Some of the substrates exhibiting measurable activity with CrSMT1 have been identified in plants. For example, 3-mercaptohexanol and methylated or non-methylated derivatives serve in wine as important aroma components that are likely to be derived from the grapes used in wine production (Culleré et al., 2004; López et al., 2003). Moreover, these compounds have also been identified in yellow passion fruit (Tominaga and Dubourdieu, 2000; Werkhoff et al., 1998). While the biosynthesis has not been investigated, it will be important to investigate whether these or related compounds are present in C. roseus. Plant SMTs methylating small molecules have been described, but these belong to other protein families (less than 20% identity with the OMT family discussed here), and none of the activities provided useful clues for a function of CrSMT1 in C. roseus. One example of a thiol-directed MT is the SMT characterized from cabbage (Brassica oleracea L.) that posesses an interestingly high activity with benzene thiol (Attieh et al., 2000a), this compound being the best substrate found to date for CrSMT1 (Figure 2). The enzyme is involved in the methylation of compounds released upon the hydrolysis of glucosinolates, contributing to the formation of sulfur volatiles (Attieh et al., 2000b, 2002). However, this Brassica SMT is strictly confined to glucosinolate-synthesizing plants, and these natural products have not been found in C. roseus. It is noteworthy that benzene thiol was also an excellent substrate for a previously described animal thiopurine SMT (Ames et al., 1986; Woodson et al., 1983) that plays a major role in the inactivation of antineoplastic and immune-suppressant thiopurine drugs in humans (reviewed in Weinshilboum et al., 1999). Thiopurine SMT is also an example of an enzyme that, while maintaining a pronounced preference for sulfhydryl groups, does not exclude activity with hydroxylated substances as the enzyme also has some activity with 8-hydroxyguanine (Deininger et al., 1994). Thiopurines do not serve as substrates for CrSMT1 (Figure 2), indicating that the active site pocket is too small to accommodate such large molecules. Notably, DTT and 2-mercaptoethanol, two good substrates for CrSMT1, can also be detoxified via S-methylation in animals (Carrithers and Hoffman, 1994; Weinshilboum, 1979; reviewed in Weinshilboum et al., 1999), suggesting that CrSMT1 could play a role in detoxification reactions involving thiols.
Two other plant SMTs are part of the S-methylmethionine cycle, SAM L-methionine S-methyltransferase (SAM + methionine = S-methylmethionine), and S-methylmethionine homocysteine S-methyltransferase (S-methylmethionine + homocysteine = two methionines); the latter enzyme can also use SAM as a methyl donor (reviewed in Ranocha et al., 2001). S-methylmethionine is presumably present in all flowering plants, plays a role in phloem sulfur transport (Bourgis et al., 1999), is an intermediate in the biosynthesis of the osmoprotectant 3-dimethylsulfoniopropionate (Hanson et al., 1994), may influence ethylene biosynthesis (Ko et al., 2004), and is the methyl donor for thiol/selenol methyltransferases involved in selenium detoxification (Lyi et al., 2005; Neuhierl et al., 1999), and the selenium analog of S-methylmethionine is an intermediate in the detoxification of the selenium metal (Tagmount et al., 2002). None of these reactions has been investigated for C. roseus, but it seems unlikely that they are carried out by CrSMT1. The methyl group acceptor substrates are in all cases amino acids or their selenium derivatives, and the enzymes synthesizing S-methylmethionine, for example, require a free carboxyl group (James et al., 1995). With CrSMT1, acceptable substrates possessed significant hydrophobic character lacking charged groups, including carboxyl moieties. Carboxyl groups in fact strongly reduced the activities of CrSMT1 in the cases tested (Figure 2), and cysteine did not serve as a substrate for CrSMT1 [assay evaluation with liquid chromatography–mass spectrometry; not shown]. It may be noteworthy, however, that 3-(methylthio)propan-1-ol, the product of CrSMT1 with 3-mercaptopropan-1-ol, was a potent inhibitor of the enzyme synthesizing S-methylmethionine in Wollastonia biflora (James et al., 1995).
In conclusion, the combined approach using structural and functional studies to understand plant small molecule biosynthesis including methylation serves as a necessary foundation for the continued discovery of novel MTs, such as CrSMT1, that play critical roles in continually expanding, modifying and diversifying physiologically and ecologically important plant natural products. An increased understanding of the molecular basis for MT substrate specificity and evolutionary divergence, coupled with a broader appreciation for previously ignored putative MT substrates, will permit one to more rapidly and precisely assess the functional characteristics of novel enzymes discovered using molecular genetic approaches.
