Abstract
We have used the orthogonal carbodiimide condensation and Copper-catalyzed azide-alkyne “click” cycloaddition (CuAAC) reactions to prepare self-assembled monolayers that present distinct peptides to stem cells in a bio-inert background. The approach involved first forming mixed SAMs with three components: i) an azide-terminated hexaethylene glycol alkanethiolate (HS---EG6---N3), ii) a carboxylate-terminated hexaethylene glycol alkanethiolate (HS---EG6---COOH), and iii) a triethylene glycol alkanethiolate (HS---EG3). An acetylene-bearing peptide and an amine-terminated peptide were then immobilized to these substrates using a “click” CuAAC reaction and a carbodiimide condensation reaction, respectively. Polarization-modulated infrared reflectance-absorbance spectroscopic analysis demonstrated formation of well-ordered, close-packed SAMs, chemoselective conjugation of amine-terminated peptides to surface carboxylate groups, and subsequent conjugation of acetylene-terminated peptides to the azide groups on SAMs. Varying the mole fraction of HS---EG6---N3, HS---EG6---COOH, and HS---EG3 during SAM formation allowed for control over the densities of each peptide on the substrate. Substrates presenting varying surface densities of RGESP (a non-functional peptide), RGDSP (a cell adhesion peptide) or TYRSRKY (a heparin/heparan sulfate-binding peptide) were then used to characterize the relationship between peptide surface density and human mesenchymal stem cell (hMSC) adhesion. Results demonstrate that RGESP does not influence RGDSP-mediated adhesion of hMSCs, which indicates that a second peptide with distinct bio-activity can be immobilized alongside RGDSP to characterize the influence of two peptides on hMSC behavior. Our results also demonstrate that RGDSP and TYRSRKY act synergistically to promote hMSC adhesion in the absence of serum. Interestingly, heparin sequestered by TYRSRKY inhibits cell adhesion on substrates presenting RGDSP = 0.1% and > 0.1% TYRSRKY or RGDSP = 1% and > 0.5% TYRSRKY. Taken together, these results indicate that two peptides can be controllably presented to stem cells on the same otherwise bio-inert SAM substrate, and that multiple, distinct extracellular moieties act in concert to regulate hMSC adhesion.
Introduction
Self-assembled monolayers (SAMs) provide chemically well-defined substrates that can be tailored for specific biochemical applications, such as cell culture1, 2, characterization of enzyme reaction kinetics3, and biosensing4. SAMs are particularly advantageous as substrates for cell culture, as standard culture formats such as protein-coated tissue culture-treated polystyrene substrates offer limited control over a cell's interaction with its extracellular matrix (ECM). The inherent complexity and multivalency associated with cell-ECM interactions5, 6 emphasizes the need to develop well-defined substrates, upon which cells interact with controllable densities of specific ligands derived from the native ECM. Toward that end, we and others have recently used bio-inert SAMs as a platform to immobilize a single ECM-derived peptide and characterize its effect on cell adhesion7-10. These previous studies have resulted in well-defined correlations between peptide density and cell adhesion measures, including attachment, spreading, and focal adhesion density.
A key SAM property that has facilitated characterization of cell adhesion in previous studies is the presence of reactive moieties, which allow for covalent immobilization of peptides onto the substrate. To date, multiple covalent mechanisms have been used to immobilize polypeptides onto SAMs including carbodiimide condensation11 and Michael-type addition12. An advantage of these mechanisms is that they rely on functional groups common to peptides. However, the reliance on these functional groups introduces a pragmatic limitation: it is difficult to immobilize multiple, distinct peptides in a controllable manner on a single substrate due to cross-reactivity. Thus, it is difficult to characterize the concerted influence of multiple ligands on cell behavior in a well-defined environment.
Recent studies have addressed the inability to immobilize multiple, distinct biomolecules on a single SAM by using complimentary DNA interactions to immobilize oligonucleotides and oligonucleotide-bearing antibodies on a SAM substrate13. Additionally, we recently demonstrated that SAMs formed from a thiol-terminated oligonucletoide and a carboxylic acid terminated alkanethiolate allow for DNA immobilization and peptide conjugation on the same SAM substrate14. While effective, these previous approaches are limited to biomolecules bearing an oligonucleotide sequence, which introduces multiple synthetic challenges including limited control over number or location of oligonucleotide subunits conjugated onto polypeptides presenting multiple reactive moities (e.g. COOH or NH2), as well as the potential for changes to polypeptide bio-activity as a result of oligonucleotide conjugation. Additionally, the susceptibility of DNA and RNA to nuclease-mediated degradation introduces limitations to the efficacy of oligonucleotide-presenting materials in cell culture applications. Based on these limitations, more recent efforts have focused on strategies that rely solely on covalent mechanisms. For example, Brozik and co-workers developed a method to immobilize two distinct biomolecules on a SAM by electrochemically introducing a functional group at precise spatial locations before each conjugation step15. However, the use of a single covalent mechanism does not directly address the limitation of cross-reactivity, and as such, only allows for immobilization of a single biomolecule within each spatial location on the SAM. Therefore, there remains a need to develop substrates that can present multiple, distinct ligands to cells to address the complexity associated with cell-ECM interactions.
This paper provides the first account of orthogonal, covalent immobilization of two distinct biomolecules throughout a SAM substrate that is otherwise bio-inert. Specifically, we demonstrate step-wise conjugation of amine- and acetylene-terminated biomolecules to mixed SAMs presenting carboxylate and azide functionalities in an otherwise bio-inert oligo(ethylene glycol) (OEG) background. The low reactivity of the azide group under most common reaction conditions allows for chemoselective activation of carboxylate groups and conjugation of amine-terminated biomolecules via carbodiimide condensation. Subsequently, unreacted azide groups on the same substrate are available for conjugation of acetylene-terminated biomolecules via Copper(I)-catalyzed azide-alkyne “click” cycloaddition (CuAAC). Importantly, this approach is likely to be applicable to any biomolecule combinations that can be modified to include a primary amine and an acetylene, and may therefore be broadly applicable for biomolecule immobilization.
