Abstract
Fluorescence microscopy provides a powerful method for localization of structures in biological specimens. However, aspects of the image formation process such as noise and blur from the microscope's point-spread function combine to produce an unintuitive image transformation on the true structure of the fluorescing molecules in the specimen, hindering qualitative and quantitative analysis of even simple structures in unprocessed images. We introduce FluoroSim, an interactive fluorescence microscope simulator that can be used to train scientists who use fluorescence microscopy to understand the artifacts that arise from the image formation process, to determine the appropriateness of fluorescence microscopy as an imaging modality in an experiment, and to test and refine hypotheses of model specimens by comparing the output of the simulator to experimental data. FluoroSim renders synthetic fluorescence images from arbitrary geometric models represented as triangle meshes. We describe three rendering algorithms on graphics processing units for computing the convolution of the specimen model with a microscope's point-spread function and report on their performance. We also discuss several cases where the microscope simulator has been used to solve real problems in biology.
1. Introduction
Fluorescence microscopy is an indispensable tool for imaging biological specimens. A traditional brightfield microscope records the image formed by the absorption and transmission of an external light source as it travels through a specimen. In contrast, a fluorescence microscope records the image from light emitted by fluorescing molecules, called fluorophores, attached to or embedded within a specimen. When illuminated with light of a specific excitation frequency, these molecules fluoresce, emitting light at a lower frequency. A dichroic mirror filters out the excitation frequency, allowing only the light at the fluorescence frequency to be registered by the image sensor. Figure 1 shows a conventional fluorescence microscope setup.
Figure 1.
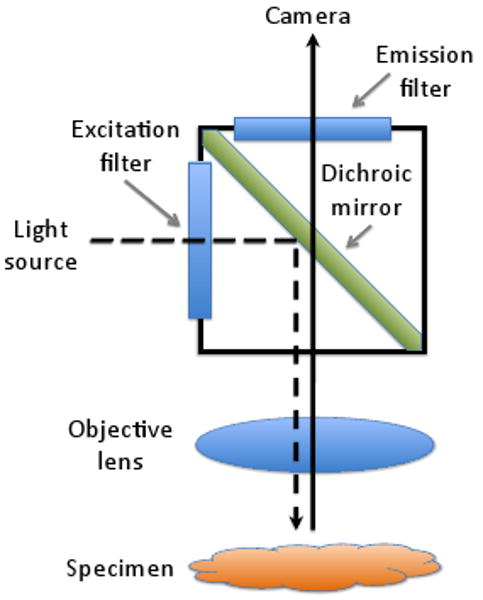
Light paths in a fluorescence microscope. A light source emits an excitation wavelength that reflects off a dichroic mirror toward the specimen where it causes fluorophores to emit a longer wavelength. The dichroic mirror allows only the emitted light to pass through to the camera. Excitation and emission filters restrict the wavelengths of light that reach the specimen and camera, respectively [Mur01].
Fluorescence microscopy has three key benefits. Scientists can label only the parts of the specimen in which they are interested with fluorophores, making tasks such as determining the location and structure of specific subcellular components much easier than with conventional bright field microscopy. Additionally, fluorescence microscopy enables in vivo experiments impossible with other higher resolution imaging modalities that require conditions fatal to the specimen, such as the vacuum required in a transmission electron microscope. Finally, fluorescence microscopy allows optical sectioning of specimens by adjusting the focal plane through a series of positions along the optical axis (conventionally denoted as the z-axis), forming a stack of 2D images that constitutes a 3D image of the specimen.
While the benefits of fluorescence microscopy have propelled it into widespread use in microbiological research, artifacts from the image formation process pose challenges for qualitative and quantitative analysis. A 3D point-spread function (PSF) characterizes the optical response of a fluorescence microscope to a point source of light. The PSF can be thought of as a blurring kernel, producing the fuzzy images characteristic of fluorescence microscopy. An xy-slice in the PSF represents how light passes from a point source emitter through the focal plane associated with that slice (see Figure 2a-d for examples of PSFs from widefield and confocal microscopes). To the extent that the PSF is shift-invariant, a model of 2D fluorescence image formation is the summation of light contributions from each fluorophore point source in the specimen to the focal plane at which the image is formed (see Figure 2e). Image formation for a full 3D stack is approximated well as a convolution of the fluorophores with the PSF [Cas79], The PSF can be calculated from theory [GL92], but it is preferable to measure the PSF from the microscope under the same conditions used to acquire experimental images. A measured PSF can be obtained by imaging a fluorescent bead smaller than the spatial extent of a pixel in the resulting image.
Figure 2.
