Abstract
NphB is a soluble prenyltransferase from Streptomyces sp. strain CL190 that attaches a geranyl group to a 1,3,6,8-tetrahydroxynaphthalene-derived polyketide during the biosynthesis of anti-oxidant naphterpin. Here we report multiple chemoenzymatic syntheses of various prenylated compounds from aromatic substrates including flavonoids using two prenyltransferases NphB and SCO7190, a NphB homolog from Streptomyces coelicolor A3(2), as biocatalysts. NphB catalyzes carbon–carbon-based and carbon–oxygen-based geranylation of a diverse collection of hydroxyl-containing aromatic acceptors. Thus, this simple method using the prenyltransferases can be used to explore novel prenylated aromatic compounds with biological activities. Kinetic studies with NphB reveal that the prenylation reaction follows a sequential ordered mechanism.
Keywords: Enzyme catalysis, Prenylation, Flavonoid, Polyketide, Kinetics
1. Introduction
To date, natural products with one or more prenyl groups have been isolated predominantly from higher plants. These compounds, originating from multiple natural product classes, often possess anti-microbial, anti-oxidant, anti-inflammatory, anti-viral, and anti-cancer activities.1 For example, several prenylated flavonoids including 8-geranyl chrysin and 6-dimethylallyl galangin counteract multi-drug resistance.2 Recent structure–activity relationships revealed that the activities of these prenylated compounds correlate with their enhanced interaction with plasma membrane ABC (ATP-binding cassette) transporters compared with their non-prenylated parent compounds.3 Given their enhanced and distinct activities, prenylated flavonoids show promise as lead compounds for the development of non-toxic anti-microbials and nutraceuticals in plants and new pharmacological agents for the treatment of animal diseases.1 However, prenylated compounds often exist at trace levels in natural sources, and are often not amenable to cost effective synthesis. Given the recent identification of soluble prenyltransfe-rases with relaxed substrate specificity displaying regiospecificity in prenyl group transfer and prenyl chain selectivity,4 these biocata-lysts can serve as an alternate production platform particularly when combined with structure-based enzyme engineering.
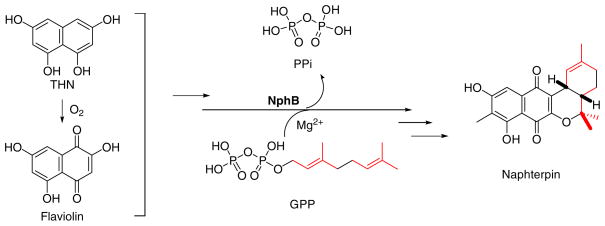
The anti-oxidant naphterpin is a natural product biosynthesized from the polyketide 1,3,6,8-tetrahydroxynaphthalene (THN) generated from malonyl-CoA by a type III polyketide synthase, and tailored by installation of a geranyl moiety that undergoes subsequent cyclization.5–7 We identified a soluble prenyltransfer-ase, Orf2, now renamed NphB, in the naphterpin biosynthetic gene cluster from Streptomyces sp. strain CL190, that could catalyze the transfer of a geranyl group to an unidentified THN-derived metab-olite (Scheme 1).4 In addition, we solved the X-ray crystal structure of NphB, an architecturally new prenyltransferase.4,8 Further, we demonstrated that NphB accepts a diverse collection of hydroxyl-containing aromatic substrates, including several flavonoids. Although substrate specificity, regiospecificity, and a physiological substrate of NphB have not been fully elucidated, it is clear that the prenyltransferase may serve as a biocatalyst for the production of prenylated compounds both in vivo and in vitro. Here, we report multiple chemoenzymatic syntheses of prenylated aromatic compounds by using two prenyltransferases NphB and SCO7190, a NphB homolog from Streptomyces coelicolor A3(2). In addition, kinetic studies provide insight into the reaction mechanism of the prenylation reaction.
Scheme 1.
Putative reaction catalyzed by NphB in naphterpin biosynthesis.
2. Results and discussion
2.1. Chemoenzymatic syntheses of prenylated dihydroxy naphthalene, prenylated flavonoids, and prenylated plant polyketides
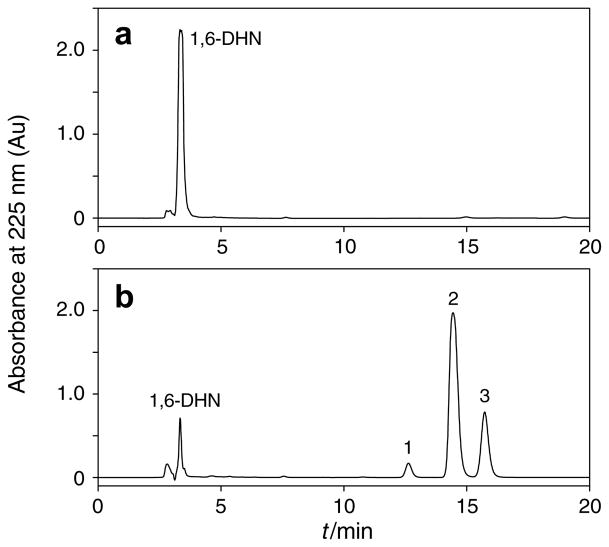
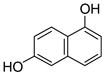
Recombinant NphB and SCO7190 were overproduced in Escherichia coli and purified to homogeneity.4 As prenyl acceptors, we focused on 1,3-dihydroxynaphthalene (DHN), 1,6-DHN 1, 2,7-DHN 2, 4-hydroxyphenylpyruvate, 2,5,7- trihydroxynaphtho-quinone (flaviolin), and the eight plant polyketides, olivetol 3, resve-ratrol 4, piceatannol, pterostilbene and the (iso)flavonoid sub-class including naringenin 5, apigenin 6, genistein 7, and daidzein 8. All substrates, except for 1,3-DHN, 4-hydroxyphenylpyruvate, flaviolin, piceatannol, and pterostilben underwent facile geranylation, yielding between one and three products (Fig. 1 and Tables 1–4).
Figure 1.
HPLC analysis of geranylated products formed from 1,6-DHN by the action of NphB. (a) No NphB added. (b) After a 36-h incubation with NphB. Peak 1, 4-geranyl 1,6-DHN 9; peak 2, 5-geranyl 1,6-DHN 10; peak 3, 2-geranyl 1,6-DHN 11.
Table 1.
Geranylated DHNs synthesized by NphB
| Substrate | Km (mM) | Product | ||
|---|---|---|---|---|
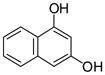 1,3-DHN |
No product | |||
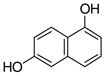 1 |
1.05 ± 0.17 |
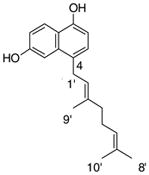 9 |
0.32 ± 0.02 .30 |
|
 1 |
0.54 ± 0.07 |
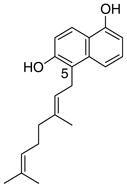 10 |
4.2 ± 0.2 7.7 |
|
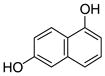
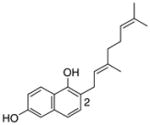
 1 |
0.51 ± 0.08 |
 11 |
1.1 ± 0.1 2.2 |
|
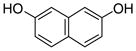 2 |
0.34 ± 0.06 |
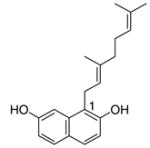 12 |
0.47 ± 0.01 1.4 |
|
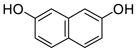 2 |
N.D.a |
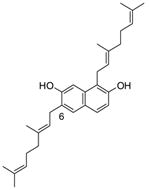 13 |
N.D.a | |
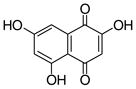 flaviolin |
Negligible products |
Not determined.