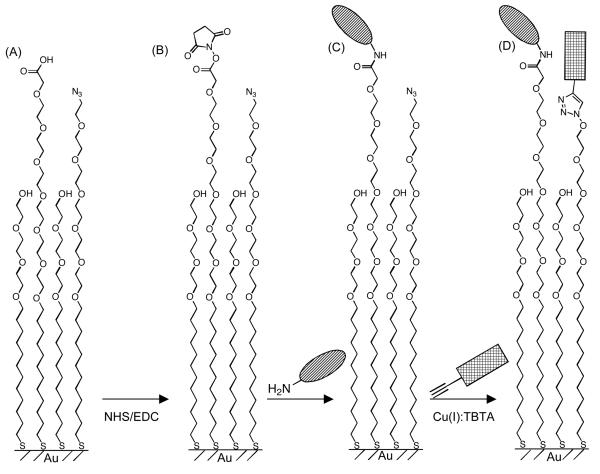
Design Rationale
We generated substrates presenting two functionally distinct peptides at co-varying molar ratios. First, ternary mixed SAMs presenting azide and carboxylate groups were prepared by incubating gold substrates overnight in an ethanolic solution containing a tri(ethylene glycol) alkanethiolate (HS---EG3), an azide-terminated hexa(ethylene glycol) alkanethiolate (HS---EG6---N3), and a carboxylate-terminated hexa(ethylene glycol) alkanethiolate (HS---EG6---COOH) (Fig. 1A). Next, SAM carboxylate groups were converted to NHS-esters (Fig. 1B), which were subsequently reacted with an amine-terminated peptide, resulting in peptide immobilization on the SAM (Fig 1C). Finally, an acetylene-terminated peptide was conjugated to SAM azide groups via a ‘click’ cycloaddition reaction (Fig. 1D). These substrates were then used to characterize human mesenchymal stem cell (hMSC) adhesion as a function of peptide surface density. Specifically, cell adhesion was first characterized on substrates presenting an integrin-binding ligand, RGDSP, and a non-functional mutant ligand, RGESP. Here, RGDSP was chosen due to the well-defined correlation between cell adhesion and the density of RGDSP, as we and others have demonstrated in recent studies7-10, which allows for direct comparison of cell adhesion on the substrates described herein to more traditional SAM-based cell culture substrates. Additionally, cell adhesion was characterized on substrates presenting RGDSP and a proteoglycan-binding peptide, TYRSRKY. TYRSRKY was chosen based on previous results from Park and co-workers demonstrating cell surface heparin sulfate proteoglycan-mediated adhesion of hMSCs onto poly(lysine) coated substrates presenting TYRSRKY16.
Figure 1.
(A) Schematic representation of ternary SAMs presenting orthogonally reactive carboxylate and azide moieties immediately after SAM formation, (B) after NHS activation of carboxylate groups, (C) after conjugating an amine-terminated peptide via carbodiimide condensation, and (D) after ‘click’ CuAAC between surface azide moieties and an alkyne-bearing peptide.
Experimental
Materials and Reagents
Gold substrates (5nm Cr, 100nm Au or 2nm Ti, 10nm Au) were from Evaporated Metal Films (Ithaca, NY). 11-tri(ethylene glycol)-undecane-1-thiol (HS---EG3), Piperidine, dimethylformamide (DMF), triisoproylsilane (TIPS), acetone, 99.999% cuprous bromide (CuBr), dimethylsulfoxide (DMSO), Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), and sodium ascorbate (Na-Asc) were from Sigma-Aldrich (St. Louis, MO). 11-carboxylic acid-hexa(ethylene glycol)-undecane-1-thiol (HS---EG6---COOH) and 11-azidohexa(ethylene glycol)-undecane-1-thiol (HS---EG6---N3) was purchased from Prochimia (Sopot, Poland). Fmoc-protected amino acids and Rink amide MBHA peptide synthesis resin were from NovaBiochem (San Diego, CA). Hydroxybenzotriazole (HOBt) was from Advanced Chemtech (Louisville, KY). Diisopropylcarbodiimide (DIC) and Fmoc-(R)-3-amino-5-hexynoic acid were from Anaspec (San Jose, CA). Trifluoroacetic acid (TFA) and diethyl ether were from Fisher Scientific (Fairlawn, NJ). Absolute ethanol was from AAPER Alcohol and Chemical Co. (Shelbyville, KY). Human mesenchymal stem cells (hMSCs) were from Cambrex (North Brunswick, NJ). 1x minimum essential medium, alpha was from CellGro (Mannassas, VA). MSC-qualified fetal bovine serum was from Invitrogen (Carlsbad, CA). 0.05% Trypsin and penicillin/streptomycin were from Hyclone (Logan, UT). Actin cytoskeleton staining kit and FITC-conjugated secondary antibody were from Chemicon (Billerica, MA).
Peptide synthesis
Peptides were synthesized using standard Fmoc solid phase peptide synthesis on a 316c automated peptide synthesizer (CSbio, Menlo Park, CA). Rink amide MBHA resin was used as the solid phase, and HOBt and DIC were used for amino acid activation and coupling. After coupling the final amino acid, incubation of resin in TFA, TIPS, and deionized (DI) water (95:2.5:2.5) for 4 hours released the peptide from the resin and removed protecting groups. The peptide was then extracted from the TFA/TIPS/H2O cocktail by precipitation with cold diethyl ether. Lyophilized peptides were analyzed on a Bruker Reflex II MALDI-TOF mass spectrometer (Billerica, MA) using dihydroxybenzoic acid (DHB) (10mg/mL) as matrix in acetonitrile:DI water (7:3).
SAM formation
Gold substrates were cut, sonicated in ethanol for 3 minutes, washed with ethanol, and dried under a stream of nitrogen prior to monolayer formation. Monolayers were formed by incubating clean gold substrates in an ethanolic solution of HS---EG3, HS---EG6---N3 and HS---EG6---COOH at various molar ratios (2mM total thiol concentration) overnight. After monolayer formation, gold substrates were removed from the ethanolic solution, washed with ethanol, and dried under a stream of nitrogen.
