Abstract
Mitochondrial DNA polymerase, POLG, is the sole DNA polymerase found in animal mitochondria. In humans, POLGα W748S in cis with an E1143G mutation has been linked to a new type of recessive ataxia, MIRAS, which is the most common inherited ataxia in Finland. We investigated the biochemical phenotypes of the W748S amino acid change, using recombinant human POLG. We measured processive and non-processive DNA polymerase activity, DNA binding affinity, enzyme processivity, and subunit interaction with recombinant POLGβ. In addition, we studied the effects of the W748S and E1143G mutations in primary human cell cultures using retroviral transduction. Here, we examined cell viability, mitochondrial DNA copy number, and products of mitochondrial translation. Our results indicate that the W748S mutant POLGα does not exhibit a clear biochemical phenotype, making it indistinguishable from wild type POLGα and as such, fail to replicate previously published results. Furthermore, results from the cell models were concurrent with the findings from patients, and support our biochemical findings.
Keywords: DNA polymerase γ, POLG, POLG1, ataxia, MIRAS, Alpers
Introduction
The minimal mitochondrial DNA (mtDNA) replisome comprises mitochondrial DNA polymerase (POLG)[1], helicase (Twinkle)[2] and mitochondrial single-stranded DNA-binding protein (mtSSB) [3, 4]. Of the 16 known mammalian DNA polymerases, POLG is the only one found in animal mitochondria, making it solely responsible for replication and repair of mtDNA. In mammals, the functional POLG holoenzyme is a heterotrimer, which consists of one catalytic subunit (POLGα) and an accessory subunit dimer (POLGβ) [5, 6].
The catalytic subunit POLGα contains DNA polymerase and exonuclease domains that are separated by a spacer region. The spacer region contains four conserved DNA sequence blocks (γ1- γ4). In recent studies the spacer region has been established to play a role in DNA template binding and positioning, and in interaction with the accessory subunit dimer POLGβ and likely mtSSB [7]. Mutations in the human POLGα spacer region have been linked to wide variety of inherited neurodegenerative phenotypes, such as ataxia, infantile Alpers syndrome and sensory ataxic neuropathy [8-11]. We and collaborators have reported a new neurodegenerative disorder with autosomal recessive ataxia, mitochondrial recessive ataxia syndrome (MIRAS)[10, 12, 13].In fact among all neurological diseases caused by mutations in POLGα, the most severe manifestations are caused by mutations in the spacer region in combination with polymorphisms and mutations in the DNA polymerase domain, which affect the central nervous system and sometimes also the liver.
MIRAS is caused by two amino acid changes in POLGα: W748S in cis with E1143G. W748 is located in the conserved γ4 region within the spacer domain, whereas E1143 localizes to the conserved pol C region in the polymerase domain. This allele is the most common genetic cause of inherited ataxia in Finland, with a carrier frequency of 1:125. The MIRAS allele is also found in patients from Australia, Norway and New Zealand[14]. MIRAS exhibits a wide variety of clinical features, resembling other ataxias caused by mitochondrial dysfunction (Friedrich's Ataxia, IOSCA) [15-17]. The age of onset varies from 5 to 50 years and the patients have ataxia and cerebellar dysarthria, which are also present in other mitochondrial ataxias. Symptoms that are not frequently present in MIRAS patients, such as muscle weakness and pes cavus (highly arched foot), distinguish MIRAS from other mitochondrial ataxias [12]. MIRAS patients exhibit tissue-specific reduced mtDNA levels, and mtDNA deletions in brain and liver. In addition, the patients exhibit a clear cell-specific respiratory chain Complex I defect in large neurons in the cerebellum[17]. However, when based solely on the clinical features, MIRAS is indistinguishable from other spino-cerebellar ataxias.Therefore, the diagnosis must be based on molecular genetic analysis.
The biochemical effects of specific POLGα spacer region mutations have been investigated with recombinant D. melanogaster and human POLGα [7, 18–20]. A deletion of the conserved γ4 region in D. melanogaster POLGα weakens the interaction with POLGβ and decreases DNA polymerase activity [7]. We and others have reported that a mutation A467T in POLGα that maps to the conserved γ1 region causes low DNA polymerase activity, diminished interaction with POLGβ, and decreased processivity of the enzyme. Another spacer region mutant, R627Q/W, exhibits normal DNA polymerase activity, and slightly higher DNA binding capacity and processivity [20]. Previously, Chan et al. have reported that the W748S amino acid substitution results in low DNA polymerase activity, DNA binding and slightly reduced processivity, and that all of these defects are rescued partially when W748S is present in cis with common variant E1143G [19]. These diverse results establish clearly that mutations in the spacer region of the POLGα do not cause any uniform biochemical phenotype.
The phenotype of the patients who are homozygous for W748S in cis with E1143G varies greatly, ranging from late-onset polyneuropathy to acute encephalopathy in adolescence [10, 11, 21, 22]. This finding suggests a considerable role for modifying factors in the severity of the disease manifestation. Given the varying clinical phenotype of MIRAS and diverse biochemical phenotypes of other spacer region mutants, we asked if a rigorous biochemical characterization of recombinant human POLGα carrying a W748S amino acid substitution could provide a new insight into the pathogenesis of MIRAS, and verify previously published results. Because POLG has the highest polymerase activity on templates with high primer density, we used both natural and synthetic sequence DNA templates with low and high primer density to measure processive and non-processive DNA synthesis[1]. We also measured DNA binding and subunit interactions.