Experimental procedures
Plant material
The cell suspension culture of Madagascar periwinkle (Catharanthus roseus L.G. Don, line CP3a) and its maintenance in MX growth medium in continuous dark with subcultures every week have been described previously (Vetter et al., 1992).
Homology-based PCR
The preparation of cDNA and EST libraries in phage lambda NM1149 followed published methods (Vetter et al., 1992). The PCR reactions for OMT-specific sequences were carried out with a degenerate primer (5′-T-[TG]-G-[AC]-I-[CT]-A-T-G-T-T-G-G-[AT]-GG-I-G-A-T-A-T-G-T-T-T-G-3′) based upon motif 3 conserved in OMTs (Ibrahim, 1997; Ibrahim et al., 1998). The PCR reactions were carried out either with phage lysates with phage primers flanking the cDNA inserts, or with a 5′/3′-RACE kit (Roche Diagnostics, Penzberg, Germany). Overlapping clones for the complete coding region were obtained by 5′ and 3′-RACEs with specific primers designed from the known sequences. The DNA was sequenced on both strands to verify the absolute integrity of the amplified clones.
Expression and purification of recombinant protein
For expression in E. coli, the protein coding regions were amplified with a 5′-primer (5′-AAGGATCCAGTTCCCACGAAGAGAAACC-3′) creating a BamHI site directly before the second codon, and a 3′-primer (5′-TAGTCGACGTTTATTTATAAAATTCCATGACC-3′) providing a SalI site after the stop codon. The BamHI/SalI fragments were inserted into vector pHIS8-3 (Ferrer et al., 1999) to obtain a protein with an N-terminal octahistidine-tag. The proteins were affinity purified with the His-trap™ purification kit from Pharmacia Biotech (Freiburg, Germany) as recommended by the manufacturer. Histagged protein bound to the Ni-NTA column was eluted with 0.5 M imidazole which was subsequently removed by passing the proteins through a PD10 column (Amersham Biosciences, München, Germany). Routine storage of the protein preparations was in 0.1 M Tris–HCl (pH 8), 4 mM DTT, and 0.1 M NaCl at −70°C. Later experiments used protein that was purified over the Talon Metal Affinity Resin (BD Biosciences, Heidelberg, Germany) and stored in 0.05 M phosphate buffer (pH 7) with 0.3 M NaCl and 5% glycerol at −70°C. Protein concentrations were determined as described previously (Bradford, 1976).
Enzyme assays
The initial incubations with hydroxyl-containing substrates (including flavonoids, caffeic acid, or caffeoyl-CoA) were performed as described previously (Schröder et al., 2002). The assays contained 50 mM Tris–HCl (pH 7.5), 2 mM DTT, 0.1 mM substrate, 40 μM unlabelled SAM, 9.3 μM S-adenosyl-L-[methyl-14C]methionine (55 000 dpm; 54 μCi μmol−1), and 5–10 μg purified recombinant protein in a final volume of 50 μl. The reactions were stopped after 30 or 60 min at 30°C by acidification (2 μl of 1 M HCl), and the EtOAc-extracted products were quantified after TLC separation (Cacace et al., 2003; Schröder et al., 2002). All reactions were carried out at least in duplicate. These conditions were also used in the early experiments analyzing DTT as substrate. The 16-hydroxytabersonine O-methyltransferase activity measurements have been described previously (Cacace et al., 2003).
The standard incubations to compare the activities against sulfhydryl substrates were carried out as follows. They contained (in a final volume of 50 μl) 50 mM Tris–HCl (pH 8.1), 50 μM unlabelled S-adenosyl-L-methionine, 41.7 nM S-adenosyl-L-[methyl-3H]methionine (3H-SAM; 232 000 dpm), and 6 μg of purified CrSMT1. All reactions were started through addition of the substrates (final 5 mM), and stopped by mixing the resultant incubations with 2 μl of 1 N HCl after 60 min at 20°C. The assays were extracted twice with 0.1 ml of EtOAc, the extract was mixed with 3 ml of scintillation cocktail (Ultima Gold XR; PerkinElmer, Boston, MA, USA), and the radioactivity was quantified in a scintillation counter (Rackbeta 1219; LKB, Turku, Finland). All reactions were independently replicated at least three times.