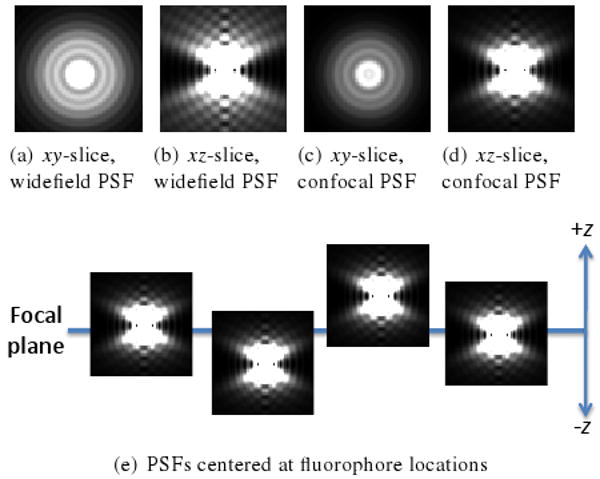
PSFs from widefield fluorescence microscope (a-b) and confocal fluorescence microscope (c-d) calculated with XCOSM [MC02]. Intensities are scaled to emphasize the overall shape of the PSFs. (e) Model of fluorescence microscope image formation. PSFs centered on fluorophores intersect the focal plane. Summing the PSF xy-slices at the intersections yields the simulated fluorescence image.
In a widefield fluorescence microscope, the PSF has roughly an hourglass shape. Focus decreases with increasing z distance from the center of the hourglass, resulting in an overall widening and dimming of the PSF. In fact, all of the light from the specimen, even from out-of-focus fluorophores, is collected in each image [MKCC99], causing the characteristic blur found in widefield fluorescence images. Confocal microscopes reduce image blurriness by using a pinhole aperture to block much of the out-of-focus light. The PSF from a confocal microscope is thereby truncated, having an approximately elliptical shape with primary axis along z.
Besides blur from the PSF, noise further distorts fluorescence images. Noise is introduced in several parts of the imaging process: shot noise arises from fluctuations in the number of photons emitted from the fluorophores and detected by a charge-coupled device (CCD), background noise comes from stray photons in the system, and read-out noise is inherent in CCDs [SGGR06], The combination of these noise sources fits an approximately Gaussian distribution.
Figure 3 shows examples of the unintuitive transformation fluorescence microscopy induces on objects such as spheres and tubes. The surprising results from these simple specimen geometries make clear that understanding the imaging process is vital for scientists who use fluorescence microscopy in the lab. Toward that end, we have developed Fluoro-Sim, an interactive visual problem-solving environment that generates images of geometric specimen models as they would appear in a fluorescence microscope (available for download at http://www.cs.unc.edu/Research/nano/cismm/download/microscopesimulator/). We see several applications for FluoroSim:
Figure 3.
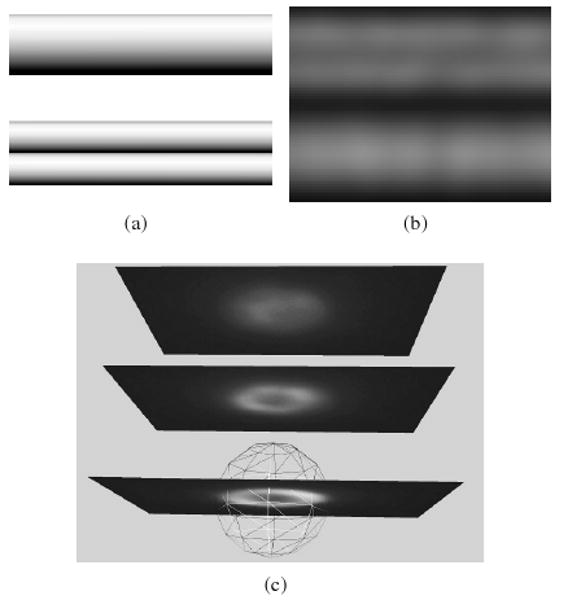
Examples of unintuitive images in fluorescence microscopy. (a) Models of surface-labeled small tubes. The side-by-side tubes are half the diameter of the larger tube. (b) Simulated noise-free fluorescence image of the tubes convolved with a calculated widefield PSF generated by XCOSM [MC02]. The two tubes can easily be misconstrued as a single tube while the single tube appears as two tubes. (c) A surface-labeled bead (in wireframe) with 1 micron radius superimposed with experimental images at focal planes with 2 micron spacing. The top-most image could be interpretted as the top of the sphere, but it is four times further away from the sphere center than the true top.
Training inexperienced scientists to understand the imaging artifacts in fluorescence microscopy.
Testing and refining hypotheses of specimen model geometry by comparing simulated images from a specimen model to experimental images from a real specimen.