Peptide immobilization on SAMs
Immediately after SAM formation, SAM substrates were immersed in an aqueous solution containing 100 mM N-Hydroxysuccinimide (NHS) and 250 mM 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) for 10 minutes to convert the surface carboxylate groups to amine-reactive NHS-esters. After 10 minutes, the substrates were washed briefly with DI H2O and ethanol and dried under a stream of nitrogen. NHS-ester-terminated SAMs were then incubated in a 1x PBS solution containing 500 mM amine-terminated RGESP or TYRSRKY (pH 7.5) for 60 minutes. After 60 minutes, gold substrates were washed sequentially with DI water, 0.1% sodium dodecyl sulfate in water, DI water, and ethanol, followed by drying under a stream of nitrogen. CuBr and Na-Asc were dissolved in DMSO at a concentration of 2mM by sonicating for 10 minutes. TBTA was then dissolved in this solution at a concentration of 2mM by sonicating for an additional 10 minutes. Lyophilized acetylene-bearing RGDSP was dissolved in HEPES (0.1 M, pH 8.5) to achieve a peptide concentration of 2mM. The DMSO solution containing CuBr, Na-Asc, and TBTA and the HEPES solution containing RGDSP were then mixed at a 1:1 ratio by vortexing, followed by sonication for 10 minutes. Azide-terminated gold substrates were immersed in this solution and allowed to incubate at room temperature for 60 minutes. At the reaction endpoint, gold substrates were washed sequentially with DI water, 0.1% sodium dodecyl sulfate in water, DI water, and ethanol, followed by drying under a stream of nitrogen.
PM-IRRAS analysis of SAMs
Infrared spectra of SAMs on gold films were recorded using a Nicolet Magna-IR 860 FT-IR spectrometer with photoelastic modulator (PEM-90, Hinds Instruments, Hillsboro, OR), synchronous sampling demodulator (SSD-100, GWC Technologies, Madison, WI), and a liquid nitrogen-cooled mercury cadmium telluride detector. All spectra were obtained at an incident angle of 83° with modulation centered at 1500cm−1 and 2500cm−1. For each sample, 500 scans were taken at a resolution of 4 cm−1 per modulation center. Data were collected as differential reflectance vs. wave number.
Binding of serum-derived heparin on SAMs
SAMs presenting 1% TYRSRKY or 1% scrambled, non-functional peptide SKTYYRR were prepared using previously described methods. Briefly, SAMs comprised of 1% HS---EG6---COOH and 99% HS---EG3 were immersed in an aqueous solution of 100 mM NHS/250 mM EDC for 10 minutes, followed by incubation in a 1x PBS (pH 7.4) solution containing 500 mM TYRSRKY or SKTYYRR. Immediately following the peptide immobilization steps, SAMs were incubated in a 50:50 (v/v) solution of 1x PBS (pH 7.4) and FBS for 20 minutes. After the serum incubation step, SAMs were rinsed briefly with DI H2O and were dried under a stream of nitrogen. The molecular composition of biomolecules bound on the SAM was then analyzed using PM-IRRAS.
hMSC Adhesion
To maintain multipotency, hMSCs were expanded at low density on tissue culture treated polystyrene plates. At passage 6, cells were harvested from the plate, suspended in medium supplemented with 10% fetal bovine serum, and counted using a hemacytometer. Cells were collected as a pellet by centrifugation at 1100 rpm for 5 minutes, the media was decanted off of the pellet, and the cells were suspended in fresh αMEM at a density of 20,000 cells/250 μL. SAM preparation and peptide conjugation were performed using the protocols described above. Immediately after peptide conjugation, SAMs were placed into 1 mL of 1x PBS (pH 7.4) in a 12-well tissue culture plate to prevent degradation of the monolayer due to air oxidation. PBS was aspirated from the wells and replaced with 1.25 mL of αMEM, followed by addition of 250 μL of the cell suspension directly over the SAM substrate in each well. Plates were then gently rocked for 10 seconds to evenly distribute cells over the substrate surface. Substrates were then incubated for a specified time frame (12 hrs for RGESP/RGDSP SAMs, 4 hrs for RGDSP/TYRSRKY) in a humid environment at 37°C, 5% CO2 to allow hMSC attachment. At the end of the attachment period, the hMSC growth media was aspirated from the well and the substrates were gently washed with sterile 1x PBS to remove any loosely bound cells. The 1x PBS solution was then replaced with fresh medium. Brightfield photomicrographs of cells were then collected using an Olympus IX51 inverted microscope.
Immunocytochemistry of hMSC cytoskeleton
hMSCs were seeded on the SAMs as described previously. After washing away loosely bound cells using 1x PBS, cytoskeletal immunostaining of hMSCs was performed by following the protocol supplied by the manufacturer. Briefly, a 4% paraformaldehyde solution in 1x PBS was added to the wells for 15 minutes to fix the cells, followed by a 5 minute incubation in a 1x PBS solution containing 0.05% Tween-20 to permeabilize the cells. Wells were subsequently blocked to prevent non-specific antibody adsorption using a 1x PBS solution containing 0.1 wt% bovine serum albumin. After blocking, a 1x PBS solution containing an anti-Vinculin primary antibody was added to each well and allowed to incubate at room temperature for 60 minutes. The wells were then washed gently three times using a 1x PBS solution containing 0.1 wt% bovine serum albumin. Immediately after washing, a 1x PBS solution containing a fluorescein-tagged mouse anti-human IgG secondary antibody and a TRITC-tagged anti-Phalloidin antibody was added to each well and allowed to incubate at room temperature for 45 minutes. Substrates were then washed using the method described previously. Cytoskeletal staining was analyzed using an Olympus IX51 inverted epifluorescent microscope equipped with FITC and TRITC filter cube sets.