Studies on the biochemical properties of the purified POLGα W748S protein suggest that the mechanisms underlying MIRAS are not explained solely by a catalytic enzyme defect. We extended these studies to examine primary cells transduced with retroviruses carrying W748S, E1143G, W748S+E1143G or wildtype transgenes. Expression levels of transduced POLGα were measured by quantitative RT-PCR and, relative mtDNA amounts by quantitative PCR. Pulse labeling of mitochondrial translation products was used to study possible effects on mitochondrial translation efficiency.
Results
Overexpression and purification of W748S mutant POLGα
We expressed and purified recombinant proteins carrying either wild-type or W748S POLGα, using the baculovirus overexpression system. The exonuclease activity of both of the recombinant polymerases was eliminated by substitution of two catalytic aspartates for alanine in the exonuclease I motif, as described previously [23]. Both catalytic cores were expressed and subjected to two independent purifications, and all experiments were performed with enzyme from both preparations. In all purifications, approximately 2.2 liters of Sf9 insect cells were infected with baculoviruses carrying the exonuclease-deficient human POLGα cDNA, and the recombinant proteins were purified 180-fold, to near homogeneity. About 0.6 mg of recombinant protein was purified per liter of infected cells, with a final yield of 6%. We found no signs of structural variation or instability in the mutant enzyme, based on chromatographic profiles and peaks in velocity sedimentation. All of the final preparations were of similar purity (Figure 1).
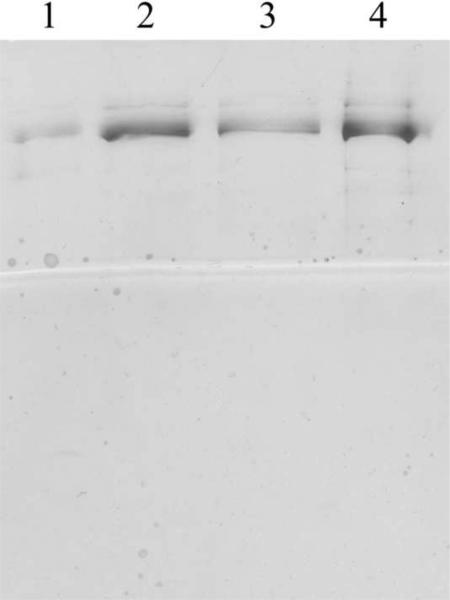
Figure 1.
Purified human POLGα protein. Recombinant wild type (lane 1, 100 ng; lane 2, 200ng) and W748S mutant of human POLGα protein (lane 3, 100 ng; lane 4, 200ng). Silver stained 10% SDS–PAGE gel. The size of the purified mutant protein was compared to the previously purified wild type enzyme. No difference in the stability of the mutant enzyme was detected in the purification procedure, compared to the wildtype.
Functional Analysis of the recombinant W748S mutant POLGα enzyme
We measured the intrinsic DNA polymerase activity of the W748S mutant POLGα catalytic core by examining non-processive and processive polymerase activity. Non-processive DNA polymerase activity was measured using a gapped, double-stranded natural sequence DNA substrate with high primer density, DNase I-activated DNA. Specific activities obtained under conditions of substrate excess were 9,335 U/mg for the wild-type human POLGα used as control, and 7,200 - 25,781 U/mg for the W748S mutant POLGα. Steady state kinetic analysis of DNA synthesis activity on this substrate in comparison with the wild-type enzyme yielded no significant differences; the Km values were 66.1 μM for wild type and 66.5 μM for W748S mutant POLGα enzyme (Table 1).
Table 1.
Biochemical properties of wild type and mutant forms of human POLG
| Wild type POLGα | W748S mutant POLGα | A467T mutant POLGα | |
|---|---|---|---|
| Specific activity (units/ mg) | |||
| Gapped double-stranded DNA | 8100- 9300 | 7200- 26 000 | 3900 |
| Poly r(A)- oligo d(T) | 712 ± 121 | 852 ± 209 | |
| Singly primed M13 DNA | 123 ± 14 | 143 ± 17 | |
| Km (μM dNTP) | |||
| Gapped double-stranded DNA | 66.1 ± 6.3 | 66.5 ± 4.8 | |
| Singly primed M13 DNA | 0.77 ± 0.07 | 0.75 ± 0.06 | |
| Average processive unit (nt) | |||
| POLGα | 52.8 ± 13.6 | 48.2 ± 12.7 | 55.8 ± 13.4 |
| POLG holoenzyme | 245 ± 34.0 | 257 ± 32.8 | 144 ±26.9 |
| DNA binding affinity, Kd (nM) | 6.8 ± 0.37 | 11.1 ± 0.52 |
Next we evaluated processive DNA polymerase activity with both natural and synthetic DNA substrates. We used singly primed M13 DNA as the natural sequence DNA substrate, and under conditions of substrate excess the specific activities were 123 U/mg and 143 U/mg for wild type and W748S mutant, respectively. Likewise, we found no differences when we used the POLG-specific synthetic substrate poly (rA)·oligo(dT) as template-primer. Here specific activities were 712 U/mg and 852 U/mg for wild type and W748S mutant POLGα respectively (Table 1).