Planning experiments by determining whether fluorescence microscopy is a suitable modality for distinguishing among multiple hypotheses.
Generating fluorescence images of known geometry to test new image processing and analysis algorithms.
Automatically registering a parameterized specimen model to experimental images obtained in the lab to determine the model parameters that best explain the image.
We aim for interactive rendering rates for several reasons. Interactivity allows a scientist to manipulate a specimen model and watch in real time how changes in orientation and position relative to the focal plane appear in the resulting fluorescence image. The real-time feedback improves the scientist's intuition for artifacts in fluorescence microscopy. Interactivity also encourages a scientist to quickly try out interesting scenarios involving specimen models, possibly leading to insight on future experiments. Finally, in future work, we plan to use the simulator for registering specimen models to 3D fluorescence images; many 3D fluorescence image stacks will be generated during registration, so reducing the time to generate each 2D image will ultimately improve registration speed.
To make fluorescence image simulation interactive, FluoroSim incorporates convolution rendering algorithms we have developed to run on graphics processing units (GPUs). These algorithms compute the convolution of an arbitrary model geometry with the PSF of the microscope at interactive frame rates for a single focal plane.
2. Related Work
Convolving a model of a fluorophore distribution with a PSF is a fundamental building block in iterative deconvolution algorithms that attempt to remove blurring induced by the PSF. Such image restoration methods attempt to invert the convolution by searching for the underlying fluorophore distribution in the specimen [MKCC99]. In these algorithms, the specimen model is represented implicitly by an image containing an estimate of the fluorophore density in each voxel.
Aside from deconvolution, convolving a representation of fluorophores with a PSF is an integral part of the image simulation technique called model convolution, on which our work builds. In this technique, the distribution of fluorophores is modeled either explicitly or as the result of a simulation of biological processes [SGP*04] [GOB07]. Fink et al. used simulated fluorescence images of subcellular component models represented by constructive solid geometry to estimate fluorophore densities inside real cells [FML98]. Lehmussola et al. developed a parametric random shape model for generating simulated fluorescence microscope images of populations of cells to test image analysis algorithms [LRS*07]. Sprague et al. used model convolution to evaluate models of kinetochore-attached microtubule dynamics in yeast during metaphase by statistical comparison of simulated and experimental images [SPM*03].
FluoroSim advances the model convolution technique in several ways. First, FluoroSim supports convolution of triangle mesh models widely used in 3D modeling. Second, FluoroSim incorporates algorithms we have developed for the GPU that enable interactive convolution of models. Finally, the FluoroSim modeling environment enables the creation and manipulation of specimen models with real-time updates of the simulated image.
3. Image Generation
FluoroSim generates 2D fluorescence images at a single focal plane. This approach has two performance advantages over generating full 3D images. First, GPUs are designed specifically for rendering 2D images, so rendering a single focal plane at a time fits their capabilities well. Second, it is efficient for mimicking real microscopes during the exploration phase prior to acquisition of a stack; an entire 3D convolution need not be computed to extract and display a single section. When a full 3D specimen image is desired, the focal plane position can be adjusted along the z-dimension in incremental steps just as in optical sectioning microscopy.
3.1. Geometric Representation and Virtual Fluorophore Placement
FluoroSim uses a standard triangle mesh for representing geometric specimen models. This representation is ubiquitous in geometric modeling and graphics applications and is well supported in the Visualization Toolkit (VTK) [SML06] and TetGen [SG05], software libraries on which FluoroSim is built. FluoroSim's modeling environment allows placement and manipulation of predefined primitive models such as spheres and tubes, and it also loads OBJ, PLY, and VTK files created in external modeling tools.
In biological applications, fluorophores are attached to a specimen's surface or embedded within the specimen material. FluoroSim emulates both kinds of specimen labeling with methods for generating uniform random samplings of geometry surfaces and the volumes contained inside surfaces. Figure 4 shows examples of each kind of labeling.
Figure 4.
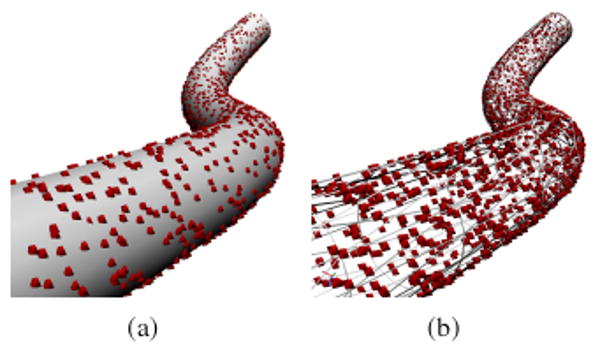
Uniform random sampling of a curvilinear tube. Spheres represent fluorophores (not to scale) generated from (a) surface labeling and (b) volume labeling. The surface of the tube on the right is represented in wireframe to make the internal fluorophores visible.