Table 4.
Prenylated products synthesized by SCO7190
| Substrate | Product |
|---|---|
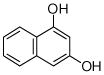 1,3-DHN |
No product |
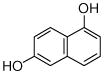 1 |
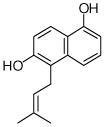 25 |
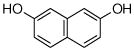 2 |
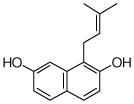 26 |
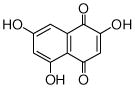 flaviolin |
No product |
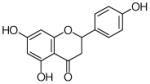 5 |
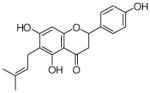 27 |
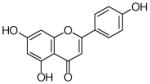 6 |
No product |
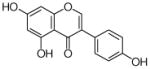 7 |
No product |
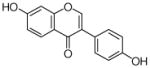 8 |
No product |
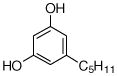 3 |
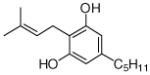 28 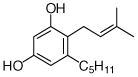 29 |
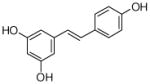 4 |
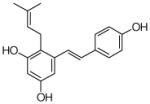 30 |
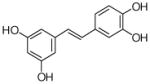 piceatonal |
No product |
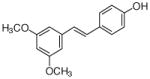 pterostilbene |
No product |
Most NphB reaction products contain a geranyl group in the ortho position with respect to a neighboring hydroxy moiety. Only 1,6-DHN underwent geranylation at the para position, albeit at a relatively low catalytic efficiency (Table 1). Interestingly, NphB appended two geranyl groups to 2,7-DHN to yield 1,6-digeranyl 2,7-DHN 13 (Table 1), which was formed from 1-geranyl 2,7-DHN 12 (Supplemental figure 1). With all flavonoids tested, NphB catalyzed the ger-anylation of a hydroxy group leading to the formation of an O-prenyl linkage (Table 2). In addition to this O-geranylation, NphB geranylat-ed the C6 position of naringenin 5 and apigenin 6 and the C8 carbon of daidzein 8 to yield 8-geranyl daidzein 20. NphB also geranylated the plant polyketides, olivetol 3 and resveratrol 4 (Table 3).
Table 2.
Geranylated flavonoids synthesized by NphB
| Substrate | Km (mM) | Product | ||
|---|---|---|---|---|
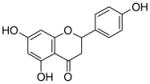 5 |
1.6±0.2 |
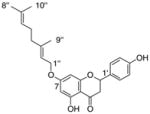 14 |
23±0.1 1.4 |
|
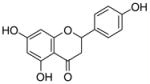 5 |
2.1±0.5 |
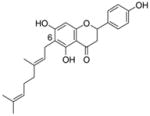 15 |
0.67±0.08 0.31 |
|
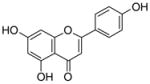 6 |
0.58±0.06 |
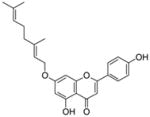 16 |
0.33±0.01 0.57 |
|
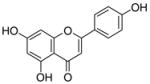 6 |
0.53±0.06 |
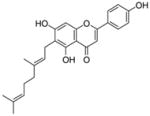 17 |
0.60±0.03 1.1 |
|
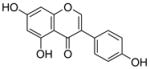 7 |
0.33±0.01 |
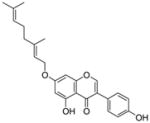 18 |
0.48±0.01 1.5 |
|
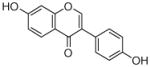
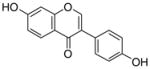 8 |
0.29±0.05 |
 19 |
0.17±0.01 0.59 |
|
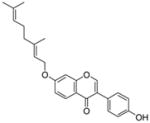
 8 |
N.D.a |
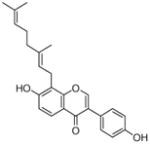 20 |
N.D.a |
Not determined.
Table 3.
Geranylated polyketides synthesized by NphB
| Substrate | Km (mM) | Product | ||
|---|---|---|---|---|
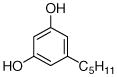
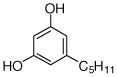 3 |
0.64±0.16 |
 21 |
0.043±0.003 0.067 |
|
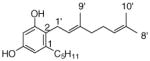
 3 |
0.52±0.03 |
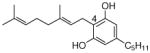 22 |
0.027±0.001 0.052 |
|
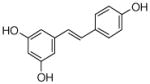 4 |
0.44±0.06 |
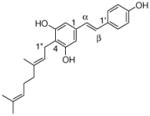 23 |
0.053±0.002 0.12 |
|
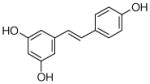 4 |
N.D.a |
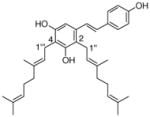 24 |
N.D.a | |
 piceatonal |
No product | |||
 pterostilbene |
No product |
Not determined.
In contrast, SCO7190 only catalyzed carbon–carbon-based pre-nylation by attaching a dimethylallyl moiety to 1,6-DHN 1, 2,7-DHN 2, naringenin 5, olivetol 3, and resveratrol 4 (Table 4). Given the structural diversity of the substrates employed and the large number of possible C- and O-accepting atoms, the enzyme assays revealed significantly relaxed substrate specificity while often preserving regioselectivity for prenyl group transfer.
2.2. Kinetic study of the NphB-catalyzed reaction
In the case of NphB incubation with 1,6-DHN and Mg2+, three ger-anylated products, 2-geranyl, 4-geranyl, and 5-geranyl 1,6-DHNs 9–11, formed (Fig. 1), indicating that 1,6-DHN must adopt at least three productive binding orientations in the NphB’s active site. We previously reported the crystal structure of NphB complexed with a non-hydrolysable geranyl diphosphate (GPP) analog, geranyl S-thiodi-phosphate (GSPP) and 1,6-DHN (PDB ID, 1zb6).4 Recently, Merz and co-workers reported simulation studies suggesting two alternative binding orientations for 1,6-DHN.9 In the light of these results, we then estimated apparent Km value for each putative binding orientation of 1,6-DHN. The kcat value for the corresponding product was also estimated from the concentration dependence of the initial velocity of the NphB reaction under conditions of saturating GPP. Apparent steady-state kinetic constants were estimated for each 1,6-DHN product by quantitatively measuring the amount of each geranylated 1,6-DHN using HPLC analysis (Table 1).
When 1,6-DHN was used as a substrate, the lowest Km values were calculated for 2-geranyl 1,6-DHN 11 (0.51 ± 0.08 mM) and for 5-geranyl 1,6-DHN 10 (0.54 ± 0.07 mM) biosynthesis. In our crystal structure of NphB complexed with GSPP and 1,6-DHN (Fig. 2),4 the 1,6-DHN molecule orients in the active site so that its C5 position is closest to the C1 position of GSPP. This binding orientation correlates with the Km value obtained for the orientation of 1,6-DHN leading to the formation of 5-geranyl 1,6-DHN.
Figure 2.