We evaluated enzyme processivity directly by analyzing DNA product strand length by denaturing gel electrophoresis under conditions of large substrate excess, such that the association-dissociation cycle was limited to once per enzyme molecule, and then calculated the average processive unit. Both wild type and W748S mutant POLGα showed similar processivities with average processive units of 52.8 and 48.2 nt, respectively (Figure 2, Table 1).
Figure 2.
Processivity of mutant and wild type human POLGα subunit. DNA synthesis by mutant POLGα was measured at 30 mM KCl on singly primed M13 DNA as described in Materials and Methods. DNA product strands were isolated, denatured and electrophoresed as duplicates in a 6% denaturing polyacrylamide gel. Lanes 1 and 2, wild type POLGα; lanes 3 and 4, W748S mutant POLGα. Size marker: NEB 2-log DNA-ladder (molecular weights shown). The processivity of the mutant catalytic core enzyme was similar to the wild type enzyme.
Processive DNA polymerase activity of recombinant POLG holoenzymes
Though we observed no differences in the activities of the wild type and mutant catalytic cores, we evaluated possible defects in the POLG holoenzymes by reconstitution with the accessory subunit, POLGβ. We measured processive DNA synthesis on singly-primed M13DNA at 100 mM KCl using the POLG holoenzyme reconstituted either with the wild type or the mutant catalytic core, in the presence of a 4-fold excess (as monomer) of the wild type POLGβ. We obtained similar Km values of 0.77 μM for the wild type and 0.75 μM for the W748S mutant (Table 1).
Products of processive DNA synthesis were also analysed as with the catalytic cores, except that the reaction conditions were optimized for POLG holoenzyme activity in the presence and absence of a 4-fold excess of wild type POLGβ as monomer. POLG holoenzyme processivities were similar, with average processive units of 245 nt and 258 nt for the wild type and mutant enzymes, respectively (Figure 3, Table 1).
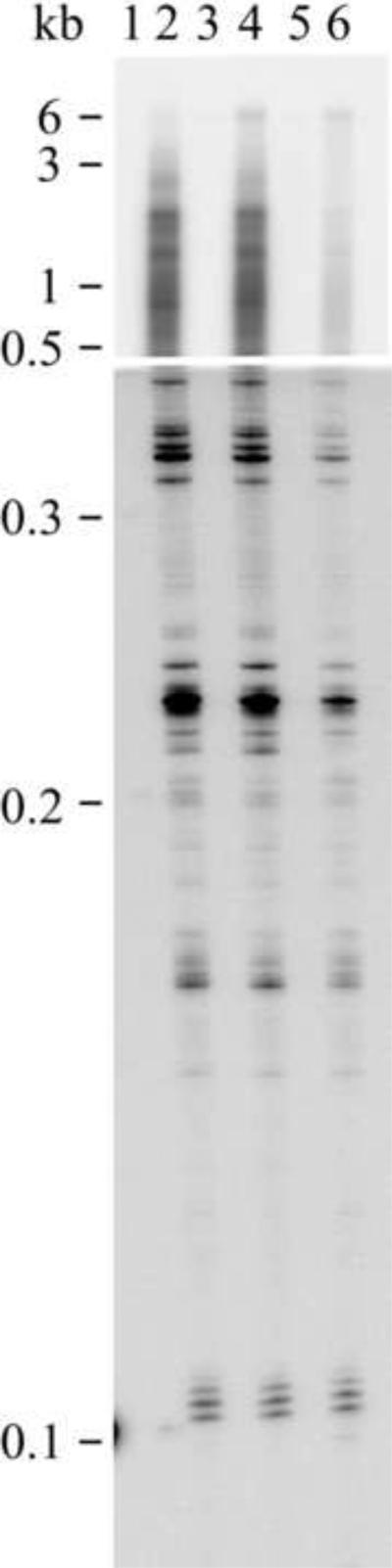
Figure 3.
Processivity of reconstituted mutant and wild-type POLG holoenzyme. Human POLG was reconstituted using a 4-fold molar excess of the accessory POLGβ subunit over POLGα, and DNA synthesis was measured at 100 mM KCl on singly-primed M13 DNA. DNA product strands were isolated, denatured and electrophoresed in denaturing 1.5% agarose (upper panel) and 6% polyacrylamide (lower panel) gels. Wild type POLGα in the absence (lane 1) and presence (lane 2) of POLGβ, lanes 3 and 4, W748S mutant of POLGα in the absence (lane 3) and presence (lane 4) of POLGβ, A467T mutant of POLGα in the absence (lane 5) and presence (lane 6) of POLGβ. Size marker: NEB 2-log DNA-ladder (molecular weights shown). As reported previously, the A467T mutant enzyme produced low-molecular-weight products similar to wildtype, but not longer products[20]. However, the W748S enzyme processivity was similar to wild type also concerning the longest products.