In surface labeling, sampling a fluorophore location according to a uniform random distribution involves selecting a random triangle in the surface mesh with probability equal to the ratio of triangle area to total surface mesh area, followed by sampling a random point in that triangle. We compute an array containing the triangle probabilities and then compute its prefix sum, which represents the cumulative distribution function (CDF) over the triangle probabilities. Given a pseudo-random number r in the range [0,1], we invert the CDF to find the index of the randomly selected triangle. Inversion entails searching for the index of the value in the CDF array closest to but less than r; the CDF array is sorted so an efficient binary search is used. The resulting index indicates the randomly selected triangle. From this triangle, we draw a uniform random sample with a method described by Turk [Tur90]. For volume labeling, we first use TetGen [SG05] to tetrahedralize a triangle mesh; fluorophore generation follows the same approach but operates on tetrahedra in the volume mesh.
FluoroSim gives users the ability to change the fluorophore density on a model and regenerate fluorophore samplings. For specimens with known fluorophore density, generating multiple images with different fluorophore samplings can reveal how the appearance of multiple real specimens in experimental fluorescence images might differ due to variability inherent in how fluorophores bind to the specimen. For specimens with unknown fluorophore density, we start with a low density and increase it until the appearance of generated images across successive samplings does not change significantly. In most cases, sampling tens of thousands of fluorophores is sufficient. This approach is equivalent to averaging images produced by successive fluorophore samplings, resulting in an expectation image of the specimen model. If the intensity range of the expectation image is different from the intensity range in experimental images, the discrepancy can be corrected by rescaling intensities in the expectation image.
3.2. Convolution Rendering on the GPU
We implemented three convolution rendering algorithms for fluorescence microscope simulation as extensions to the VTK library. The algorithms are partially expressed as programs in the OpenGL Shading Language and NVIDIA's CUDA Toolkit. Each algorithm takes a list of 3D points representing fluorophore locations and a 3D image representing the PSF of the microscope. For all algorithms, we store the PSF in a 3D texture because it provides an easy mechanism for looking up trilinearly interpolated values in the PSF. The camera model is orthographic because of the small depth of field in fluorescence microscopes, and the view direction is fixed parallel to the z-axis, looking in the -z direction.
3.2.1. Billboarding Algorithm
In this approach, we use a standard method for producing volumetric effects in rasterized computer graphics. The basic approach involves drawing rectangular billboards aligned with the image plane that are textured with images. By enabling the GPU's blending mode, the billboards can be blended together in interesting ways to produce volumetric effects such as fog. In the case of fluorescence microscope simulation, one billboard is drawn for every fluorophore so that the billboard's center is at the projected (x,y) location of the fluorophore in image space. The billboard's extent matches the extent of the PSF in x and y in image space, and the texture on each billboard is an xy-slice through the PSF (see Figure 2a for an example) at a specific z-depth. The z-depth is the difference between the fluorophores position's z-component and the focal plane position; the center of the PSF, where the point-source is most in focus, corresponds to a z-depth of 0. Rendering the billboards with 32-bit floating-point additive blending yields the sum of the fluorophore contributions.
In our OpenGL implementation, each corner of the billboard has a 3D texture coordinate used to look up the xy-slice from the PSF texture. In particular, the four texture coordinates are (0,0,z), (0,1,z), (1,1,z), and (1,0,z), where z is the difference computed above scaled and biased to fit in the texture coordinate range.
3.2.2. Per-Pixel Gather Algorithm
Modern GPUs have many programmable streaming processors that support typical computational patterns such as accessing arbitrary memory locations and executing dynamic length loops. To exploit these streaming processors, we have implemented a fragment program that, for every pixel in the output image, iterates through the list of fluorophore locations and sums the light contribution from each fluorophore to the pixel.
A fluorophore's contribution to a pixel is determined by computing a 3D offset from the fluorophore location to the pixel center in world space and using that vector, after appropriate scaling and biasing, as an index into the 3D PSF texture. The value returned by the texture lookup is computed by hardware-accelerated trilinear interpolation of the eight PSF voxels that surround the center of the image pixel. Fluorophores falling outside the boundary of the PSF centered at the pixel contribute no light to the pixel.