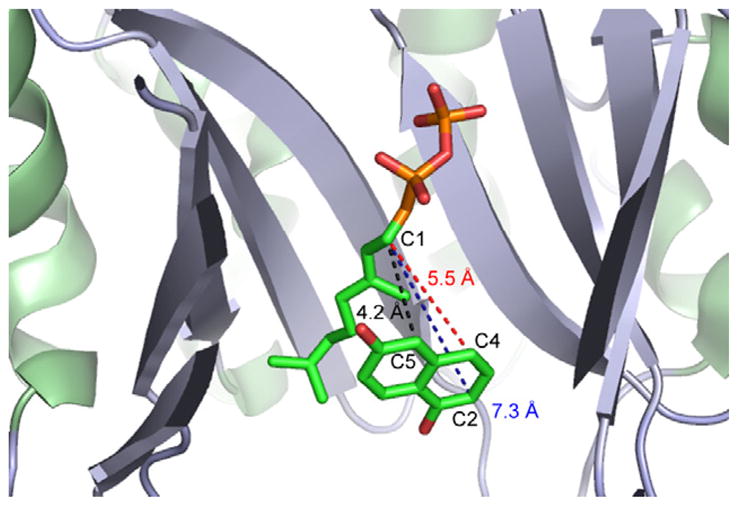
Crystal structure of NphB complexed with GSPP and 1,6-DHN (PDB ID, 1z6b).4 C5 of 1,6-DHN is the closest atom from C1 of GSPP. The distance is 4.2 Å. On the other hand, the distance between C1 of GSPP and C2 of 1,6-DHN is 7.3 Å. The distance between C1 of GSPP and C4 of 1,6-DHN is 5.5 Å.
Our kinetic data (Tables 1–3) indicate that all substrates tested thus far with NphB possess Km values ranging from 0.29 to 2.1 mM. These steady-state measurements bode well for the possible use of NphB or mutants thereof for the biocatalytic production of regio-specifically prenylated aromatic compounds of natural and synthetic origin.
2.3. Reaction mechanism of NphB
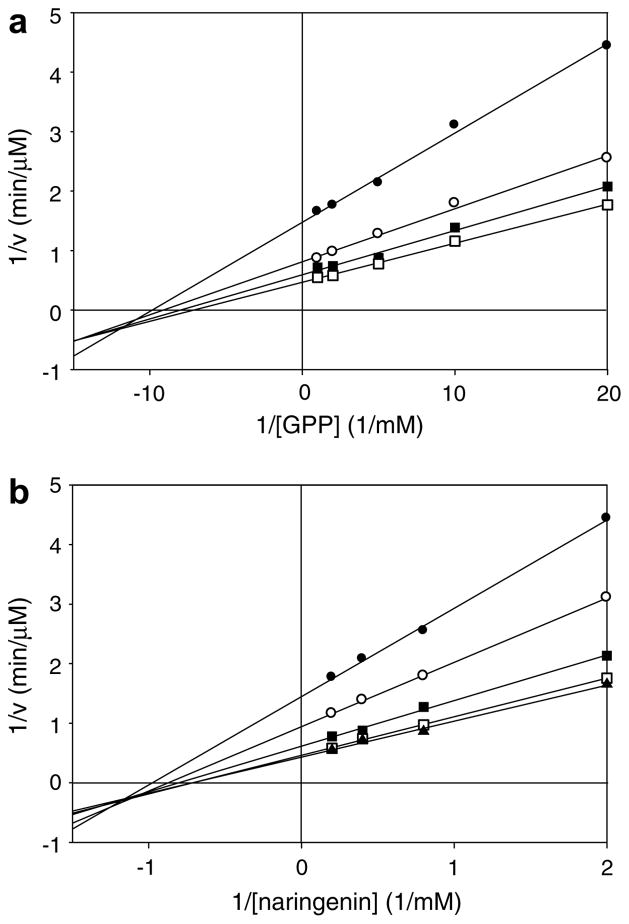
To gain insight into the kinetic mechanism of NphB, double-reciprocal plots (1/initial velocity vs 1/substrate concentration) of GPP and naringenin turnover were calculated (Fig. 3). In both plots, the lines intersect at a common point to the left of the 1/initial-velocity axis, indicating that the NphB reaction follows a sequential mechanism, where both GPP and naringenin must bind in succession at the enzyme’s active site before product formation can occur. However, these plots fail to elucidate the binding order of the two substrates. We solved the 3D structures of NphB complexed with GPP alone (PDB ID, 1zcw) and with GSPP and 1,6-DHN.4 However, we repeatedly failed to obtain NphB structures complexed with any aromatic acceptor alone (i.e., 1,6-DHN or res-veratrol). This observation suggests that 1,6-DHN cannot bind to NphB in the absence of bound GPP. Thus, GPP most likely binds first in the sequential ordered mechanism of the NphB-catalyzed reaction (Supplemental figure 2).
Figure 3.
Double-reciprocal plots of GPP and naringenin turnover catalyzed by NphB. (a) Lineweaver–Burk plots for the NphB reaction with 0.5 mM (●), 1.25 mM (○), 2.5 mM (■), 5 mM (□) of naringenin and varied [GPP]. (b) Lineweaver–Burk plots for the NphB reaction with 0.05 mM (●), 0.1 mM (○), 0.2 mM (■), 0.5 mM (□), 1 mM (▲) of GPP and varied [naringenin]. 1/initial velocity versus 1/substrate concentration was fit to the Lineweaver–Burk plot using a software SigmaPlot 10 (Systat Software, Point Richmond, CA).
We have proposed an electrophilic aromatic substitution reaction in NphB, where ionic cleavage of the carbon–oxygen bond in GPP and the resulting formation of a geranyl carbocation occur before prenylation of the electron rich aromatic acceptor.4 Together with the results of the present kinetic study, we posit that the NphB reaction proceeds as follows. (1) GPP binds to the NphB active site and orients through diphosphate–cation interactions (Mg2+ and/or basic side chains), (2) the aromatic acceptor then binds triggering, (3) the formation of a geranyl carbocation through pyrophosphate loss whereupon (4) the carbocation performs an electrophilic attack on the aromatic acceptor, terminated by (5) acceptor neutralization through proton loss.
2.4. Time course of the NphB-catalyzed reaction
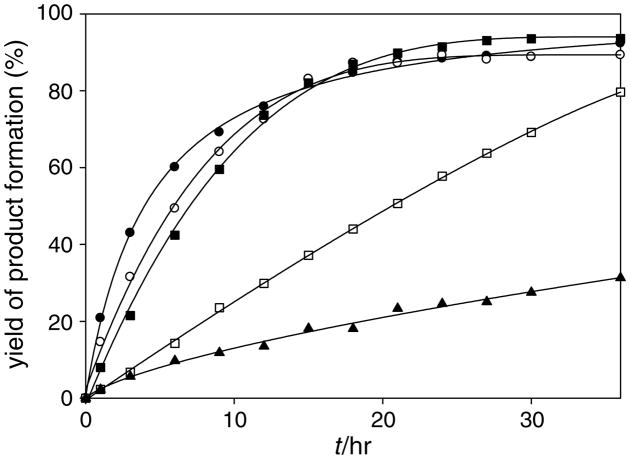
Finally, we determined yields of geranylated product formation (Fig. 4). Naringenin 5, apigenin 6, and genistein 7 underwent nearly complete geranylation during a 36-h incubation. On the other hand, the yield of the geranylated products of daidzein 8 and olive-tol 3 ranged from 20% to 80% over the same time period. Time courses of the NphB reactions indicate that NphB maintains stability for the entire 36-h incubation.
Figure 4.
Time course of the NphB-catalyzed reaction. Reactions were carried out in 50 mM Tris–HCl (pH 9.0), containing 5 mM MgCl2, 0.5 mM aromatic substrate, 5 mM GPP, 1 mg/ml NphB at 25 °C in a total volume of 60 μl. Naringenin (●), apigenin (○), genistein (■), daidzein (□), and olivetol (▲).