DNA binding affinity of W748S mutant POLG holoenzyme
We examined the DNA-binding affinity of W748S POLGα using a quantitative gel electrophoretic mobility-shift assay (EMSA) with a 21/45-mer DNA as a primer-template. The DNA-binding affinity was measured with and without wild type POLGβ, at 100 mM or 30 mM KCl, respectively. Reconstituted mutant POLG holoenzyme showed slightly reduced DNA binding, with a Kd value of 11.1 nM as compared to 6.8 nM for wild type POLG holoenzyme (Table 1).
Effect of W748S in cis with E1143G mutant POLGα overexpression in cells
We subcloned a human POLGα cDNA with or without mutations W748S and E1143G into a retroviral vector. When introduced into the host cell genome, the retroviral vectors induced a high level of overexpression of POLGα, confirmed with quantitative RT-PCR to be 7-19 fold higher as compared to endogenous expression levels. We also measured in parallel the expression levels of TWINKLE, the mtDNA helicase, which was not affected by overexpression of the mutant or wild type POLGα. Likewise, no changes in relative mtDNA levels were detected, nor was mitochondrial translation affected by mutant or wild type POLGα overexpression, as measured by pulse labeling of mitochondrial proteins.
Predicted structural consequences of POLGα amino acid changes
We utilized the recently published structure of human POLGα[24] to generate a modeled complex with docked template DNA. We identified the relevant amino acids W748, R627 and E1143 in our model and marked them for clarity. W748 and R627 are both located in the IP-domain of the spacer region, not in direct contact with DNA. Notably, the E1143 residue locates to a different domain of POLGα; the distance between W748 and E1143 is ~61.5 Å.
Discussion
Spacer region mutations of POLGα result typically in recessive nervous system disorders, whereas catalytic site mutations cause dominant myopathies in addition to neuronal defects [20, 25]. The basis for the tissue-specificity is unknown. We investigated here whether altered biochemical activities of the enzyme could explain some of the tissue-specific findings. The spacer region of human POLGα participates in DNA template positioning and in subunit interactions [18, 20]. Furthermore, in D. melanogaster, the spacer region has been shown to affect processivity and to interact functionally with mitochondrial single-strand DNA- binding protein [7]. Our results indicate that similar to another ataxia mutation, R627Q/W, the W748S mutation causing MIRAS-ataxia does not affect the catalytic activity of POLGα. These results suggest that ataxia-associated spacer region mutations of POLGα manifest in a highly tissue-specific context.
We purified recombinant W748S mutant POLGα twice in independent purifications, and did not find signs of structural instability or variability. Our initial measurements of specific DNA polymerase activity on three substrates, partially degraded DNAse I-activated calf thymus DNA, poly (rA)·oligo(dT) and singly-primed M13 DNA, failed to reveal differences between the W748S mutant and wild type POLGα. This prompted us to study the DNA polymerase activity further using steady-state kinetics on the DNAse I-activated DNA substrate, yet again we observed no difference between the wild type or W748S mutant enzyme, thus corroborating our initial data. In contrast, Chan et al. have reported that the W748S mutant POLGα has only 2.3% of wild-type POLGα polymerase activity using poly(rA)·oligo(dT) template-primer as a substrate [19]. At the same time, with regard to the A467T mutant POLGα, our results agree with those of Chan et al., where both laboratories have shown a clear reduction of DNA polymerase activity [18, 20]. In the case of the W748S mutation, we feel that an artifactual gain of mutant enzyme activity is unlikely; rather, we show clearly that the presence of the W748S substitution in the POLGα polypeptide does not affect the catalytic properties of the enzyme.
One of the major findings in the studies on A467T mutant POLGα was that the interaction between the catalytic core and accessory subunit is affected [18, 20]. This prompted us to evaluate a possible defect in subunit interactions in W748S mutant POLGα using steady state kinetic assays. Our results indicate that the W748S mutant POLGα is stimulated by wild type POLGβ in the same fashion as wild type POLGα, arguing strongly that subunit interaction is not affected by the W748S POLGα mutation. These results are consistent with the earlier study, which shows that the subunit interaction of W748S mutant POLGα with the accessory subunit is similar to that of wild type POLGα [19].
The accessory subunit of POLG contributes to DNA polymerase activity of the holoenzyme, as well as to DNA binding by allowing it to bind its substrates more tightly [5]. We studied the ability of W748S mutant and wild type POLG holoenzyme to bind DNA using an electrophoretic mobility shift assay. W748S mutant POLG holoenzyme shows a 1.6-fold reduction in DNA binding as compared to wild type POLG holoenzyme. The DNA binding ability of W748S mutant POLGα was also studied by Chan et al., who reported that the W748S mutation caused an eight-fold decrease in DNA binding by the catalytic core alone [19]. Together these results might suggest that impairment of the DNA binding capability of W748S mutant POLGα is compensated in the POLG holoenzyme by its interaction with POLGβ. However, we find it unlikely, because we found that enzyme processivity was not affected by the W748S amino acid change regardless of the presence or absence of POLGβ. Because processivity is influenced by both DNA binding ability and polymerase activity[1], and neither W748S mutant POLGα nor its reconstituted holoenzyme exhibit lower processivity as compared to the wild type forms, we conclude that the small decrease we observe in the DNA binding ability of the W748S mutant POLG holoenzyme is apparently irrelevant for processivity of the POLG holoenzyme. In contrast, Chan et al. reported that the W748S mutant POLGα core and the mutant holoenzyme processivities are impaired as compared to the wild type forms, and conclude that this impairment is caused by the weak DNA binding and low polymerase activity of W748S mutant POLGα [19]. POLGβ stimulates the processivity of wild type POLGα as much as 50-fold, to a value over 1000 nt, mainly by increasing DNA binding affinity [1, 6]. Our current data show stimulation that result in products up to 3 kb and the discrepancy between the processivity analyses in the two report might relate to the fact that Chan et al. visualized products up to 200 nt, not taking into account the expected effect of the accessory subunit [19].