To determine SAM kinetic constants, benzene thiol or furfuryl thiol concentrations were kept constant at 5 mM, while SAM concentrations were varied from 0.1 to 1000 μM (16.7 nM 3H-SAM; 110 000 dpm). For kinetic studies with sulfhydryl compounds, SAM was held constant at 0.3 mM and benzene thiol and furfuryl thiol concentrations ranged from 20 to 10 000 μM and from 6 to 2000 μM, respectively. For each point, at least three independent samples were incubated at 20°C for 10 min, after which they were extracted and quantified as described above.
Synthesis and analysis of methylated reference compounds
The sulfhydryl compound (0.1 mmol) was mixed with 0.1 M NaOH (1 ml) in 5 ml of methanol and treated with 0.1 mmol iodomethane for 4–12 h. The solution was neutralized with 0.1 M HCl, mixed with 5 ml of water, and extracted 2–3 times with diethyl ether (5–10 ml). The organic phase was dried over Na2SO4, concentrated and analyzed by GC-MS. The monomethyl thioether of DTT was purified by preparative RP18 chromatography with water/acetonitrile mixtures and characterized by 1H NMR spectroscopy: 1H NMR CDCl3 δ 3.81 (1H, CHOH-CH2-SH, m), 3.63 (1H, CHOH-CH2-SCH3, m), 3.10 (2H, 2 OH, broad s), 2.76 (2H, CH2-SCH3, m), 2.71 (2H, CH2-SH, m), 2.15 (3H, SCH3, s) and 1.57 (1H, SH, t, J = 8 Hz).
Analytical techniques
Capillary gas chromatography–mass spectrometry (GCMS)
GC-MS analysis was performed with a Thermo Finnigan Trace DSQ mass spectrometer coupled (Thermo Finnigan, Bremen, Germany) to a Thermo Finnigan Trace GC with a split injector (1:20) equipped with XCALIBUR software (version 1.4). The GC was equipped with a BPX5 20 M fused silica capillary column (30 m × 0.25 mm inner diameter; thickness of the film = 0.25 μm). The GC parameters were as follows: initial temperature of 40°C for 3 min, increased to 250°C at 5°C min−1 intervals. The helium gas flow rate was 3 ml min−1. The EI-MS ionization voltage was 70 eV (electron impact ionization) and the ion source and interface temperature were kept at 230°C and 240°C, respectively. Compounds were identified by comparing their mass spectra and retention indices to the National Institute of Standards and Technology mass spectra library and reference compounds.
Liquid chromatography-electrospray ionization-UV-tandem mass spectrometry (LC-ESI-UV-MSn)
A Bruker esquire 3000 plus mass spectrometer (Bruker Instruments, Karlsruhe, Germany), equipped with an Agilent 1100 HPLC system (Agilent, Waldbronn, Germany), composed of an Agilent 1100 quaternary pump and an Agilent 1100 variable wavelength detector, was used for LC-ESI-UV-MSn analysis. The HPLC column was a Eurospher C18 column (Grom Analytik & HPLC GmbH, Rottenburg, Germany), particle size 5 μm, 10 cm × 2 mm. The voltage of the capillary was 3074 V and the end plate was set to −500 V. The capillary exit was −109.8 V and the Octopole RF amplitude was 120 Vpp. The temperature of the dry gas (N2) was 300°C at a flow of 10 l min−1. The full scan mass spectra were measured from m/z 50 to 500 until the ion charge control target reached 20 000 or 200 ms, whichever was reached first. Tandem mass spectrometry was performed using helium as the collision gas, and the collision energy was set at 1.00 V. All mass spectra were acquired in the negative ionization mode. Auto-tandem mass spectrometry was used to break down the most abundant [M-H]− or the [M + HCOO]− ion of the different compounds. The LC parameters were from 0% acetonitrile and 100% water (acidified with 0.05% formic acid) to 50% acetonitrile and 50% acidic water in 35 min, then in 2.5 min to 100% acetonitrile, with these conditions maintained for 2.5 min, and finally back to 100% water and 0% acetonitrile in 5 min at a flow rate of 0.200 ml. The detection wavelength was 280 nm.
Nuclear magnetic resonance (NMR) spectroscopy
NMR spectra of the monomethyl thioether of DTT were recorded at 25°C using a Bruker 400 spectrometer (Bruker, Rheinstetten, Germany) at transmitter frequencies of 400 MHz for 1H. Samples were dissolved in CDCl3 (1.5 mg in 0.5 ml) and spectra were acquired and analyzed with the standard Bruker software (XWINNMR).