3. Conclusion
The present study describes multiple chemoenzymatic syntheses of various prenylated compounds from aromatic substrates including flavonoids using a newly described structural and functional class of small molecule prenyltransferase isolated from bacteria. In particular, one such prenyltransferase, NphB possesses synthetically useful relaxed substrate specificities while maintaining in some cases regioselectivity for prenyl group transfer. As a prenyltransferase belonging to the same class as that of NphB, Fnq26 of furanonaphthoquinone biosynthesis has been identi-fied.10 However, Fnq26 possesses substantially different substrate specificities than that of NphB, although the two enzymes share 42% of sequence identity. For example, Fnq26 catalyzes the gerany-lation of a hydroxy group of 1,3-DHN and 4-hydroxybenzoic acid, both of which are not geranylated by NphB. In addition, interestingly, Fnq26 is able to catalyze attachment of the C3 carbon of the ionized geranyl group rather than the C1 carbon to flaviolin. In contrast, Fnq26 does not accept 1,6-DHN and 2,7-DHN as a substrate. Finally, the flavanone naringenin undergoes limited Fnq26 directed prenylation. Thus, in order to more easily predict and engineer the substrate selectivity/promiscuity and regioselectivity of newly identified prenyltransferases of the NphB/Fnq26 class, it will be important to carry out a combination of structural studies and additional biochemical experiments.
The potential of employing chemoenzymatic approaches as described above provides a convenient starting point to explore novel prenylation chemistry and the bioactivity of aromatic compounds using structure-based enzyme engineering. In addition, such rationally based engineering of NphB or related prenyltransferases will provide another powerful synthetic tool to expand the diversity and bioactivities of many synthetic and natural compounds through enzyme directed regiospecific prenylation.
4. Experimental
4.1. Enzyme assay
Assays for the NphB-catalyzed reaction were carried out in 50 mM Tris–HCl (pH 9.0), containing 5 mM MgCl2, 5 mM aromatic substrate, 5 mM geranyl diphosphate (GPP), 1 mg/ml NphB, in a total volume of 60 μl. The reaction mixtures were incubated at 25 °C for overnight, and then extracted with 200 μl ethyl acetate. The extracts were evaporated in a centrifugal vacuum concentrator, and the residues were resolved in 20 μl of methanol. The methanol solution was spotted on silica plates (Merck 20 cm × 20 cm silica gel 60 F254, 0.2 mm). The plates were developed with chloroform: methanol (15:1 v/v) and visualized by UV254. The reaction products were also analyzed by high performance liquid chromatography (JASCO, Tokyo) equipped with a MD-2010 Plus photodiode array with PEGASIL ODS (4.6 × 250 mm, Senshu, Tokyo). Assays for SCO7190-catalyzed reaction were carried out in 50 mM Tris–HCl (pH 9.0), containing 5 mM aromatic substrate, 5 mM dimethylallyl diphosphate (DMAPP), 1 mg/ml SCO7190, in a total volume of 60 μl. All the subsequent steps were identical to those of the NphB assay.
4.2. Product analysis
Large-scale productions of prenyl products in a total volume of 10 ml were carried out in the same condition as that for the enzyme assay described above. The reaction mixtures were incubated at 25 °C for overnight, and then extracted with 10 ml ethyl acetate three times. The extracts were evaporated and the residues were resolved in 1 ml of methanol. Prenylated products were purified by high performance liquid chromatography with PEGASIL ODS (20 × 250 mm, Senshu, Tokyo) employing an isocratic elution of 80% methanol, monitored at 254 nm. The chemical structures of the reaction products were elucidated by analyzing 1H NMR, 13C NMR, HMBC spectra, and HRESI-MS data.
4.3. Steady-state kinetic parameters
Steady-state kinetic parameters were determined by the Michaelis–Menten Equation using a software SigmaPlot 10.0 and an Enzyme Kinetic Module 1.3 (Systat Software, Point Richmond, CA). The concentrations of GPP were 0.05–2.5 mM, while the concentration of naringenin was 5 mM. Under the condition, no substrate inhibition of NphB was observed and Km value for GPP was calculated to be 130 μM. For DHNs, the concentrations were 0.5, 1.0, 2.5, and 5 mM, while the concentration of GPP was 2.5 mM. Incubation times were 3, 6, 9, and 15 min. For naringenin, apigenin, and olivetol, the concentrations were 0.5, 1.25, 2.5, and 5.0 mM. For genistein, the concentrations were 0.3, 0.5, 1.0, and 2.5 mM. For resveratrol, the concentrations were 0.15, 0.3, 0.5, and 1.0 mM. For daidzein, the concentrations were 0.15, 0.3, 0.25, and 0.5 mM. The incubation times for these reactions were 10, 20, 30, and 40 min. To gain insight into mechanism of the NphB-catalyzed reaction, Lineweaver–Burk plots (Fig. 3) were created with 0.5, 1.25, 2.5, and 5 mM of naringenin and 0.05, 0.1, 0.2, 0.5, and 1 mM of GPP. The incubation times were 5, 17, 29, and 41 min. We confirmed that all the reactions proceeded in a time-dependent manner under each substrate concentration.
All the reactions were performed in 50 mM Tris–HCl (pH 9.0), containing 5 mM MgCl2, each aromatic substrate, 2.5 mM GPP, 0.5 mg/ml NphB, in a total volume of 1 ml. The reaction mixture was initiated by adding NphB and incubated at 25 °C. Aliquots (200 μl) were removed from the reaction mixture at appropriate incubation times mentioned above. Each aliquot was extracted with an equal volume of ethyl acetate twice. The organic layer was collected and evaporated. The residual material was dissolved in 60 μl of methanol, and the formation of each product was quantified by HPLC using the standard curve. A plot of product versus time was used to determine initial velocity.
The reactions to determine yield of geranylated products formation (Fig. 4) were carried out in 50 mM Tris–HCl (pH 9.0), containing 5 mM MgCl2, 0.5 mM aromatic substrate, 5 mM GPP, 1 mg/ml NphB, in a total volume of 60 μl, because 5 mM daidzein is not soluble in the buffer, but 0.5 mM daidzein is soluble.
4.4. 1H, 13C NMR, and HRMS data of the prenylated products
The structures were analyzed by their mass spectral data [HRMS (ESI−); JEOL JMS-T100LC] and nuclear magnetic resonance (NMR) spectral data (600 MHz, JEOL ECA-600).
4.4.1. 4-Geranyl 1,6-DHN (9; product was synthesized from 1,6-DHN and GPP by recombinant NphB)
HRMS (ESI−) calcd for C20H23O2 (M−), 295.16980; found 295.16781. 1H NMR (CD3OD) δ: 1.50 (s, 3H, H-10′), 1.54 (s, 3H, H-8′), 1.84 (s, 3H, H-9′), 1.94 (m, 2H, H-4′), 2.03 (m, 2H, H-5′), 3.67 (d, J = 6.2 Hz, 2H, H-1′), 5.00 (m, 1H, H-6′), 5.14 (m, 1H, H-2′), 6.51 (d, J = 7.6 Hz, 1H, H-2), 6.97 (m, 2H, H-3, H-7), 7.17 (s, 1H, H-5), 8.05 (d, J = 8.9 Hz, 1H, H-8).