There are some implementation challenges with this approach. We store the list of fluorophore locations as a 32-bit floating-point RGB texture on the GPU where the red, green, and blue components correspond to x, y, and z components, respectively. While a 1D texture provides a natural way to store a list of points, each texture dimension is limited to only several thousand texture elements. Because potentially millions of fluorophores may be generated from a specimen model, we store the 3D fluorophore locations in a 2D texture and add appropriate 2D indexing calculations to the fragment program. Furthermore, on older GPUs, the number of instructions that can be executed is finite, limiting the number of fluorophores that can be processed in one fragment program invocation. Our solution is a multi-pass approach where each pass gathers the light contributions from a subset of the fluorophores and adds the results to the framebuffer with additive blending.
Two optimizations increase the speed of this algorithm. In the first optimization, a screen-space bounding rectangle of the projected fluorophores dilated by a rectangle half the screen-space extent of the PSF limits the computation domain. The bounding rectangle is large enough to ensure that all pixels potentially having fluorophore contributions are processed while excluding pixels that cannot receive contributions from fluorophores. The second optimization comes from considering that fluorophores on a specimen model are likely to be close together while specimen models may be far apart. A multi-pass approach is used where a separate bounding rectangle is computed and processed for each specimen model. This optimization potentially reduces the total number of pixels covered by the bounding rectangles, particularly for small specimens separated by large distances in the rendered image. It also reduces the ratio of fluorophores that contribute to a pixel to those that do not because fluorophores far from the pixel are not examined in the fragment program.
3.2.3. Fourier Domain Algorithm
The previous two convolution methods operate in the spatial domain. Using the Fast Fourier Transform (FFT), convolution via point-wise multiplication in the Fourier domain is asymptotically faster than the spatial domain methods. We have implemented a Fourier domain based algorithm that bins fluorophores into an image, then convolves that image with the PSF. Rather than compute a full 3D convolution, we follow Sprague et al. and implement a method that sums together partial 2D convolutions of subsets of the fluorophores to produce a 2D image at a particular focal plane depth [SPM*03]. The algorithm proceeds as follows:
With additive blending enabled, rasterize as points the fluorophores within a thin slab normal to the z-axis into a fluorophore image.
Render the PSF slice corresponding to the slab into a second image.
Convolve the two images using the FFT algorithm and component-wise multiplication in the Fourier domain.
Add the convolution result to an accumulation image.
Repeat for all slabs in z within the z-extent of the PSF.
On the GPU we compute Step 1 by adjusting camera clipping planes and rendering all the fluorophores into a texture target with additive floating-point blending. A slab thickness 0.25 times the z-spacing of the PSF offers a tradeoff between accuracy and speed; a smaller fraction of the z-spacing would produce more accurate results at the cost of computing the convolution of more slabs. We compute Step 2 by rendering the PSF into a second texture target, cyclically shifting the PSF slice so that its center is at the image origin. As in the billboarding algorithm, the PSF slice is determined by converting the world space difference between the slab and the focal plane into the third coordinate of the 3D PSF texture lookup. Step 3 makes use of the CUFFT library function that computes the FFT on the GPU as well as a custom CUDA kernel function for computing the point-wise multiplication in the Fourier domain. Finally, Step 4 is computed by rendering the convolution result into an accumulation texture with additive blending.
When specimen models are small relative to the extent of the PSF in the z-dimension, many of the slabs will contain no fluorophores and hence there is no need to compute the convolution. To prevent unnecessary computation, we check how many fluorophores were rendered into the slab image with OpenGL occlusion queries. If none are rendered, we can skip the convolution for that slab.
Because rasterizing the fluorophores quantizes the fluorophore locations, the method is less accurate than the two spatial domain methods. To increase accuracy, thinner slabs and higher resolution of the fluorophore and PSF images could be used at the cost of increased memory and computation.
3.3. Gaussian Noise Generation
To indicate expected variability due to noise in fluorescence images, we implemented Gaussian noise generation on the GPU. We generate uniform random numbers in parallel using the method described in [TW08]. For each pixel, two of the four 32-bit pseudo-random numbers generated with this method are used to generate a sample from a normalized Gaussian distribution using the Box-Muller transform [BM58]. The generated noise is added to the image following the convolution rendering step.
4. Performance Results
We tested the performance of the three convolution rendering algorithms under three modeling scenarios:
Rendering an image of a single small specimen model where the spatial extent of the image is small relative to the spatial extent of the PSF image.
Rendering an image of a single small specimen model where the spatial extent of the image is large relative to the spatial extent of the PSF image.
Rendering a large image of three small specimens separated in x and y by distances greater than the spatial extent of the PSF and separated by one micron in z.
The specimen models were surface-labeled one micron spheres, the PSF was 150×150×41 pixels with pixel size 65×65×100nm, and spatial extent of the rendered 2D image pixels was 65 × 65nm. Noise generation was disabled. All tests were run on a PC with an Intel 2.33 GHz Core 2 Duo processor, 4 GB RAM, and a single NVIDIA GeForce 8800 GTX GPU.