Homology modeling and automated substrate docking
The homology model of CrSMT1 was built using the MODELER software package (Fiser et al., 2000; Marti-Renom et al., 2000). The structure of MsCOMT in complex with 5-hydroxyferulic acid (PDB code 1KYZ) was used as the structural template for homology modeling. The CrSMT1 amino acid sequence was aligned to MsCOMT using CLUSTAL W (1.82) and then homology modeling was carried out using MODELER. Five models were generated and ranked by Model Rank bundled in the MODELER package. The top-ranked model was chosen for further analysis, checked for suitable geometric parameters using PROCHECK (Laskowski et al., 1993) and visualized using O (Jones et al., 1991). For substrate docking analysis, Genetic Optimization for Ligand Docking (GOLD) (Otwinowski and Minor, 1997) was employed for automated docking of the substrate furfuryl thiol into the modeled CrSMT1 active site. The parameters controlling the precise operation of the genetic algorithm were as follows: population size (100), selection pressure (1.100 000), number of operations (100 000), number of islands (5), niche size (2), cross-over weight (95), mutate weight (95), and migrate weight (10). The default parameter values for van der Waal’s and hydrogen bonding were used throughout the docking process. The size of the active site was defined within 15 Å around the NE2 atom of the potential catalytic residue His 267. Ten docking calculations were run and the GOLD score was used to identify the lowest energy docking results.
Acknowledgments
The work in Freiburg was supported by a grant from the Deutsche Forschungsgemeinschaft to GS and JS. Financial support by Degussa to HC and WS is gratefully acknowledged. Work in the Noel Laboratory (JPN and CJL) at the Salk Institute was supported by the National Science Foundation under Grant No. 0236027 to JPN. JPN is an investigator of the Howard Hughes Medical Institute. Work in the Brookhaven National Laboratory was supported by a grant from the Laboratory Directed Research and Development Program (LDRD) to CJL. We thank Martin Steinhaus and Peter Schieberle (Garching) for the acquisition of NMR spectra and Efraim Lewinsohn (Israel) for providing chavicol.
Footnotes
Database accession number of CrSMT1: DQ084384.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames MM, Selassie CD, Woodson LC, Van Loon JA, Hansch C, Weinshilboum RM. Thiopurine methyltransferase: structure–activity relationships for benzoic acid inhibitors and thiophenol substrates. J Med Chem. 1986;29:354–358. doi: 10.1021/jm00153a009. [DOI] [PubMed] [Google Scholar]
- Attieh J, Kleppinger-Sparace KF, Nunes C, Sparace SA, Saini HS. Evidence implicating a novel thiol methyltransferase in the detoxification of glucosinolate hydrolysis products in Brassica oleracea L. Plant Cell Environ. 2000a;23:165–174. [Google Scholar]
- Attieh J, Sparace SA, Saini HS. Purification and properties of multiple isoforms of a novel thiol methyltransferase involved in the production of volatile sulfur compounds from Brassica oleracea. Arch Biochem Biophys. 2000b;380:257–266. doi: 10.1006/abbi.2000.1896. [DOI] [PubMed] [Google Scholar]
- Attieh J, Djiana R, Koonjul P, Étienne C, Sparace SA, Saini HS. Cloning and functional expression of two plant thiol methyltransferases: a new class of enzymes involved in the biosynthesis of sulfur volatiles. Plant Mol Biol. 2002;50:511–521. doi: 10.1023/a:1019865829534. [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, et al. S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1497. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burga L, Wellmann F, Lukacin R, Schwab W, Schröder J, Matern U. Unusual pseudosubstrate specificity of a novel 3,5-dimethoxyphenol O-methyltransferase cloned from Ruta graveolens L. Arch Biochem Biophys. 2005;440:54–64. doi: 10.1016/j.abb.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Cacace S, Schröder G, Wehinger E, Strack D, Schmidt J, Schröder J. A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry. 2003;62:127–137. doi: 10.1016/s0031-9422(02)00483-1. [DOI] [PubMed] [Google Scholar]
- Carew DP, Krueger RJ. Anthocyanidins of Catharanthus roseus callus cultures. Phytochemistry. 1976;15:442. [Google Scholar]
- Carrithers SL, Hoffman JL. Sequential methylation of 2-mercaptoethanol to the dimethyl sulfonium ion, 2-(dimethylthio) ethanol, in vivo and in vitro. Biochem Pharmacol. 1994;48:1017–1024. doi: 10.1016/0006-2952(94)90373-5. [DOI] [PubMed] [Google Scholar]