4.4.2. 5-Geranyl 1,6-DHN (10; product was synthesized from 1,6-DHN and GPP by recombinant NphB)4
HRMS (ESI−) calcd for C20H23O2 (M−), 295.16980; found 295.17000. 1H NMR (CD3OD) δ: 1.50 (s, 3H, H-10′), 1.54 (s, 3H, H-8′), 1.84 (s, 3H, H-9′), 1.94 (m, 2H, H-4′), 2.03 (m, 2H, H-5′), 3.67 (d, J = 6.2 Hz, 2H, H-1′), 5.00 (m, 1H, H-6′), 5.14 (m, 1H, H-2′), 6.58 (d, J = 7.6 Hz, 1H, H-2), 6.97 (d, J = 8.9 Hz, 1H, H-7), 7.16 (dd, J = 8.3, 7.6 Hz, 1H, H-3), 7.27 (d, J = 8.3 Hz, 1H, H-4), 7.91 (d, J = 8.9 Hz, 1H, H-8).
4.4.3. 2-Geranyl 1,6-DHN (11; product was synthesized from 1,6-DHN and GPP by recombinant NphB)4
HRMS (ESI−) calcd for C20H23O2 (M−), 295.16980; found 295.16691. 1H NMR (CD3OD) δ: 1.50 (s, 3H, H-10′), 1.54 (s, 3H, H-8′), 1.84 (s, 3H, H-9′), 1.94 (m, 2H, H-4′), 2.03 (m, 2H, H-5′), 3.67 (d, J = 6.2 Hz, 2H, H-1′), 5.00 (m, 1H, H-6′), 5.14 (m, 1H, H-2′), 6.98 (m, 2H, H-3, H-7), 7.05 (m, 2H, H-4, H-5), 8.05 (m, 1H, H-8).
4.4.4. 1-Geranyl 2,7-DHN (12; product was synthesized from 2,7-DHN and GPP by recombinant NphB)
HRMS (ESI−) calcd for C20H23O2 (M−), 295.16980; found 295.16599. 1H NMR (CD3OD) δ: 1.50 (s, 3H, H-10′), 1.53 (s, 3H, H-8′), 1.85 (s, 3H, H-9′), 1.95 (m, 2H, H-4′), 2.03 (m, 2H, H-5′), 3.62 (d, J = 6.2 Hz, 2H, H-1′), 5.01 (m, 1H, H-6′), 5.15 (m, 1H, H-2′), 6.81 (d, J = 8.9 Hz, 1H, H-6), 6.86 (d, J = 8.9 Hz, 1H, H-3), 7.10 (s, 1H, H-8), 7.42 (d, J = 8.9 Hz, 1H, H-5), 7.55 (d, J = 8.9 Hz, 1H, H-4).
4.4.5. 1,6-Digeranyl 2,7-DHN (13; product was synthesized from 2,7-DHN and GPP by recombinant NphB)
HRMS (ESI−) calcd for C30H39O2 (M−), 431.29500; found 431.29322. 1H NMR (CD3OD) δ: 1.45 (s, 3H, H-10″), 1.47 (s, 3H, H-8″), 1.58 (s, 3H, H-10′), 1.68 (s, 3H, H-8′), 1.69 (s, 3H, H-9″), 1.83 (s, 3H, H-9′), 1.92 (m, 2H, H-4′), 2.00 (m, 2H, H-5′), 2.04 (m, 2H, H-4″), 2.10 (m, 2H, H-5″), 3.59 (m, 2H, H-1″), 3.60 (m, 2H, H-1′), 5.01 (m, 2H, H-6′), 5.38 (m, 2H, H-6″), 6.86 (d, J = 8.3 Hz, 1H, H-3), 7.11 (s, 1H, H-8), 7.35 (m, 1H, H-4), 7.35 (s, 1H, H-5).
4.4.6 7-O-Geranyl naringenin (14; product was synthesized from naringenin and GPP by recombinant NphB)4
HRMS (ESI−) calcd for C25H27O5 (M−), 407.18515; found 407.18732. 1H NMR (DMSO-d6) δ: 1.50 (s, 3H, H-10″), 1.57 (s, 3H, H-8″), 1.63 (s, 3H, H-9″), 1.97 (m, 2H, H-4″), 2.01 (m, 2H, H-5″), 2.66 (dd, J = 17.2, 2.8 Hz, 1H, H-3eq), 3.25 (dd, J = 17.2, 13.0 Hz, 1H, H-3ax), 4.53 (d, J = 6.2 Hz, 2H, H-1″), 5.00 (m, 1H, H-6″), 5.32 (m, 1H, H-2″), 5.42 (dd, J = 13.1, 2.8 Hz, 1H, H-2), 6.01 (s, 1H, H-8), 6.03 (s, 1H, H-6), 6.75 (d, J = 6.9 Hz, 2H, H-3′, H-5′), 7.27 (d, J = 6.9 Hz, 1H, H-2′, H-6′), 9.67 (br, s, C-4′-OH), 12.05 (br, s, C-5-OH). 13C NMR (DMSO-d6) δ: 16.7 (C-9″), 18.1 (C-10″), 26.0 (C-8″), 26.2 (C-5″), 39.5 (C-4″), 42.5 (C-3), 65.7 (C-1″), 79.1 (C-2), 94.8 (C-8), 95.7 (C-6), 103.0 (C-4a), 115.7 (C-3′), 115.7 (C-5′), 119.3 (C-2″), 124.2 (C-6″), 128.9 (C-2′), 128.9 (C-6′), 129.2 (C-1′), 131.7 (C-7″), 141.8 (C-3″), 158.3 (C-4′), 161.0 (C-8a), 163.6 (C-5), 167.2 (C-7), 197.4 (C-4).
4.4.7. 6-Geranyl naringenin (15; product was synthesized from naringenin and GPP by recombinant NphB)4
HRMS (ESI−) calcd for C25H27O5 (M−), 407.18515; found 407.18454. 1H NMR (DMSO-d6) δ: 1.48 (s, 3H, H-10″), 1.55 (s, 3H, H-8″), 1.65 (s, 3H, H-9″), 1.85 (m, 2H, H-4″), 1.94 (m, 2H, H-5″), 2.63 (dd, J = 17.2, 2.8 Hz, 1H, H-3eq), 3.07 (d, J = 6.9 Hz, 2H, H-1″), 3.16 (dd, J = 17.2, 13.1 Hz, 1H, H-3ax), 4.99 (m, 1H, H-6″), 5.08 (m, 1H, H-2″), 5.34 (dd, J = 13.1, 2.8 Hz, 1H, H-2), 5.91 (s, 1H, H-8), 6.75 (d, J = 8.3 Hz, 2H, H-3′, H-5′), 7.26 (d, J = 8.3 Hz, 2H, H-2′, H-6′), 9.64 (br, s, C-4′-OH), 12.39 (s, 1H, C-5-OH). 13C NMR (DMSO-d6) δ: 16.4 (C-9″), 18.0 (C-10″), 21.1 (C-1″), 26.0 (C-8″), 26.7 (C-5″), 39.9 (C-4″), 42.6 (C-3), 78.8 (C-2), 95.0 (C-8), 101.9 (C-4a), 108.1 (C-6), 115.7 (C-3′), 115.7 (C-5′), 123.0 (C-2″), 124.6 (C-6″), 128.8 (C-2′), 128.8 (C-6′), 129.6 (C-1′), 131.2 (C-7″), 134.3 (C-3″), 158.2 (C-4′), 161.0 (C-8a), 161.1 (C-5), 165.4 (C-7), 196.8 (C-4).