Results and Discussion
SAM Formation and Sequential Peptide Immobilization
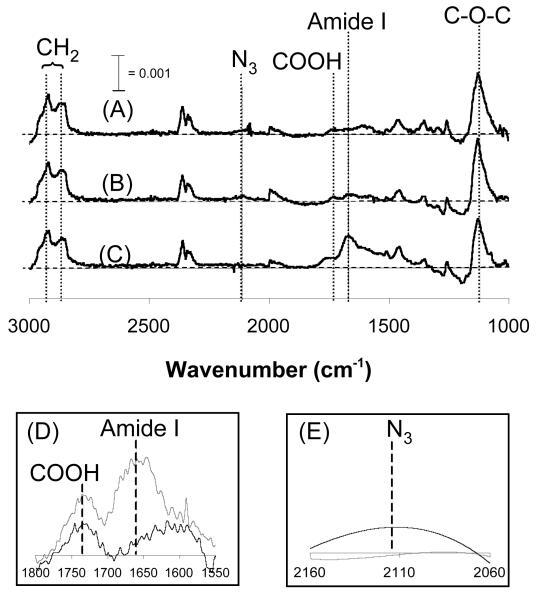
A PM-IRRAS spectrum collected immediately after SAM formation (Fig. 2A) demonstrated a well-ordered, close-packed monolayer presenting azide and carboxylate moieties. Specifically, peaks corresponding to the methylene symmetric and asymmetric stretch (λ = 2850 and 2920 cm−1, respectively), the C-O-C of OEG (λ = 1130 cm−1), the azide moiety (λ = 2110 cm−1), and the carbonyl stretch of the carboxylate moiety (λ = 1730 cm−1) are located at wavenumbers consistent with previously published IR spectra collected from well-ordered SAMs7, 17, 18.
Figure 2.
(A) PM-IRRAS analysis of a 5% HS--EG6---COOH, 5% HS---EG6---N3, 90% HS---EG3 SAM immediately after SAM formation, (B) after conjugating an RGESP via carbodiimide condensation, and (C) after conjugating RGDSP via ‘click’ CuAAC. (D) PM-IRRAS spectrum centered around λ = 1666 cm−1 before ( ) and after (
) and after ( ) carbodiimide condensation18. (E) PM-IRRAS spectrum centered around λ = 2110 cm−1 before (
) carbodiimide condensation18. (E) PM-IRRAS spectrum centered around λ = 2110 cm−1 before ( ) and after (
) and after ( ) CuAAC. Raw data was smoothed using a moving average with a period of 10.
) CuAAC. Raw data was smoothed using a moving average with a period of 10.
Comparison of the PM-IRRAS spectrum collected after SAM formation (Fig. 2A) with the spectrum collected after RGESP conjugation (Fig. 2B, D) demonstrated chemoselective conjugation of RGESP to the carboxylate moiety. Specifically, the emergence of the amide I peak (λ = 1666 cm−1)18 (Fig. 2B, D) indicated that RGESP was present on the substrate at the end of the 60-minute reaction19. Additionally, the remaining presence of the azide peak (λ = 2110 cm−1) (Fig. 2A, B) in both spectra indicated that no chemical transformation of the azide group had occurred, demonstrating that conjugation of amine-terminated RGESP was chemoselective to surface carboxylate groups.
Comparison of the PM-IRRAS spectrum collected after RGESP conjugation (Fig. 2B) with the spectrum collected after RGESP and RGDSP conjugation (Fig. 2C, E) demonstrated that acetylene-bearing RGDSP reacted with surface azide groups after RGESP conjugation. In particular, the total absorbance of the amide I peak (λ = 1666 cm−1) (Fig. 2C) increased, while the peak corresponding to the azide moiety (λ = 2110 cm−1) (Fig. 2C, E) was absent in the spectrum collected after RGDSP conjugation, similar to IR spectra previously collected from binary SAMs formed from HS---EG6---N3 and HS---EG37. Taken together, our results demonstrate that two distinct peptides can be conjugated in a controllable manner to SAMs presenting orthogonally reactive moieties through the immobilization approach outlined in Figure 1.
Correlation Between Reactive Moiety Density and Peptide Density
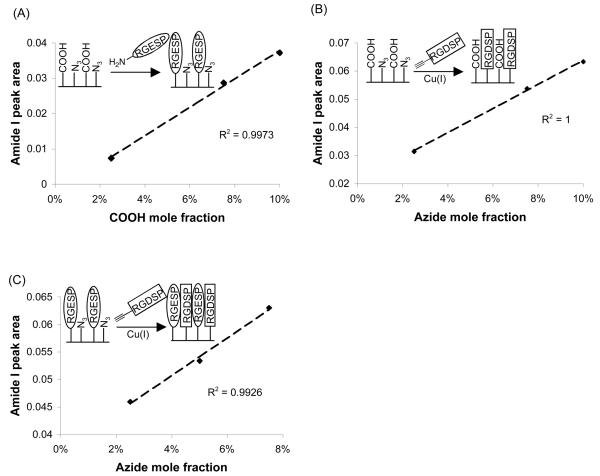
A plot of the COOH mole fraction in ethanol during SAM formation versus the amide I peak area after conjugation of RGESP to COOH demonstrated a linear correlation (Fig. 3A). Additionally, a plot of the N3 mole fraction in ethanol during SAM formation versus the amide I peak area after conjugation of RGDSP to N3 demonstrated a linear correlation (Fig. 3B). The observed linear correlations indicated that both the carbodiimide condensation reaction and click cycloaddition proceeded with similarly high efficiency at each functional group surface density studied20. Importantly, a plot of the N3 mole fraction in ethanol during SAM formation versus the amide I peak area after RGDSP conjugation via CuAAC on RGESP-presenting SAMs also demonstrated a linear correlation (Fig. 3C). This result indicated that the presence of RGESP on the substrate does not inhibit the nearly quantitative reaction previously observed between acetylene-terminated RGDSP and surface azide groups (Fig. 3B & reference 7).
Figure 3.