- Chiron H, Drouet A, Claudot AC, Eckerskorn C, Trost M, Heller W, Ernst D, Sandermann H., Jr Molecular cloning and functional expression of a stress-induced multifunctional O-methyltransferase with pinosylvin methyltransferase activity from Scots pine (Pinus sylvestris L.) Plant Mol Biol. 2000;44:733–745. doi: 10.1023/a:1026507707186. [DOI] [PubMed] [Google Scholar]
- Culleré L, Escudero A, Cacho J, Ferreira V. Gas chromatography-olfactometry and chemical quantitative study of the aroma of six premium quality spanish aged red wines. J Agric Food Chem. 2004;52:1653–1660. doi: 10.1021/jf0350820. [DOI] [PubMed] [Google Scholar]
- De Luca V, Balsevich J, Tyler RT, Eilert U, Panchuk BD, Kurz WGW. Biosynthesis of indole alkaloids. Developmental regulation of the biosynthetic pathway from tabersonine to vindoline in Catharanthus roseus cultivar. Little delicata. J Plant Physiol. 1986;125:147–156. [Google Scholar]
- Deininger M, Szumlanski CL, Otterness DM, Van LJ, Ferber W, Weinshilboum RM. Purine substrates for human thiopurine methyltransferase. Biochem Pharmacol. 1994;48:2135–2138. doi: 10.1016/0006-2952(94)90515-0. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 Available at: World Wide Web http://pymol.sourceforge.net/
- Fahn W, Laussermair E, Deus-Neumann B, Stöckigt J. Late enzymes of vindoline biosynthesisS-Adenosyl-L-methionine: 11-O-demethyl-17-O-deacetylvindoline 11-O-methyltransferase and unspecific acetylesterase. Plant Cell Rep. 1985;4:337–340. doi: 10.1007/BF00269893. [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Zubieta C, Dixon RA, Noel JP. Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 2005;137:1009–1017. doi: 10.1104/pp.104.048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick S, Kutchan TM. Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J. 1999;17:329–339. doi: 10.1046/j.1365-313x.1999.00379.x. [DOI] [PubMed] [Google Scholar]
- Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E. Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14:505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A, Gulick PJ, Ibrahim RK. Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch Biochem Biophys. 1998;351:243–249. doi: 10.1006/abbi.1997.0554. [DOI] [PubMed] [Google Scholar]
- Hanson AD, Rivoval J, Paquet L, Gage DA. Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.) DC. Evidence that S-methylmethionine is an intermediate. Plant Physiol. 1994;105:103–110. doi: 10.1104/pp.105.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibdah M, Zhang XH, Schmidt J, Vogt T. A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J Biol Chem. 2003;278:43961–43972. doi: 10.1074/jbc.M304932200. [DOI] [PubMed] [Google Scholar]
- Ibrahim RK. Plant O-methyltransferase signatures. Trends Plant Sci. 1997;2:249–250. [Google Scholar]
- Ibrahim RK, Bruneau A, Bantignies B. Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol. 1998;36:1–10. doi: 10.1023/a:1005939803300. [DOI] [PubMed] [Google Scholar]
- James F, Nolte KD, Hanson AD. Purification and properties of S-adenosyl-L-methionine: L-methionine S-methyltransferase from Wollastonia biflora leaves. J Biol Chem. 1995;270:22344–22350. doi: 10.1074/jbc.270.38.22344. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved method for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Ko S, Eliot AC, Kirsch JF. S-Methylmethionine is both a substrate and an inactivator of 1-aminocyclopropane-1-carboxylate synthase. Arch Biochem Biophys. 2004;421:85–90. doi: 10.1016/j.abb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Krezel A, Lesniak W, Jezowska-Bojczuk M, Mlynarz P, Brasun J, Kozlowski H, Bal W. Coordination of heavy metals by dithiothreitol, a commonly used thiol group protectant. J Inorg Biochem. 2001;84:77–88. doi: 10.1016/s0162-0134(00)00212-9. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- López R, Ortín N, Pérez-Trujillo JP, Cacho J, Ferreira V. Impact odorants of different young white wines from the Canary Islands. J Agric Food Chem. 2003;51:3419–3425. doi: 10.1021/jf026045w. [DOI] [PubMed] [Google Scholar]