4.4.8. 7-O-Geranyl apigenin (16; product was synthesized from apigenin and GPP by recombinant NphB)
HRMS (ESI−) calcd for C25H25O5 (M−), 405.17027; found 405.16686. 1H NMR (DMSO-d6) δ: 1.52 (s, 3H, H-10″), 1.57 (s, 3H, H-8″), 1.69 (s, 3H, H-9″), 2.02 (m, 2H, H-4″), 2.04 (m, 2H, H-5″), 4.63 (d, J = 6.9 Hz, 2H, H-1″), 4.99 (m, 1H, H-6″), 5.17 (t, J = 6.9 Hz, 1H, H-2″), 6.30 (s, 1H, H-3), 6.72 (s, 1H, H-8), 6.76 (s, 1H, H-6), 6.85 (d, J = 8.9 Hz, 2H, H-3′, H-5′), 7.88 (d, J = 8.9 Hz, 2H, H-2′, H-6′), 12.93 (br, 1H, C-5-OH). 13C NMR (DMSO-d6) δ: 16.9 (C-9″), 18.1 (C-10″), 26.0 (C-8″), 26.3 (C-5″), 39.9 (C-4″), 65.9 (C-1″), 93.8 (C-8), 99.0 (C-4a), 103.0 (C-3), 105.1 (C-6), 116.8 (C-3′), 116.8 (C-5′), 119.4 (C-2″), 120.5 (C-1′), 124.3 (C-6″), 129.1 (C-2′), 129.1 (C-6′), 131.6 (C-7″), 141.8 (C-3″), 157.7 (C-4′), 161.7 (C-8a), 163.3 (C-2), 164.8 (C-5), 164.8 (C-7), 182.3 (C-4).
4.4.9. 6-Geranyl apigenin (17; product was synthesized from apigenin and GPP by recombinant NphB)
HRMS (ESI−) calcd for C25H25O5 (M−), 405.17027; found 405.17115. 1H NMR (DMSO-d6) δ: 1.48 (s, 3H, H-10″), 1.54 (s, 3H, H-8″), 1.70 (s, 3H, H-9″), 1.88 (m, 2H, H-4″), 1.96 (m, 2H, H-5″), 3.19 (d, J = 6.9 Hz, 2H, H-1″), 4.99 (m, 1H, H-6″), 5.17 (t, J = 6.9 Hz, 1H, H-2″), 6.44 (s, 1H, H-3), 6.67 (s, 1H, H-8), 6.86 (d, J = 8.2 Hz, 2H, H-3′, H-5′), 7.84 (d, J = 8.2 Hz, 2H, H-2′, H-6′), 13.15 (br, 1H, C-5-OH). 13C NMR (DMSO-d6) δ: 16.5 (C-9″), 18.0 (C-10″), 21.6 (C-1″), 26.0 (C-8″), 26.7 (C-5″), 39.9 (C-4″), 93.9 (C-8), 103.0 (C-3), 103.2 (C-4a), 111.7 (C-6), 116.5 (C-3′), 116.5 (C-5′), 121.7 (C-1′), 122.9 (C-2″), 124.7 (C-6″), 128.8 (C-2′), 128.8 (C-6′), 161.9 (C-2), 131.1 (C-7″), 134.4 (C-3″), 155.8 (C-4′), 158.7 (C-8a), 161.9 (C-5), 163.7 (C-7), 181.9 (C-4).
4.4.10. 7-O-Geranyl genistein (18; product was synthesized from genistein and GPP by recombinant NphB)
HRMS (ESI−) calcd for C25H25O5 (M−), 405.17027; found 405.16781. 1H NMR (DMSO-d6) δ: 1.52 (s, 3H, H-10″), 1.57 (s, 3H, H-8″), 1.68 (s, 3H, H-9″), 2.02 (m, 2H, H-4″), 2.04 (m, 2H, H-5″), 4.63 (d, J = 6.8 Hz, 2H, H-1″), 5.02 (m, 1H, H-6″), 5.39 (m, 1H, H-2″), 6.35 (s, 1H, H-8), 6.60 (s, 1H, H-6), 6.78 (d, J = 7.6 Hz, 2H, H-3′, H-5′), 7.34 (d, J = 7.6 Hz, 2H, H-2′, H-6′), 8.35 (s, 1H, H-2). 13C NMR (DMSO-d6) δ: 16.9 (C-9″), 18.1 (C-10″), 25.9 (C-8″), 26.2 (C-5″), 39.8 (C-4″), 65.9 (C-1″), 93.5 (C-8), 99.1 (C-6), 105.9 (C-4a), 115.7 (C-3′), 115.7 (C-5′), 119.2 (C-2″), 121.5 (C-1′), 123.0 (C-3), 124.2 (C-6″), 130.7 (C-2′), 130.7 (C-6′), 131.7 (C-7″), 142.0 (C-3″), 154.8 (C-2), 158.0 (C-8a), 158.0 (C-4′), 162.3 (C-5), 164.9 (C-7), 180.9 (C-4).
4.4.11. 7-O-Geranyl daidzein (19; product was synthesized from daidzein and GPP by recombinant NphB)
HRMS (ESI−) calcd for C25H25O4 (M−), 389.17528; found 389.17653. 1H NMR (DMSO-d6) δ: 1.58 (s, 3H, H-10″), 1.61 (s, 3H, H-8″), 1.78 (s, 3H, H-9″), 2.09 (m, 2H, H-4″), 2.13 (m, 2H, H-5″), 4.70 (d, J = 6.2 Hz, 2H, H-1″), 5.07 (m, 1H, H-6″), 5.47 (m, 1H, H-2″), 6.83 (d, J = 4.8 Hz, 2H, H-3′, H-5′), 7.02 (s, 1H, H-8), 7.03 (d, J = 13.0 Hz, 1H, H-6), 7.36 (d, J = 4.8 Hz, 2H, H-2′, H-6′), 8.09 (d, J = 13.0 Hz, 1H, H-5), 8.17 (s, 1H, H-2). 13C NMR (DMSO-d6) δ: 14.5 (C-9″), 17.1 (C-10″), 24.6 (C-8″), 26.0 (C-5″), 39.3 (C-4″), 64.7 (C-1″), 93.5 (C-8), 99.1 (C-6), 104.9 (C-4a), 115.7 (C-3′), 115.7 (C-5′), 117.7 (C-5), 118.9 (C-2″), 122.9 (C-3), 123.7 (C-6″), 125.0 (C-1′), 130.2 (C-2′), 130.2 (C-6′), 131.5 (C-7″), 141.9 (C-3″), 153.6 (C-2), 157.5 (C-4′), 158.5 (C-8a), 164 (C-7), 176.7 (C-4).
4.4.12. 8-Geranyl daidzein (20; product was synthesized from daidzein and GPP by recombinant NphB)
HRMS (ESI−) calcd for C25H25O4 (M−), 389.1753; found 389.1794. 1H NMR (CD3OD) δ: 1.57 (s, 3H, H-10″), 1.60 (s, 3H, H-8″), 1.89 (s, 3H, H-9″), 2.04 (m, 2H, H-4″), 2.11 (m, 2H, H-5″), 3.63 (d, J = 6.2 Hz, 2H, H-1″), 5.06 (m, 1H, H-6″), 5.31 (m, 1H, H-2″), 6.90 (d, J = 7.6 Hz, 2H, H-3′, H-5′), 7.02 (d, J = 8.9 Hz, 1H, H-6), 7.43 (d, J = 7.6 Hz, 2H, H-2′, H-6′), 7.98 (d, J = 8.9 Hz, 1H, H-5), 8.27 (s, 1H, H-2). 13C NMR (CD3OD) δ: 15.0 (C-9″), 16.3 (C-10″), 21.4 (C-1″), 24.5 (C-8″), 26.2 (C-5″), 39.4 (C-4″), 114.2 (C-6), 114.9 (C-3′), 114.9 (C-5′), 115.6 (C-8), 116.9 (C-4a), 121.6 (C-2″), 123.0 (C-1′), 123.9 (C-6″), 124.0 (C-5), 124.2 (C-3), 130.1 (C-2′), 130.1 (C-6′), 130.7 (C-7″), 135.2 (C-3″), 153.3 (C-2), 156.2 (C-4′), 157.3 (C-8a), 160.5 (C-7), 177.3 (C-4).