After submission of our manuscript, Lee et al. published the three-dimensional structure of human POLG [24]. Their structure shows that the W748 residue is located in the IP-region of the spacer, and that the ataxia-associated R627Q/R mutant is also located in the same domain. Both of the mutants share the lack of a catalytic defect, and a nervous system phenotype [20]. The IP region is not in direct contact with DNA or with the catalytic domain. However, the mutation could cause local changes in the structure of the catalytic subunit that might affect DNA binding. As proposed by Lee et al., the W748 residue is likely involved in stacking interactions with H733 and F750 that maintain the conformation of that part of IP domain which is in contact with single-stranded DNA. This interpretation warrants further experimentation because the crystals do not contain a bound DNA template. Nonetheless, our results support an interpretation of indirect interaction with DNA, as we observed slightly lower DNA binding in the W748S mutant holoenzyme, which did not, however, affect the processivity of the holoenzyme. Our data suggest that the spacer IP domain mutants manifest in a highly tissue-specific context, mainly in the nervous system.
In the patients, the W748S amino acid change is always found in cis with an E1143G polymorphic variant. This can be explained either by a functional role of the variant or by a single ancestral chromosome, in which the two mutations happened to occur. Baruffini et al. reported that in S. cerevisiae, POLGα with and E1143G mutation showed low protein levels, probably because of decreased stability, which likely resulted in reduced DNA polymerase activity [26]. The actual POLGα protein level in patients has not been reliably measured, mainly due to poorly specific antibodies (commercial and private, unpublished observations) and to low endogeneous protein levels. We have previously shown that W748S+E1143G occurred in a single ancestral chromosome, and therefore all MIRAS patients with W748S+E1143G are distantly related [12, 27]. In our structural model, we show that E1143 locates to a different domain of POLGα with a distance of ~61.5 Å to W748, and no direct interaction between the two amino acids can be predicted. These data together with human genealogical data do not suggest a specific role for the E1143G variant, but an independent role in protein stability cannot be excluded.
Our aim in this study was to investigate whether the biochemical phenotype of W748S mutant POLGα could explain the drastic clinical phenotype seen in MIRAS patients. The failure to identify any substantial differences between the mutant and wild type enzymes in processive or non-processive DNA synthesis on several different DNA templates establishes that the disease is not caused by a global DNA synthetic defect. Further, cell lines highly overexpressing POLGα W748S or W748S+E1143G protein exhibited no changes, either in the level or integrity of the mtDNA nor in translation of mitochondrial proteins. The lack of a catalytic defect appears to be a common feature of nervous system phenotypes caused by POLGα spacer mutations (including R627Q/W and W748S) [12, 17]. Mutations in the mitochondrial replicative helicase Twinkle, a direct functional partner of POLG, underly the recessively-inherited infantile-onset spinocerebellar ataxia, IOSCA, which manifests at early age, strictly in the nervous system[28]. Interestingly, the IOSCA mutant enzyme also exhibits normal in vitro activities [17]. These findings emphasize that the neurons affected in MIRAS and IOSCA depend on cell-specific mtDNA maintenance mechanisms, in which the POLGα spacer plays an important role. These may involve tissue-specific protein interactions, and/ or effects of mtDNA organization or segregation.
Materials and Methods
Construction of recombinant baculoviruses
Baculovirus transfer vectors carrying mutant POLGα cDNAs described were prepared by QuikChange mutagenesis with Pfu DNA polymerase (Stratagene), according to the manufacturer's recommendations and as described previously [12, 29]. The following primer pairs were used
5'-CTG GCT GCT CGT TTT TCA AGC TTC CTC ACA AG-3' and
5'-CTT GTG AGG AAG CTT GAA AAA CGA GCA GCC AG-3'.
Transfer vectors encoding the mutant versions of POLGα were purified and baculoviruses prepared as described by Wang and Kaguni [29].
Production and purification of the recombinant POLGa subunit
The purification of POLGα was done as described previously [20].
DNA polymerase assays
DNA polymerase activity was assayed on DNase I-activated calf thymus DNA, singly primed M13 DNA or poly r(A)·oligo d(T) as previously described [30, 31]. Assays were performed at least two times, in duplicate. Steady-state kinetic values were determined with either 5-400 nM DNase I-activated calf thymus DNA or 0,125 – 8 μM singly primed M13 DNA, with or withoutadded 100 mM KCl. Poly r(A)·oligo d(T) assays were done with 0.4 mM MnCl2 instead of MgCl2, 100 μM poly r(A)·oligo d(T) and 20 μM dTTP. In stimulation assays fourfold excess of wild-type POLGβ was added to reaction mixtures. Km values were determined using Origin 7.5 software (OriginLab). One unit of standard activity is that amount of protein that catalyzes the incorporation of 1 nm of deoxynucleoside triphosphate into acid insoluble material in 60 min at 30 °C using DNase I-activated calf thymus DNA as the substrate.