Table 1 shows timings for the three scenarios with models labeled by different numbers of fluorophores. In all tests, the per-pixel gather algorithm is over twice as fast as the billboarding algorithm. For 800,000 fluorophores, the bill-boarding algorithm failed to compute the 512×512 images because it exceeded a time limit imposed by the GPU driver. The Fourier domain algorithm is generally slower than the other algorithms for small numbers of fluorophores but is faster when the number of fluorophores is sufficiently high. Finally, the Fourier algorithm run-time is determined primarily by the size of the rendered image and the number of slab convolutions computed; it grows slowly as the number of fluorophores increases.
Table 1. Rendering times (in milliseconds) of the three convolution algorithms as a function of fluorophore count.
| # fluorophores | Billboarding algorithm | Gather algorithm | Fourier algorithm |
|---|---|---|---|
| 64 × 64 image of a single specimen | |||
| 50,000 | 242.1 | 112.0 | 125.7 |
| 100,000 | 483.5 | 217.5 | 133.2 |
| 200,000 | 964.8 | 429.5 | 146.7 |
| 400,000 | 1,932.0 | 852.9 | 174.3 |
| 800,000 | 3,857.8 | 1,711.5 | 231.4 |
| 512 × 512 image of a single specimen | |||
| 50,000 | 1,307.3 | 499.5 | 1,398.9 |
| 100,000 | 2,613.6 | 993.4 | 1,404.4 |
| 200,000 | 5,226.0 | 1,982.0 | 1,428.2 |
| 400,000 | 10,452.6 | 3,976.6 | 1,445.6 |
| 800,000 | failed | 7,949.0 | 1,503.7 |
| 512 × 512 image of three specimens separated in z | |||
| 50,000 | 1,307.2 | 505.3 | 4,092.2 |
| 100,000 | 2,617.1 | 1,000.1 | 4,131.7 |
| 200,000 | 5,230.6 | 1,985.6 | 4,170.4 |
| 400,000 | 10,458.8 | 3,961.5 | 4,182.5 |
| 800,000 | failed | 7,894.6 | 4,299.7 |
The billboarding approach is optimal for minimizing the number of additions performed on the GPU because every pixel affected by a fluorophore is touched exactly once when computing the contribution of that fluorophore. The per-pixel gather method is potentially suboptimal because every pixel in the screen-space bounding rectangle is touched by each fluorophore regardless of whether it contributes to the pixel. However, the GPU on which we have tested FluoroSim has fewer raster units than shading processors and they operate at a lower clock frequency, significantly reducing the rate at which the additive blending used in the billboarding method can be performed. This explains the lower performance of billboarding algorithm compared to the per-pixel gather algorithm.
5. Applications
We have used FluoroSim to answer questions in several real-world applications. All images in these applications were generated by the per-pixel gather algorithm with no Gaussian noise. In all applications, we used the same PSF which was calculated by XCOSM [MC02] with the microscope parameters used to take the experimental images in Figures 5c and 5e.
Figure 5.
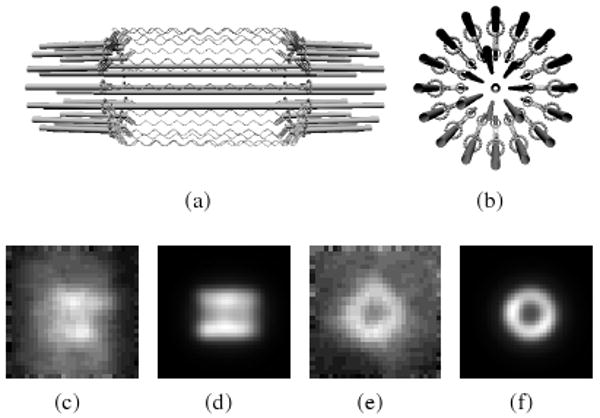
The mitotic spindle model [YHP*08]. (a) Side-on view; (b) end-on view. Helices represent the chromatin labeled with green fluorescing protein. Experimental and noise-free simulated images in side-on orientation (c-d) and end-on orientation (e-f). Differences in background between experimental and simulated images are caused by external chromatin not accounted for in the model. Structural variation in real specimens account for shape discrepancies.
Is a model of the mitotic spindle in yeast plausible?
During cell division, the mitotic spindle is a structure that ensures both the mother and daughter cells receive a copy of the DNA. We created a model of the mitotic spindle in S. cerevisiae featuring a hypothesized cylindrical arrangement of chromatin surrounding interpolar microtubules that form the backbone of the spindle apparatus [YHP*08]. Our collaborators (authors Haase and Bloom) take the qualitative match of the simulated and experimental images (see Figure 5) as evidence that supports the model.