(A) Correlation between the mole fraction of HS---EG6---COOH in ethanol during SAM formation and the area under the amide I peak (λ = 1666 cm−1) after coupling RGESP via carbodiimide condensation. (B) Correlation between the mole fraction of HS---EG6---N3 in ethanol during SAM formation and the area under the amide I peak (λ = 1666 cm−1) after coupling RGDSP via CuAAC. (C) Correlation between the HS---EG6---N3 mole fraction in ethanol and the area under the amide I peak after RGESP conjugation via carbodiimide condensation and RGDSP conjugation via CuAAC. For (A) and (B), where χ equals mole fraction in solution during SAM formation, χCOOH + χN3 = 0.1, χEG3 = 0.9 during SAM formation. For (C), χCOOH = 0.025, χN3 = 0.025, 0.05, 0.075, χEG3 = 1 − (χCOOH + χN3).
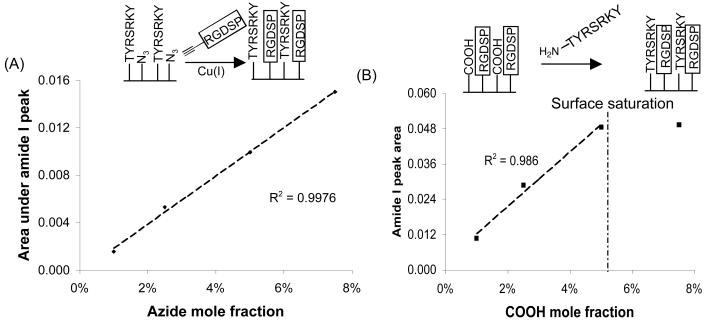
Interestingly, similar trends were observed after immobilization of amine-terminated TYRSRKY and acetylene-bearing RGDSP to SAM COOH and N3 groups, respectively (Fig. 4). Specifically, when the surface density of TYRSRKY was maintained at 2.5% of total alkanethiolate and the surface density of N3 was varied from 1-7.5% of total alkanethiolate, a plot of N3 mole fraction versus the area under the amide I peak after RGDSP immobilization provided a linear correlation (Fig. 4A). Moreover, when the surface density of RGDSP was maintained at 2.5% of total alkanethiolate and the surface density of COOH was varied from 1-7.5% of total alkanethiolate, a plot of COOH mole fraction versus the area under the amide I peak after TYRSRKY immobilization provided a linear correlation over the range of 1-5% COOH, with surface saturation observed between 5-7.5% COOH (Fig. 4B)21. Taken together, our results demonstrate that the surface density of distinct peptides on a SAM can be controlled by varying the mole fraction of alkanethiolates bearing orthogonally-reactive terminal groups.
Figure 4.
(A) Correlation between the mole fraction of HS---EG6---N3 in ethanol during SAM formation and the area under the amide I peak (λ = 1666 cm−1) after coupling RGDSP via CuAAC. (B) Correlation between the mole fraction of HS---EG6---COOH in ethanol during SAM formation and the area under the amide I peak (λ = 1666 cm−1) after coupling TYRSRKY via carbodiimide condensation. For (A), χCOOH = 0.025, χN3 = 0.01, 0.025, 0.05, 0.075, χEG3 = 1 − (χCOOH + χN3). For (B), χN3 = 0.025, χCOOH = 0.01, 0.025, 0.05, 0.075, χEG3 = 1 − (χCOOH + χN3).
Binding of serum-derived heparin proteoglycans on TYRSRKY SAMs
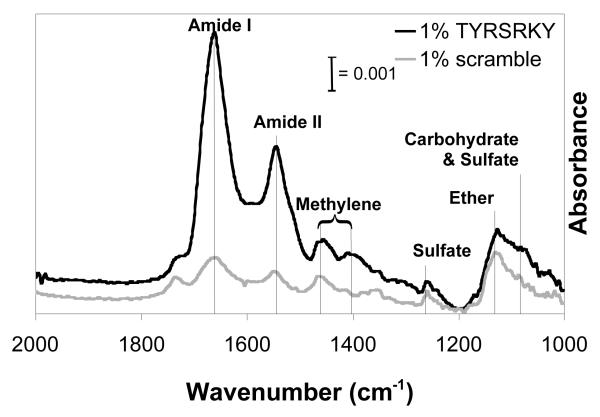
Park and co-workers have recently demonstrated that the peptide sequence TYRSRKY, derived from the heparin/heparan sulfate-binding domain of FGF-2, binds specifically to heparin when immobilized on a solid substrate16. Here we characterized binding of mast cell-derived heparin proteoglycans present in FBS onto SAMs presenting 1% TYRSRKY or 1% scrambled, non-functional peptide SKTYYRR using PM-IRRAS. IR spectra collected from 1% TYRSRKY SAMs after incubation in a 50% FBS solution demonstrate a significant increase in amide I (λ = 1666 cm−1), amide II (λ = 1550 cm−1), methylene (λ = 1460, 1400 cm−1), sulfate (λ = 1260, 1080 cm−1), and carbohydrate (λ = 1100 cm−1) absorbance when compared to 1% SKTYYRR SAMs after incubation in a 50% FBS solution (Fig. 5). The increase in absorbance due to amide I/II content is consistent with IR spectra previously collected from protein monolayers22, and herein is attributed to the heparin proteoglycan protein core. Additionally, the increase in absorbance due to sulfate and carbohydrate groups is at wavenumbers consistent with IR spectra previously collected from aqueous solutions of heparin23, and herein is attributed to heparin glycosaminoglycans. Taken together, these results indicate that SAMs presenting TYRSRKY sequester heparin proteoglycans from complex biomolecule mixtures, such as FBS. Moreover, this result suggests that SAMs presenting 1% TYRSRKY may allow for characterization of the influence of sequestered heparin proteoglycans on hMSC behavior.
Figure 5.
PM-IRRAS analysis of 1% TYRSRKY( ) or 1% scrambled peptide (
) or 1% scrambled peptide ( ) SAMs after immersion in 50% fetal bovine serum for 20 minutes. λamide I= 1666 cm−1, λamide II = 1550 cm−1, λmethylene = 1460, 1400 cm−1, λsulfate = 1260, 1080 cm−1, λether = 1130 cm−1, λcarbohydrate = 1100 cm−1.