Gel electrophoretic mobility-shift assay
DNA binding affinity was assayed by a quantitative gel electrophoretic mobility-shift assay, as described previously [5], with the following modifications: the amounts of POLGα were 9 – 576 nmol, and POLGβ was added in fourfold molar excess over POLGα, as in other assays. The data were analyzed by scanning the Phosphor Screen using the Typhoon 9400 phosphorimager (GE Healthcare, Piscataway, NJ), and the volume of each band was determined by computer integration analysis using ImageQuant version 5.2 software (GE Healthcare, Piscataway, NJ). Kd values were determined using Origin 7.5 software (OriginLab), from the fraction of DNA bound, samples derived from at least two independent experiments.
Analysis of products of processive DNA synthesis by gel electrophoresis
Processivity of the POLG enzymes was measured as described previously [5], but with the following modifications: 10 μM singly primed M13 DNA, and 20 ng (0.14 pmol) of POLGα was used in assays and human POLGβ was added in fourfold excess (0.56 pmol) over POLGα. The data were analyzed by scanning the Phosphor Screen using the Typhoon 9400 phosphorimager (GE Healthcare, Piscataway, NJ), and the density of distinct bands from duplicate samples was determined by computer integration analysis using ImageQuant version 5.2 software (GE Healthcare, Piscataway, NJ); the density of the bands was normalized to the nucleotide level to correct for the uniform labeling of the DNA products, and the resulting DNA product strand lengths are presented as apu (in nt) in Table 1.
Generation of POLG-retroviral vectors with and without mutation(s)
The cDNA of human POLGα cloned in pcDNA™ 3.1/myc-His Awas a kind gift from Docent JN Spelbrink. The cDNA of human POLGα was mutagenised by PCR site-directed mutagenesis with primer pairs
5’-CCCTGGCTGCTCGTTTTTCAAGCTGCCTCAC-3’,
5’- GTGAGGCAGCTTGAAAAACGAGCAGCCAGG3’ and
5’- CGCTACCTGGTGCGGGGGGAGGACCGCTACCG-3’,
5’- CGGTAGCGGTCCTCCCCCCGCACCAGGTAGCG-3’
to produce nucleotide changes G2243C with primers 1 and 2 and A3428G with primers 3 and 4 for W748S and E1143G respectively. Altered aminoacid is indicated in bold font. In the PCR-SDM reactions were 28,8-60 ng of template plasmid DNAs, 1 U of Phusion High fidelity DNA polymerase (Finnzymes), 20 μM of primers 1 and 2 or 3 and 4 (Sigma-Proligo), 200 μM of dNTP mix (Roche Diagnostics GmbH), 3% of dimethyl sulfoxide (Roche Diagnostics GmbH) in 50μl of 1x Phusion HF buffer. Two step PCR amplification was as follows: primary denaturation at 99°C for 30 sec, denaturation at 99°C for 10 sec, annealing and extension at 70°C or 72°C for 8 min for W748S and E1143G mutations respectively and the final extension at 72°C for 10 min. Denaturation and annealing/extension steps were repeated for 30 times. Mutated PCR products were purified from parental template with DpnI restriction endonuclease (New England Biolabs). Inserts POLGα, POLGα W748S, POLGα E1143G and POLGα W748S E1143G were digested from 30 μg of plasmid in double digestion reactions with EcoRI and BamHI restriction endonucleases in EcoRI buffer (Fermentas). The digested DNA pieces were gel purified with QIAquick Gel Extraction Kit (Qiagen). To form blunt ends the purified inserts were incubated with 36 μM of dNTP mix and 1 U/ μg of DNA of DNA Polymerase I, large (Klenow) fragment in NEBuffer 2 (New England Biolabs). Samples were incubated at 25°C for 15 min after which they were gel purified. 30 μg of pBABEpuro vector was digested with 0,5 U/ 1μg of plasmid DNA of SnaBI restriction enzyme (New England Biolabs) with 1 μg/ml of bovine serum albumin (BSA) (New England Biolabs) in NEBuffer 4. pBABEpuro was dephosphorylated with 0,5 U/1 μg of plasmid DNA of calf intestinal alkaline phosphatase (CIP) (Finnzymes) for one hour at 37°C and subsequently gel purified. Blunt end ligations of different POLGα inserts to pBABEpuro were done in reactions that contained 300-375 ng of insert, 30-75 ng of vector, 2000-3000 U of T4 DNA Ligase and T4 DNA Ligase Reaction Buffer (New England Biolabs) with polyethylene glycol (PEG) 8000 (Sigma). All reactions were incubated at 15°C over night. Correct orientation of the insert was verified by sequencing and PCR.