Can fluorescence microscopy distinguish between branching and adjacent fibrin fibers?
Fibrin is a polymerized protein that forms into a mesh in blood clots. A challenge in understanding the structure of fibrin meshes and their mechanical properties is distinguishing between fibers that branch and fibers that are merely adjacent to each other. We used FluoroSim to create models of both configurations to determine whether a fluorescence microscope with this PSF can distinguish between them.
Figure 6c-d shows simulated images from the two volume-labeled model configurations where the focal plane is centered in the horizontal fiber. The images appear indistinguishable. However, intensity profiles along a line running down the center of the horizontal fiber in the simulated images show that where the two fibers meet, the branching fiber model is brighter than the adjacent fiber model. Assuming the two fibers have uniform diameters, the result is unexpected; where the fibers overlap, the two adjacent fibers have twice the number of fluorophores that are in the branching fiber model and should therefore appear brighter there. Our branching model, however, happens to have slightly more volume where the two fibers meet than would the union of two fibers, accounting for some of the increased intensity. Moreover, the additional fluorophores in that extra volume are nearer the focal plane than the top adjacent fiber, increasing their light contributions and accounting for the rest of the increased intensity. This investigation provides evidence that distinguishing between the two fiber configurations depends on the thickness of the fibrin at the branch points.
Figure 6.
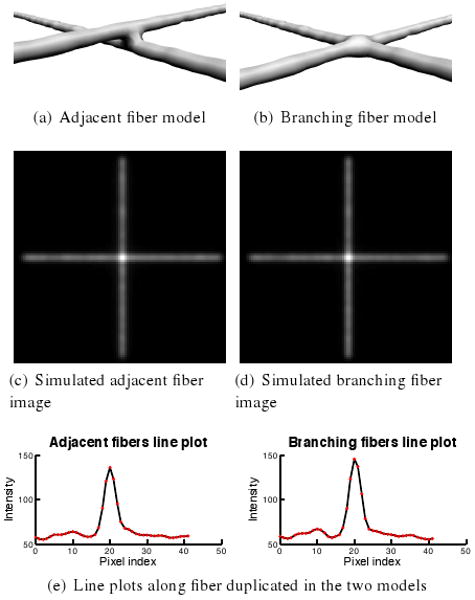
Testing whether fluorescence microscopy can distinguish between two fiber configurations in a fibrin mesh. Counterintuitively, peak intensity in adjacent fibers (136) is lower than peak intensity in branching fibers (146).
Can inter-bead distances less than the Abbe limit be determined by fitting a Gaussian to an intensity profile?
Ernst Abbe determined that diffraction-limited microscopes such as fluorescence microscopes have a limit below which two distinct point sources of light cannot be distinguished. In single molecule imaging, methods to resolve point sources separated by less than the Abbe limit have been developed [LMP*00]. However, we are not aware of any method that attempts to estimate the separation distance of two point sources by finding the standard deviation of a 1D Gaussian fit to the image intensities in a line plot. With FluoroSim, it took only minutes to find that such a method might be feasible for determining separating distances down to around 50nm for noise-free images. Figure 7 shows plots of the image intensities and the best-fitting Gaussian for six separation distances.
Figure 7.
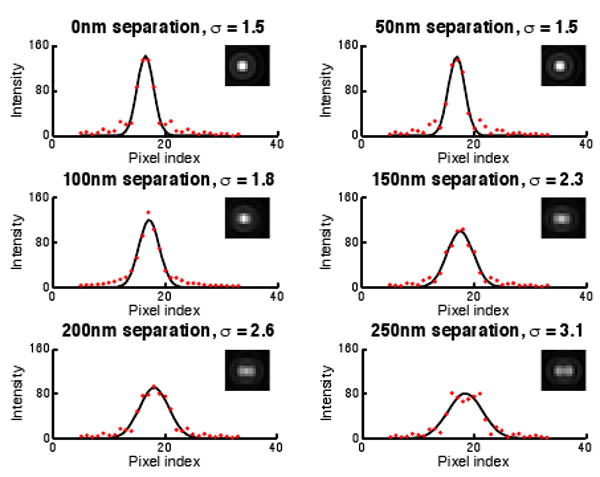
Gaussian functions fit in the least-squares sense to line profiles through simulated images of two fluorophores separated by varying sub-Abbe limit distances (inset). The standard deviation changes down to about a 50nm separation distance.
6. Conclusions and Future Work
FluoroSim is a new software tool for training, hypothesis testing, and experiment planning involving fluorescence microscopy. Our main contribution is a method for generating simulated fluorescence images from arbitrary geometric models represented by triangle meshes at interactive rates. As part of this contribution, we have described three convolution algorithms that exploit GPUs and analyzed their performance in several modeling scenarios. We also showed three biological applications where FluoroSim has been used to answer relevant questions.