) SAMs after immersion in 50% fetal bovine serum for 20 minutes. λamide I= 1666 cm−1, λamide II = 1550 cm−1, λmethylene = 1460, 1400 cm−1, λsulfate = 1260, 1080 cm−1, λether = 1130 cm−1, λcarbohydrate = 1100 cm−1.
hMSC adhesion onto SAM presenting an integrin-binding ligand and a nonfunctional ligand
We have previously demonstrated that RGDSP surface density dictates hMSC adhesion, spreading, and focal adhesion complex formation on SAMs7, 10. Here, the behavior of hMSCs on RGESP- and RGDSP-presenting SAMs was explored to demonstrate the applicability of orthogonally-reactive SAMs as cell culture substrates in a well-defined model system. Our results demonstrated that a significant number of hMSCs were present on all SAMs presenting RGDSP (i.e. RGDSP = 0.0001, 0.001, or 0.01), but were absent on the SAM presenting RGESP alone (i.e. RGDSP = 0) (Fig. 6B). This dependence of hMSC adhesion on RGDSP demonstrates that the underlying substrates are resistant to cell attachment, an important characteristic of chemically well-defined cell culture substrates.
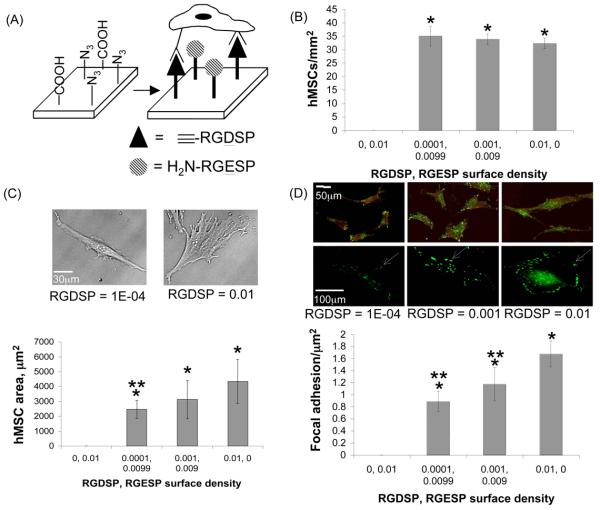
Figure 6.
hMSC adhesion on SAMs presenting RGDSP and RGESP at co-varying molar ratios. (A) Schematic representation of hMSC integrin receptors with SAM-immobilized RGDSP. (B) Quantification of hMSC number per unit area, (C) projected hMSC area, and (D) focal adhesion complex formation on ternary SAMs presenting various surface densities of RGDSP (conjugated to HS---EG6---N3) and RGESP (conjugated to HS---EG6---COOH) after overnight attachment. χCOOH + χN3 = 0.01, χEG3 = 0.99, where χ equals mole fraction in ethanol during SAM formation. * denotes significant difference compared to RGDSP = 0, ** denotes significant difference compared to RGDSP = 0.01 (p < 0.05). White arrow indicates location of a punctate focal adhesion complex within a single cell.
Analysis of projected cell area on SAMs presenting different RGDSP and RGESP surface densities demonstrated that the extent of hMSC spreading is dependent on RGDSP surface density (Fig. 6C), as expected. Specifically, hMSCs on surfaces presenting a low density of RGDSP adopt a polarized, spindle-shaped morphology (RGDSP = 0.0001), while hMSCs on surfaces presenting higher RGDSP densities adopt a more well-spread morphology24 (RGDSP = 0.001 or 0.01). Quantification of focal adhesion complexes also demonstrated a direct correlation between the number of focal adhesion complexes and the surface density of RGDSP (Fig. 6D). The observed correlation between hMSC adhesion measures – projected cell area and focal adhesion density - and RGDSP surface density is consistent with our previous results7, 10. Therefore, the co-immobilized RGESP on SAMs does not influence RGDSP-dependent hMSC adhesion.
hMSC adhesion onto SAMs presenting an integrin-binding ligand and a proteoglycan-binding ligand
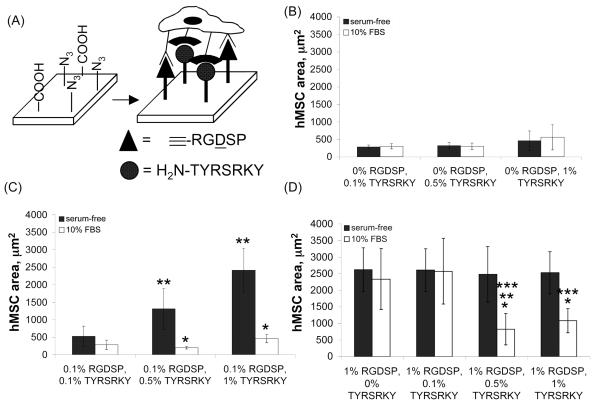
Recently, Park and co-workers have demonstrated that a peptide, TYRSRKY, derived from the heparin/heparan sulfate-binding domain of FGF-2 promotes hMSC adhesion in a glycosaminoglycan-dependent manner when immobilized on poly(lysine) coated substrates16. Moreover, Goetinck and co-workers have recently demonstrated that integrin receptors and the cell-surface proteoglycan syndecan-4 work in concert to promote cell spreading and focal adhesion complex formation on fibronectin-coated substrates25. Here, we characterized the influence of an integrin-binding ligand, RGDSP, and a proteoglycan-binding ligand, TYRSRKY, on hMSC adhesion. Analysis of projected cell area of hMSCs on SAMs presenting 0.01-1.0% TYRSRKY (Fig. 7B) demonstrated that hMSCs attach to the substrates, but adopt a rounded morphology in the presence or absence of serum. Interestingly, analysis of the projected cell area of hMSCs on SAMs presenting 0.1% RGDSP and 0.01-1.0% TYRSRKY in the absence of serum (Fig. 7C) demonstrated that a low TYRSRKY surface density promoted a rounded hMSCs morphology, whereas a high TYRSRKY density promoted a well-spread morphology24. The rounded morphology observed on SAMs presenting 0.1% RGDSP and 0.1% TYRSRKY is significantly different from the spread hMSC morphology we have previously observed on 0.1% RGDSP7 or 0.1% RGDSP, 0.9% RGESP SAMs (Fig. 6C). Integrin-mediated cell spreading is dependent on clustering of ligand-integrin subunits, typically referred to as avidity26, and we have previously observed this phenomenon on substrates presenting RGDSP at surface densities > 0.1% (Fig. 6C and reference 7). The decreased hMSC spreading at RGDSP = 0.1% and TYRSRKY = 0.1% (Fig. 7C) suggests that simultaneous co-localization of integrin receptors and cell-surface proteoglycans at the cell-material interface may inhibit ligand-integrin avidity and, in turn, decrease the extent of hMSC spreading. Moreover, the increased extent of hMSC spreading with increasing TYRSRKY surface density (Fig. 7C) suggests that above a certain threshold of proteoglycan-material binding, the limitation to hMSC spreading mediated by decreased integrin-ligand avidity is overcome by the increased extent of proteoglycan-ligand binding. Taken together, these results indicate that both the type and density of extracellular adhesion molecules in the pericellular environment are key regulators of hMSC adhesion. This observation is not surprising, as Goetinck and co-workers have recently demonstrated that integrin receptors and cell surface proteoglycans work in concert to promote cell spreading on fibronectin25. What is particularly interesting, however, is the contrast between our observations of hMSC adhesion on SAMs presenting RGDSP and a proteoglycan-binding ligand under serum-free conditions (Fig. 7C) and recent results from Bellis and co-workers characterizing hMSC adhesion on hydroxyapatite coated with RGD and a proteoglycan-binding peptide under serum-free conditions27. Their results demonstrate that hMSCs do not spread on hydroxyapatite materials coated with RGD, a proteoglycan-binding peptide, or a mixture of RGD and proteoglycan-binding peptide. Taken together, these results emphasize that the underlying biomaterial and, in turn, ligand presentation, may be important regulators of stem cell-biomaterial interactions.