4.4.13. 2-Geranyl olivetol (21; product was synthesized from olivetol and GPP by recombinant NphB)
HRMS (ESI−) calcd for C21H31O2 (M−), 389.17528; found 389.17653. 1H NMR (DMSO-d6) δ: 0.88 (t, J = 6.9 Hz, 3H, H-5′), 1.32 (m, 6H, H-2′, H-3′, H-4′), 1.55 (s, 3H, H-10″), 1.60 (s, 3H, H-8″), 1.71 (s, 3H, H-9″), 1.94 (t, J = 6.8, 7.6 Hz, 2H, H-4″), 2.04 (c, J = 6.8, 6.8, 7.6 Hz, 2H, H-5″), 2.43 (t, J = 7.6 Hz, 2H, H-1′), 3.21 (d, J = 6.2 Hz, 2H, H-1″), 5.04 (m, 2H, H-2″, H-6″), 6.10 (s, 1H, H-6), 6.12 (s, 1H, H-4). 13C NMR (DMSO-d6) δ: 13.1 (C-5′), 15.0 (C-9″), 16.4 (C-10″), 22.4 (C-1″), 23.8 (C-4′), 24.5 (C-8″), 26.4 (C-5″), 30.8 (C-3′), 31.9 (C-2′), 33.0 (C-1′), 39.5 (C-4″), 99.8 (C-4), 107.1 (C-6), 117.5 (C-2), 124.1 (C-2″), 124.8 (C-6″), 130.7 (C-7″), 132.7 (C-3″), 142.8 (C-1), 155.1 (C-5), 155.7 (C-3).
4.4.14. 4-Geranyl olivetol (22; product was synthesized from olivetol and GPP by recombinant NphB)
HRMS (ESI−) calcd for C21H31O2 (M−), 389.17528; found 389.17323. 1H NMR (DMSO-d6) δ: 0.88 (t, J = 6.9 Hz, 3H, H-5′), 1.29 (m, 6H, H-2′, H-3′, H-4′), 1.54 (s, 3H, H-10″), 1.59 (s, 3H, H-8″), 1.73 (s, 3H, H-9″), 1.93 (t, J = 6.8, 7.6 Hz, 2H, H-4″), 2.04 (c, J = 6.8, 7.6, 6.8 Hz, 2H, H-5″), 2.37 (t, J = 7.6 Hz, 2H, H-1′), 3.22 (d, J = 6.9 Hz, 2H, H-1″), 5.05 (m, 1H, H-6″), 5.21 (t, J = 6.9 Hz, 1H, H-2″), 6.11 (s, 2H, H-2, H-6). 13C NMR (DMSO-d6) δ: 13.1 (C-5′), 14.5 (C-9″), 16.4 (C-10″), 21.7 (C-1″), 22.3 (C-4′), 24.5 (C-8″), 26.4 (C-5″), 30.9 (C-3′), 31.3 (C-2′), 35.4 (C-1′), 39.6 (C-4″), 106.5 (C-2), 106.5 (C-6), 112.2 (C-4), 123.7 (C-2″), 124.3 (C-6″), 130.6 (C-7″), 133.2 (C-3″), 141.0 (C-1), 155.6 (C-3), 155.6 (C-5).
4.4.15. 4-Geranyl resveratrol (23; product was synthesized from resveratrol and GPP by recombinant NphB)
HRMS (ESI−) calcd for C24H26O3 (M−), 363.19602; found 363.19137. 1H NMR (DMSO-d6) δ: 1.53 (s, 3H, H-10″), 1.59 (s, 3H, H-8″), 1.73 (s, 3H, H-9), 1.92 (m, 2H, H-4″), 2.02 (m, 2H, H-5″), 3.25 (d, J = 7.6 Hz, 2H, H-1″), 5.20 (m, 2H, H-2″, H-6″), 6.47 (s, 2H, H-2, H-6), 6.73 (d, J = 15.8 Hz, 1H, H-β), 6.75 (d, J = 8.3 Hz, 2H, H-3 ′, H-5′), 6.89 (d, J = 15.8 Hz, 1H, H-α), 7.31 (d, J = 8.3 Hz, 2H, H-2′, H-6′). 13C NMR (DMSO-d6) δ: 15.0 (C-9″), 16.4 (C-10″), 21.9 (C-1″), 24.6 (C-8″), 26.4 (C-5″), 39.6 (C-4″), 104.5 (C-2), 104.5 (C-6), 114.6 (C-4), 115.2 (C-3′), 115.2 (C-5′), 123.2 (C-2″), 124.1 (C-6″), 125.9 (C-α), 127.0 (C-2′), 127.3 (C-β), 127.3 (C-6′), 129.4 (C-1′), 130.9 (C-7″), 133.7 (C-3″), 136.3 (C-1), 155.8 (C-3), 155.8 (C-5), 156.6 (C-4′).
4.4.16. 2,4-Digeranyl resveratrol (24; product was synthesized from resveratrol and GPP by recombinant NphB)
HRMS (ESI−) calcd for C34H43O3 (M−), 499.3212; found 499.3189. 1H NMR (CD3OD) δ: 1.49 (s, 3H, H-10″), 1.54 (s, 6H, H-8″, H-10‴), 1.60 (s, 3H, H-8‴), 1.76 (s, 3H, H-9‴), 1.77 (s, 3H, H-9″), 1.96 (t, J = 6.9 Hz, 4H, H-4″, H-4‴), 2.03 (m, 2H, H-5‴), 2.06 (m, 2H, H-5″), 3.34 (d, J = 6.9 Hz, 2H, H-1‴), 3.39 (d, J = 6.9 Hz, 2H, H-1″), 5.01 (t, J = 6.9 Hz, 1H, H-6″), 5.05 (t, J = 6.9 Hz, 1H, H-6‴), 5.09 (t, J = 5.5, 6.9 Hz, 1H, H-2″), 5.21 (t, J = 6.2, 6.9 Hz, 1H, H-2‴), 6.62 (s, 1H, H-6), 6.72 (d, J = 8.2 Hz, 2H, H-3′, H-5′), 6.76 (d, J = 16.5 Hz, 1H, H-β), 7.06 (d, J = 16.5 Hz, 1H, H-α), 7.27 (d, J = 8.2 Hz, 2H, H-2′, H-6′). 13C NMR (CD3OD) δ: 15.0 (C-9‴), 15.2 (C-9″), 16.4 (C-10″), 16.4 (C-10‴), 22.4 (C-1‴), 24.5 (C-1″),24.5 (C-8″), 24.6 (C-8‴), 26.4 (C-5″), 26.4 (C-5‴),39.5 (C-4‴), 39.6 (C-4″), 103.9 (C-6), 115.1 (C-3′), 115.1 (C-5′), 115.2 (C-4), 118.6 (C-2), 123.0 (C-2‴), 124.0 (C-6″), 124.1 (C-6‴), 124.2 (C-α), 124.6 (C-2″), 127.3 (C-2′, C-6′), 128.4 (C-β), 129.7 (C-1′), 130.8 (C-7″), 130.8 (C-7‴), 133.4 (C-3″), 134.5 (C-3‴), 135.3 (C-1), 152.9 (C-3), 153.4 (C-5), 156.8 (C-4′).