In future work, we intend to add model registration to FluoroSim. This will enable the automatic optimization of a geometric model's parameters to a fluorescence image such that the model parameters best explain the image. As part of this effort, we plan to validate the simulated images by comparing them to experimental images of specimens with known geometry such as spherical polystyrene beads. We also intend to further improve rendering speed by parallelizing image generation across multiple GPUs.
Acknowledgments
We gratefully acknowledge David Feng, Brian Eastwood, Ryan Schubert, and the anonymous reviewers for their helpful suggestions for this paper. We also thank David Borland for guidance on aspects of the OpenGL implementation.
C. W. Quammen, A. C. Richardson, and R. M. Taylor II were supported by National Institutes of Health grant P41 EB002025. J. Haase, B. D. Harrison, and K. S. Bloom were supported by National Institutes of Health grant GM32238 and National Science Foundation grant MCB 0451240.
Footnotes
The definitive version is available at http://www.eg.org/EG/DL/WS/VCBM/VCBM08.
References
- BM58.Box GEP, Muller ME. A note on the generation of random normal deviates. Annals of Mathematical Statistics. 1958;29(2):610–611. [Google Scholar]
- Cas79.Castleman KR. Digital Image Processing. Prentice-Hall; 1979. [Google Scholar]
- FML98.Fink C, Morgan F, Loew LM. Intracellular fluorescent probe concentrations by confocal microscopy. Biophysical Journal. 1998;75(4):1648–1658. doi: 10.1016/S0006-3495(98)77607-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GL92.Gibson SF, Lanni F. Experimental test of an analytical model of aberration in an oil-immersion objective lens used in 3-d microscopy. Journal of the Optical Society of America. 1992;9(1):154–166. doi: 10.1364/josaa.9.000154. [DOI] [PubMed] [Google Scholar]
- GOB07.Gardner MK, Odde DJ, Bloom K. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods. 2007;41(2):232–237. doi: 10.1016/j.ymeth.2006.08.002. [DOI] [PubMed] [Google Scholar]
- LMP*00.Lacoste TD, Michalet X, Pinaud F, Chemla DS, Alivisatos AP, Weiss S. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. Proc Natl Acad Sci U S A. 2000;97(17):9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LRS*07.Lehmussola A, Ruusuvuori P, Selinummi J, Huttunen H, Yli-Harja O. Computational framework for simulating fluorescence microscope images with cell populations. IEEE Transactions on Medical Imaging. 2007 July;26(7):1010–1016. doi: 10.1109/TMI.2007.896925. [DOI] [PubMed] [Google Scholar]
- MC02.McNally JG, Conchello JA. XCOSM. 2002 http://www.omrfcosm.omrf.org/xcosm.
- MKCC99.McNally JG, Karpova T, Cooper J, Conchello JA. Three-dimensional imaging by deconvolution microscopy. Methods. 1999;19(3):373–385. doi: 10.1006/meth.1999.0873. [DOI] [PubMed] [Google Scholar]
- Mur01.Murphy DB. Fundamentals of Light Microscopy and Electronic Imaging. Wiley-Liss; 2001. [Google Scholar]
- SG05.SI H, Gaertner K. Meshing piecewise linear complexes by constrained Delaunay tetrahedralizations; Proceedings of the 14th International Meshing Roundtable; Sept. 2005; pp. 147–163. [Google Scholar]
- SGGR06.Sheppard CJR, Gan X, Gu M, Roy M. Signal-to-noise ratio in confocal microscopes. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. Springer; 2006. pp. 442–452. [Google Scholar]
- SGP*04.Sprague BL, Gardner MK, Pearson CG, Maddox PS, Bloom K, Salmon ED, Odde DJ. Model-convolution approach to modeling fluorescent protein dynamics; Conference Record of the 38th Asilomar Conference on Signals, Systems and Computers; Nov. 2004; IEEE; pp. 1821–1825. [Google Scholar]
- SML06.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: An Object Oriented Approach to 3D Graphics. 4th. Prentice-Hall; 2006. [Google Scholar]
- SPM*03.Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophysical Journal. 2003;84(6):3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tur90.Turk G. Generating random points in triangles. In: Glassner A, editor. Graphics Gems. Academic Press; 1990. pp. 24–28. [Google Scholar]
- TW08.Tzeng S, Wei LY. Parallel white noise generation on a GPU via cryptographic hash. I3D '08: Proceedings of the 2008 Symposium on Interactive 3D Graphics and Games; 2008. [Google Scholar]
- YHP*08.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Current Biology. 2008;18(2):81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]