Figure 7.
(A) Schematic representation of hMSC adhesion on SAMs presenting RGDSP and TYRSRKY. hMSC projected area on SAMs presenting (B) 0% RGDSP and 0.1-1% TYRSRKY, (C) 0.1% RGDSP and 0.1-1% TYRSRKY, and (D) 1% RGDSP and 0-1% TYRSRKY after 4 hrs in serum-free medium (■) or medium supplemented with 10% FBS (□). * denotes significant difference compared to serum-free condition, ** denotes significant difference compared to 0.1% TYRSRKY, *** denotes significant difference compared to 0% TYRSRKY (p < 0.05).
Interestingly, analysis of projected cell area on SAMs presenting 0.1% RGDSP and 0.1-1% TYRSRKY in the presence of 10% FBS (Fig. 7C) demonstrated that hMSCs adopted a rounded morphology, regardless of the TYRSRKY surface density. Moreover, analysis of projected cell area of hMSCs on SAMs presenting 1.0% RGDSP and 0.01-1.0% TYRSRKY (Fig. 7D) demonstrated a well-spread hMSC morphology on all substrates in the absence of serum, whereas a decrease in hMSC spreading was observed at TYRSRKY > 0.5% during culture in medium supplemented with 10% FBS. These results are in stark contrast to previous data collected from surfaces presenting RGDSP7 or RGDSP and RGESP (Fig. 6C), where spread morphologies are typically observed at surface densities of RGDSP > 0.1% during culture in the presence of serum. The observed decrease in hMSC spreading on substrates presenting TYRSRKY in the presence of serum is most likely due to the sequestration of serum-derived heparin proteoglycans onto the SAM. Specifically, heparin proteoglycans sequestered from serum may compete with cell surface proteoglycans for TYRSRKY binding sites and, in turn, may decrease the extent of cell spreading mediated by material-cell surface proteoglycan interactions. Additionally, the large serum-derived heparin molecules bound on the SAM may mask RGDSP molecules and interfere with RGDSP-integrin ligation. The observed correlation between loss of hMSC spreading on TYRSRKY presenting substrates in the presence of serum-derived heparin (Fig. 7C-D) is not surprising, as it is consistent with previous results from Park and co-workers who demonstrated a change from spread to rounded hMSC morphology on TYRSRKY-presenting poly(lysine) substrates in the presence of soluble heparin glycosaminoglycans16.
Taken together, these results demonstrate that an integrin binding moiety and a proteoglycan binding moiety work in concert to influence hMSC adhesion on 2-D substrates. Additionally, these results demonstrate that soluble biomolecules present during cell culture can compete with cell surface biomolecules for material binding sites and, in turn, directly influence specific cell-material interactions. Although SAM instability can limit the long-term efficacy of these materials when characterizing the influence of immobilized biomolecules on cell function over the course of weeks28, SAMs presenting peptides are commonly used to characterize cell-material interactions over a relatively short-term (e.g. hours to days)7,8,10. Our results herein demonstrate that SAMs presenting orthogonally-reactive moieties are useful base materials to characterize the concerted influence of two biochemically-distinct peptides on stem cell adhesion, and suggest widespread applicability of these materials to characterize additional stem cell-material interactions mediated by immobilized biomolecules.
Conclusions
Orthogonally-reactive, ternary SAMs allow for controllable immobilization of two distinct peptides over the entire SAM (Fig. 2). Importantly, changing the alkanethiolate mole ratio during SAM formation allows for control over the surface density of peptide conjugated to each of the reactive functional groups present on the surface (Figs. 3 and 4). This result suggests that SAMs presenting orthogonally reactive moieties may allow for facile preparation of substrates presenting two ligands with distinct biochemical activities for widespread SAM-based applications, such as biosensing and characterization of enzyme-substrate reactions. Our results demonstrate that the integrin-binding ligand RGDSP promotes hMSC adhesion, spreading, and focal adhesion complex formation even in the presence of the non-functional peptide RGESP (Fig. 6). The observed correlation between RGDSP surface density and hMSC adhesion demonstrates that hMSCs adhere to the SAMs via specific RGDSP-integrin interactions, rather than through interactions with non-specifically adsorbed serum proteins. Additionally, our results demonstrate that RGDSP and the heparin/heparan sulfate-binding ligand TYRSRKY act in concert to regulate hMSC adhesion (Fig. 7). The observed correlation between RGDSP surface density, TYRSRKY surface density and hMSC adhesion demonstrates that multiple, distinct extracellular factors work in concert to regulate hMSC adhesion. The relative bio-inertness and feasibility of varying peptide identity and surface density afforded by our approach may allow for characterization of the integrated role of additional peptides on stem cell function to an extent that is unattainable with traditional cell culture substrates.