Production of infectious pBABEpuro virus with human polymerase γ complementary DNA –insert
Retrovirus packaging cells of both GP+E86 and PA317 cell lines were grown in D-MEM with 10% FBS (Gibco) and 1% L-Glutamate (Invitrogen). Transfections of GP+E86 packaging cells with pBABEpuro, + POLGα, + POLGα W748S /+ POLGα E1143G or + POLGα W748S+E1143G plasmids were done with Lipofectamine 2000 transfection reagent (Invitrogen) following manufacturer's instructions. For transfection GP+E86 packaging cells were grown to 90% confluency and 4 hours prior to transfection medium was changed. Plasmid DNA and Lipofectamine reagent ration in transcfection was 1:3 and 20 μg of plasmid was used to transfect one 10 cm plate. Transfected cells were passed 1:10 24-48 h after transfection and after 48 h of transfection puromycin selection was started with 3 μg/ml of puromycin (Sigma). Subsequently PA317 packaging cells were infected with filtered medium from transfected GP+E86 cells. Transfected GP+E86 cells were passed to 50% confluency 24 h prior to collection of virus containing medium. Medium from GP+E86 cells was collected and filtered through 0,45 μm syringe filter. PA317 cells were at 50% confluency when 1 ml of filtered virus containing medium and 3 ml of D-MEM, 10% FBS, 1% L-Glutamate with 4 μg/ml of polybrene (hexadimethrine bromide) (Sigma) was added to them. PA317 cells were incubated in infectious polybrene containing medium for 2 hours at 37°C in CO2-incubator after which 5 ml of D-MEM, 10% FBS, 1% L-Glutamate with 4 μg/ml of polybrene was added to them. Incubation was continued for 5 hours at 37°C in CO2-incubator. Infectious medium was removed and cells were washed once with D-MEM. Infected cells were grown at 37°C in CO2-incubator for 24 hours before starting puromycin selection with 3 μg/ml of puromycin that was raised to 4 μg/ml after 24 hours of selection.
Transduction of target primary human fibroblasts
Primary human fibroblasts were transduced with PA317 cells’ virus containing medium as described above. Cells were grown in D-MEM, 10% FBS, 1% L-Glutamate, 50μg/ml uridine (Sigma), 2μg/ml puromycin.
Protein expression analysis
Primary fibroblasts were collected using trypsin (Invitrogen). Total cellular RNA was extracted from cells with Qiagen RNeasy Mini Kit (Qiagen). 500 ng of total RNA and 0,25 μg of Random Primers (Promega) in total volume of 15 μl were incubated for 5 min at 70°C. 500 ng of total RNA, previously incubated with 0.25 μg of random primers, was reverse transcribed in 0.625 U of rRNasin inhibitor (Promega), 312.5 μM of dNTP, and 200 U of M-MLV Reverse Transcriptase in 40 μl of its buffer (M-MLV RT buffer) (Promega). Samples were incubated at 37°C for one hour. Quantitative PCR reactions consisted of 83 pg of cDNA, TaqMan Gene expression Assay mix, in final concentrations of 45 nM of primers and 12.5 nM TaqMan MGB probe in TaqMan Universal PCR Master Mix (Applied Biosystems). In addition to the mRNA levels of transduced POLGα, analysed with assay on demand Hs00160298_m1, and its transduced mutant forms, TWINKLE expression levels were also measured and analysed using assay on demand Hs00222440_m1 for human TWINKLE. Beta-actin gene ACTB was used as internal control using assay on demand Hs99999903_m1. Gene expression QPCRs were done in separate tubes on MicroAmp Optical 96-well Reaction Plate (Applied Biosystems) for each assay. PCR amplification was done with ABI Prism 7000 quantitative PCR instrument and data was analyzed with 7000 SDS Sequence Detection Software version 1.2.3 (Applied Biosystems).
Mitochodrial DNA copy number analysis
MtDNA was quantified as described elsewhere [27].
Pulse Labeling of Mitochondrial Translation Products
In vitro labeling of mitochondrial translation was performed in general as described elsewhere. [32]
3-D Modeling of the POLG-DNA Complex
Docking of the template DNA onto the structure of the catalytic subunit of POLG (PDB entry 3IKM, chain A) was performed using the VMD software [24, 33]. The structure of T7 DNA polymerase in complex with DNA (PDB entry 1T8E [34]) was used for superposition of the catalytic domains of the two enzymes.
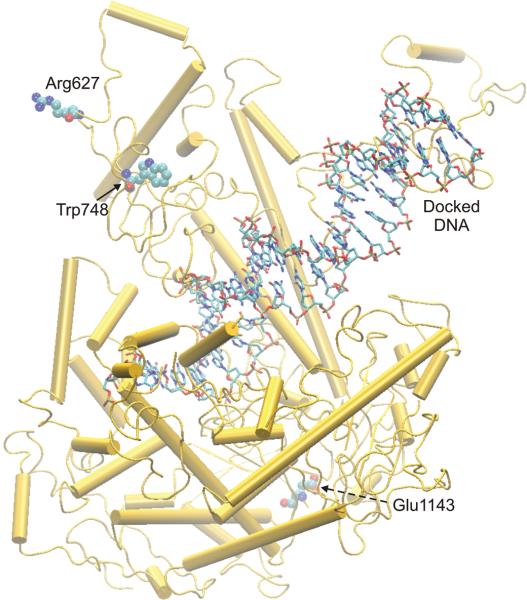
Figure 4.