- Lukacin R, Matern U, Specker S, Vogt T. Cations modulate the substrate specificity of bifunctional class I O-methyltransferase from Ammi majus. FEBS Lett. 2004;577:367–370. doi: 10.1016/j.febslet.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Lyi SM, Heller LI, Rutzke M, Welch RM, Kochian LV, Li L. Molecular and biochemical characterization of the selenocysteine Se-methyltransferase gene and Se-methylselenocysteine synthesis in Broccoli. Plant Physiol. 2005;138:409–420. doi: 10.1104/pp.104.056549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Maury S, Geoffroy P, Legrand M. Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 1999;121:215–223. doi: 10.1104/pp.121.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo J, Levy A, Akavia N, Ashri A, Palevitch D. Inheritance of corolla color and anthocyanin pigments in periwinkle (Catharanthus roseus [L.] G. Don) Z Pflanzenzüchtg. 1985;95:352–360. [Google Scholar]
- Neuhierl B, Thanbichler M, Lottspeich F, Böck A. A family of S-methylmethionine-dependent thiol/selenol methyltransferases – role in selenium tolerance and evolutionary relation. J Biol Chem. 1999;274:5407–5414. doi: 10.1074/jbc.274.9.5407. [DOI] [PubMed] [Google Scholar]
- Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer JL. Structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Adv Phytochem. 2003;37:37–58. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M. Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol. 1993;103:509–517. doi: 10.1104/pp.103.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovan A, Filippini R, Favretto D. Characterization of the anthocyanins of Catharanthus roseus (L.) G. Don in vivo and in vitro by electrospray ionization ion trap mass spectrometry. Rapid Commun Mass Spectr. 1998;12:361–367. [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li CJ, Tarczynski MC, Hanson AD. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575–584. doi: 10.1046/j.1365-313x.2001.00988.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schröder G, Wehinger E, Schröder J. Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry. 2002;59:1–8. doi: 10.1016/s0031-9422(01)00421-6. [DOI] [PubMed] [Google Scholar]
- Schröder G, Wehinger E, Lukacin R, Wellmann F, Seefelder W, Schwab W, Schröder J. Flavonoid methylation: a novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry. 2004;65:1085–1094. doi: 10.1016/j.phytochem.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Tagmount A, Berken A, Terry N. An essential role of S-adenosyl-L-methionine: L-methionine S-methyltransferase in selenium volatilization by plants. Methylation of selenomethionine to selenium-methyl-L-selenium-methionine, the precursor of volatile selenium. Plant Physiol. 2002;130:847–856. doi: 10.1104/pp.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Dubourdieu D. Identification of cysteinylated aroma precursors of certain volatile thiols in passion fruit juice. J Agric Food Chem. 2000;48:2874–2876. doi: 10.1021/jf990980a. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Vetter HP, Mangold U, Schröder G, Marner FJ, Werck-Reichhart D, Schröder J. Molecular analysis and heterologous expression of an inducible cytochrome P-450 protein from periwinkle (Catharanthus roseus L.) Plant Physiol. 1992;100:998–1007. doi: 10.1104/pp.100.2.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- Vogt T. Regiospecificity and kinetic properties of a plant natural product O-methyltransferase are determined by its N-terminal domain. FEBS Lett. 2004;561:159–162. doi: 10.1016/S0014-5793(04)00163-2. [DOI] [PubMed] [Google Scholar]
- Wein M, Lavid N, Lunkenbein S, Lewinsohn E, Schwab W, Kaldenhoff R. Isolation, cloning and expression of a multifunctional O-methyltransferase capable of forming 2,5-dimethyl-4-methoxy-3(2H)-furanone, one of the key aroma compounds in strawberry fruits. Plant J. 2002;31:755–765. doi: 10.1046/j.1365-313x.2002.01396.x. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM. Human erythrocyte thiol methyltransferase: radiochemical microassay and biochemical properties. Clin Chim Acta. 1979;97:59–71. doi: 10.1016/0009-8981(79)90025-1. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- Werkhoff P, Güntert M, Krammer G, Sommer H, Kaulen J. Vacuum headspace method in aroma research: flavor chemistry of yellow passion fruits. J Agric Food Chem. 1998;46:1076–1093. [Google Scholar]
- Woodson LC, Ames MM, Selassie CD, Hansch C, Weinshilboum RM. Thiopurine methyltransferase. Aromatic thiol substrates and inhibition by benzoic acid derivatives. Mol Pharmacol. 1983;24:471–478. [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nature Struct Biol. 2001;8:271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002;14:1265–1277. doi: 10.1105/tpc.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]