Acknowledgements
This work was funded by the National Institutes of Health (R01HL093282 and R21EB005374) and the National Science Foundation (CAREER #0745563). PM-IRRAS data were obtained at the NSF-funded University of Wisconsin Materials Research Science and Engineering Center.
References
- 1.Toworfe GK, Bhattacharyya S, Composto RJ, Adams CS, Shapiro IM, Ducheyne P. J. Tissue Eng. Regen. Med. 2009;3:26–36. doi: 10.1002/term.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derda R, Li L, Orner BP, Lewis RL, Thomson JA, Kiessling LL. ACS Chem. Biol. 2007;2:347–55. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- 3.Nayak S, Yeo WS, Mrksich M. Langmuir. 2007;23:5578–83. doi: 10.1021/la062860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaki NK, Vijayamohanan K. Biosens. Bioelectron. 2002;17:1–12. doi: 10.1016/s0956-5663(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 5.Humphries MJ. J. Cell. Sci. 1990;97(4):585–92. doi: 10.1242/jcs.97.4.585. [DOI] [PubMed] [Google Scholar]
- 6.Berrier AL, Yamada KM. J. Cell. Physiol. 2007;213(3):565–73. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 7.Hudalla GA, Murphy WL. Langmuir. 2009;25:5737–46. doi: 10.1021/la804077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. J. Am. Chem. Soc. 1998;120(26):6548–6555. [Google Scholar]
- 9.Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. Chemphyschem. 2004;5(3):383–8. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 10.Koepsel JT, Murphy WL. Langmuir. 2009;25(21):12825–34. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahiri J, Isaacs L, Tien J, Whitesides GM. Anal. Chem. 1999;71:777–90. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 12.Houseman BT, Gawalt ES, Mrksich M. Langmuir. 2003;19:1522–1531. [Google Scholar]
- 13.Ladd J, Taylor AD, Piliarik M, Homola J, Jiang S. Anal. Chem. 2008;80:4231–6. doi: 10.1021/ac800263j. [DOI] [PubMed] [Google Scholar]
- 14.Choi S, Murphy WL. Langmuir. 2008;24(13):6873–80. doi: 10.1021/la800553p. [DOI] [PubMed] [Google Scholar]
- 15.Harper JC, Polsky R, Wheeler DR, Dirk SM, Brozik SM. Langmuir. 2007;23:8285–7. doi: 10.1021/la701775g. [DOI] [PubMed] [Google Scholar]
- 16.Lee JY, Choo JE, Choi YS, Lee KY, Min DS, Pi SH, Seol YJ, Lee SJ, Jo IH, Chung CP, Park YJ. J. Biomed. Mater. Res. A. 2007;83(4):970–9. doi: 10.1002/jbm.a.31351. [DOI] [PubMed] [Google Scholar]
- 17.Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. J. Phys. Chem. B. 1998;102:426–436. [Google Scholar]
- 18.Frey BL, Corn RM. Anal. Chem. 1996;68:3187–3193. [Google Scholar]
- 19.The peak corresponding to the stretch of the carboxylate moiety present in PM-IRRAS spectra collected after RGESP conjugation is due to the carboxylate group on the side-chain of glutamic acid introduced on the surface as a result of peptide conjugation.
- 20.An additional notable observation is that the amide I peak area after CuAAC was greater than the amide I peak area after carbodiimide condensation at each alkanethiolate mole fraction characterized. This observation suggests that the molar ratio of azide to carboxylate groups in the resulting SAM is greater than the molar ratio of azide to carboxylate groups in ethanol during SAM formation for all conditions studied. This observation is consistent with previous results that demonstrated that the rate of adsorption of HS---EG6---N3 (kN3) onto gold is significantly faster than the rate of adsorption of HS---EG6---EG3 (kEG3) onto gold (ref: 7) and suggests that preferential HS---EG6---N3 adsorption onto gold may also occur in the presence of HS---EG6---COOH
- 21.The loss of linearity between 5% and 7.5% COOH is likely due to the large TYRSRKY peptide occupying all available COOH groups at a surface density of COOH between 5-7.5%. Assuming that the alkanethiolates are well-packed and homogenously mixed on the Au substrate, the alkanethiolate packing would be estimated at 7.76 pmol/mm2. Assuming a partial specific volume of 0.73 cm3/g, we can estimate a hard-sphere peptide volume of 1385 A3. Based on the Stokes radius calculated from the estimated peptide volume, and assuming homogeneous alkanethiolate packing, the maximal packing of the peptide should be observed at ~ 11.4% COOH. In contrast, our experiments indicate a maximum packing density between 5-7.5% COOH. The discrepancy could be explained by variation from theoretical calculations in the peptide Stokes radius or heterogeneous SAM packing.
- 22.Boucher J, Trudel E, Methot M, Desmeules P, Salesse C. Coll. Surf. B: Biointerfaces. 2007;58(2):73–90. doi: 10.1016/j.colsurfb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Cabassi F, Casu B, Perlin AS. Carbohydrate Res. 1978;63:1–11. [Google Scholar]
- 24.Deans RJ, Moseley AB. Exp. Hematol. 2000;28:875–84. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 25.Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, Hynes RO, Goetinck PF. Proc. Natl. Acad. Sci. 1999;96(6):2805–10. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Amaro R, Sanchez-Madrid F. Crit. Rev. Immunol. 1999;19(5-6):389–429. [PubMed] [Google Scholar]
- 27.Sawyer AA, Hennessy KM, Bellis SL. Biomaterials. 2007;28(3):383–92. doi: 10.1016/j.biomaterials.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 28.Luk Y-Y, Kato M, Mrksich M. Langmuir. 2000;16(24):9604–9608. [Google Scholar]