4.4.17. 5-Dimethylallyl 1,6-DHN (25; product was synthesized from 1,6-DHN and DMAPP by recombinant SCO7190)
HRMS (ESI−) calcd for C15H15O2 (M−), 227.10720; found 227.10785. 1H NMR (CD3OD) δ: 1.57 (s, 3H, H-5′), 1.77 (s, 3H, H-4′), 3.56 (d, J = 6.9 Hz, 2H, H-1′), 5.07 (m, 1H, H-2′), 6.58 (d, J = 8.9 Hz, 1H, H-7), 6.59 (d, J = 6.8 Hz, 1H, H-2), 7.16 (dd, J = 8.2, 7.6 Hz, 1H, H-3), 7.18 (d, J = 8.2 Hz, 1H, H-4).
4.4.18. 1-Dimethylallyl 2,7-DHN (26; product was synthesized from 2,7-DHN and DMAPP by recombinant SCO7190)
HRMS (ESI−) calcd for C15H15O2 (M−), 227.10720; found 227.10488. 1H NMR (CD3OD) δ: 1.59 (s, 3H, H-5′), 1.78 (s, 3H, H-4′), 3.48 (d, J = 6.2 Hz, 2H, H-1′), 5.06 (m, 1H, H-2′), 6.86 (d, J = 8.9 Hz, 1H, H-3′), 6.87 (d, J = 8.9 Hz, 1H, H-6), 6.97 (s, 1H, H-8), 7.41 (d, J = 8.9 Hz, 1H, H-5), 7.53 (d, J = 8.9 Hz, 1H, H-4).
4.4.19. 6-Dimethylallyl naringenin (27; product was synthesized from naringenin and DMAPP by recombinant SCO7190)
HRMS (ESI−) calcd for C20H19O5 (M−), 339.12325; found 339.12554. 1H NMR (CD3OD) δ: 1.54 (s, 3H, H-5″), 1.62 (s, 3H, H-4″), 2.58 (dd, J = 13.1, 2.8 Hz, 1H, H-3eq), 3.01 (d, J = 6.9 Hz, 2H, H-1″), 3.09 (dd, J = 17.2, 13.1 Hz, 1H, H-3ax), 5.04 (t, J = 6.2, 6.9 Hz, 1H, H-2″), 5.29 (dd, J = 13.1, 2.8 Hz, 1H, H-2), 5.81 (s, 1H, H-8), 6.73 (d, J = 8.3 Hz, 2H, H-3′, H-5′), 7.25 (d, J = 8.3 Hz, 2H, H-2′, H-6′), 12.36 (s, 1H, C-5-OH). 13C NMR (CD3OD) δ: 18.1 (C-4″), 21.1 (C-5″), 26.0 (C-1″), 42.4 (C-3), 78.7 (C-2), 95.4 (C-8), 108.3 (C-4a), 108.4 (C-6), 115.7 (C-3′), 115.7 (C-5′), 123.3 (C-2″), 128.8 (C-2′), 128.8 (C-6′), 129.7 (C-1′), 130.7 (C-3″), 157.9 (C-4′), 160.9 (C-8a), 161.0 (C-5), 165.4 (C-7), 196.8 (C-4).
4.4.20. 4-Dimethylallyl olivetol (28; product was synthesized from olivetol and DMAPP by recombinant SCO7190)
HRMS (ESI−) calcd for C16H23O2 (M−), 247.16980; found 247.17005. 1H NMR (CD3OD) δ: 0.88 (t, J = 6.9,7.5 Hz, 3H, H-5′), 1.29 (m, 6H, H-2′, H-3′, H-4′), 1.62 (s, 3H, H-5″), 1.72 (s, 3H, H-4″), 2.37 (t, J = 6.9, 7.6 Hz, 2H, H-1′), 3.21 (d, J = 6.8 Hz, 2H, H-1″), 5.18 (m, 2H, H-2″), 6.11 (s, 1H, H-2, H-6).
4.4.21. 2-Dimethylallyl olivetol (29; product was synthesized from olivetol and DMAPP by recombinant SCO7190)
HRMS (ESI−) calcd for C16H23O2 (M−), 247.16980; found 247.17123. 1H NMR (CD3OD) δ: 0.89 (t, J = 6.9 Hz, 3H, H-5′), 1.32 (m, 6H, H-2′, H-3′, H-4′), 1.63 (s, 3H, H-5″), 1.71 (s, 3H, H-4″), 2.42 (m, 2H, H-1′), 3.20 (d, J = 6.8 Hz, 2H, H-1″), 5.03 (m, 2H, H-2″), 6.09 (s, 1H, H-6), 6.11 (s, 1H, H-4).
4.4.22. 2-Dimethylallyl resveratrol (30; product was synthesized from resveratrol and DMAPP by recombinant SCO7190)
1H NMR (CD3OD) δ: 1.64 (s, 3H, H-5″), 1.78 (s, 3H, H-4″), 3.34 (d, J = 7.6 Hz, 2H, H-1″), 5.08 (m, 1H, H-2″), 6.20 (s, 1H, H-4), 6.53 (s, 1H, H-6), 6.74 (d, J = 8.3 Hz, 2H, H-3′, H-5′), 6.80 (d, J = 16.5 Hz, 1H, H-β), 7.10 (d, J = 16.5 Hz, 1H, H-α), 7.29 (d, J = 8.3 Hz, 2H, H-2′, H-6′).
Supplementary Material
Acknowledgments
This work was supported by METI and Grant-in-Aid for Scien-tific Research on Priority Areas ‘Applied Genome’ (18018008) from MEXT, Japan (T. Kuzuyama) and HHMI (J.P.N.).
Footnotes
Supplemental figure 1 and supplemental figure 2 can be found in the Supporting Information. Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmc.2008.07.052.
References and notes
- 1.Di Pietro A, Conseil G, Perez-Victoria JM, Dayan G, Baubichon-Cortay H, Trompier D, Steinfels E, Jault JM, de Wet H, Maitrejean M, Comte G, Boumendjel A, Mariotte AM, Dumontet C, McIntosh DB, Goffeau A, Castanys S, Gamarro F, Barron D. Cell Mol Life Sci. 2002;59:307. doi: 10.1007/s00018-002-8424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed-Belkacem A, Pozza A, Munoz-Martinez F, Bates SE, Castanys S, Gamarro F, Di Pietro A, Perez-Victoria JM. Cancer Res. 2005;65:4852. doi: 10.1158/0008-5472.CAN-04-1817. [DOI] [PubMed] [Google Scholar]
- 3.Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G. Curr Med Chem. 2005;12:717. doi: 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- 4.Kuzuyama T, Noel JP, Richard SB. Nature. 2005;435:983. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin-ya K, Imai S, Furihata K, Hayakawa Y, Kato Y, Vanduyne GD, Clardy J, Seto H. J Antibiot. 1990;43:444. doi: 10.7164/antibiotics.43.444. [DOI] [PubMed] [Google Scholar]
- 6.Shin-ya K, Furihata K, Hayakawa Y, Seto H. Tetrahedron Lett. 1990;31:6025. [Google Scholar]
- 7.Seto H, Watanabe H, Furihata K. Tetrahedron Lett. 1996;37:7979. [Google Scholar]
- 8.Tello M, Kuzuyama T, Heide L, Noel JP, Richard SB. Cell Mol Life Sci. 2008;65:1459. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui G, Li X, Merz KM., Jr Biochemistry. 2007;46:1303. doi: 10.1021/bi062076z. [DOI] [PubMed] [Google Scholar]
- 10.Haagen Y, Unsöld I, Westrich L, Gust B, Richard SB, Noel JP, Heide L. FEBS Lett. 2007;581:2889. doi: 10.1016/j.febslet.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.