Three-dimensional structural model of the catalytic subunit of human POLG with docked DNA template. Locations of ataxia-mutants R627, W748 and E1143 within the catalytic subunit of human POLGα are indicated (Protein Data Bank entry 3IKM, chain A[24]) relative to DNA docked into the structure. The figure was prepared using the Visual Molecular Dynamics software (VMD) [33].
Acknowledgements
The authors would wish to thank the Helsingin Sanomain 100 year foundation for E.P. to visit the L.S.K. laboratory; Helsinki Biomedical Graduate School for A.L.; the Academy of Finland, the Sigrid Juselius Foundation, and the Jane and Aatos Erkko Foundation, University of Helsinki (A.S.); and grant GM45295 from the National Institutes of Health (L.S.K.).
Abbreviations
- MIRAS
Mitochondrial Recessive Ataxia Syndrome
- pol
DNA Polymerase
- apu
Average Processive Unit
- POLG
DNA Polymerase Gamma Holoenzyme
- POLGα
Catalytic Subunit of DNA Polymerase Gamma
- POLGβ
Accessory Subunit of DNA Polymerase Gamma
- dsDNA
Double Stranded DNA
- EMSA
Electrophoretic Mobility-Shift Assay
- Twinkle
Mitochondrial DNA Helicase
- mtSSB
Mitochondrial DNA Single Strand Binding Protein
- mtDNA
Mitochondrial DNA
- IOSCA
Infantile Onset Spinocerebellar Ataxia
- PCR
Polymerase Chain Reaction
- RT-PCR
Real Time Polymerase Chain Reaction
- nt
nucleotide
- CNS
Central Nervous System
- CT-DNA
Calf Thymus DNA
- tRNA
Transfer RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 2.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature genetics. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 3.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- 5.Fan L, Kim S, Farr C, Schaefer K, Randolph K, Tainer J, Kaguni L. A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase. Journal of molecular biology. 2006;358:1229–1243. doi: 10.1016/j.jmb.2006.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. The Journal of biological chemistry. 2005;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 7.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J Biol Chem. 2005;280:2491–2497. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 8.Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi GP, Zeviani M. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- 9.Van Goethem G, Dermaut B, Löfgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 10.Van Goethem G, Luoma P, Rantamäki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Löfgren A, Martin JJ, De Jonghe P, Suomalainen A, Udd B, Van Broeckhoven C. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 11.Van Goethem G, Martin JJ, Dermaut B, Löfgren A, Wibail A, Ververken D, Tack P, Dehaene I, Van Zandijcke M, Moonen M, Ceuterick C, De Jonghe P, Van Broeckhoven C. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul Disord. 2003;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- 12.Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, Kaakkola S, Majamaa K, Varilo T, Udd B, Kaariainen H, Bindoff LA, Suomalainen A. Mitochondrial DNA Polymerase W748S Mutation: A Common Cause of Autosomal Recessive Ataxia with Ancient European Origin. Am J Hum Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rantamäki M, Krahe R, Paetau A, Cormand B, Mononen I, Udd B. Adult-onset autosomal recessive ataxia with thalamic lesions in a Finnish family. Neurology. 2001;57:1043–1049. doi: 10.1212/wnl.57.6.1043. [DOI] [PubMed] [Google Scholar]
- 14.Hakonen AH, Isohanni P, Paetau A, Herva R, Suomalainen A, Lönnqvist T. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain : a journal of neurology. 2007;130:3032–3040. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- 15.Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Cañizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science (New York, N.Y.) 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 16.Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS letters. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 17.Hakonen AH, Goffart S, Marjavaara S, Paetau A, Cooper H, Mattila K, Lampinen M, Sajantila A, Lönnqvist T, Spelbrink JN, Suomalainen A. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. 2008;17:3822–3835. doi: 10.1093/hmg/ddn280. 10.1093/hmg/ddl424. [DOI] [PubMed] [Google Scholar]
- 18.Chan SSL, Longley MJ, Copeland WC. The Common A467T Mutation in the Human Mitochondrial DNA Polymerase (POLG) Compromises Catalytic Efficiency and Interaction with the Accessory Subunit. J Biol Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 19.Chan SSL, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase gamma by the E1143G polymorphismin mitochondrial disorders. Human molecular genetics. 2006;15:3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luoma PT, Luo N, Löscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxiamyopathy syndrome. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 21.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, McKiernan PJ, Gray G, Naviaux RK. POLG mutations in Alpers syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- 23.Longley M, Ropp P, Lim S, Copeland W. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y-S, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, Majamaa K, Somer H, Suomalainen A. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 26.Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Hakonen AH, Davidzon G, Salemi R, Bindoff LA, Van Goethem G, Dimauro S, Thorburn DR, Suomalainen A. Abundance of the POLG disease mutations in Europe, Australia, New Zealand, and the United States explained by single ancient European founders. European journal of human genetics : EJHG. 2007;15:779–783. doi: 10.1038/sj.ejhg.5201831. [DOI] [PubMed] [Google Scholar]
- 28.Nikali K, Suomalainen A, Saharinen J, Kuokkanen M, Spelbrink JN, Lönnqvist T, Peltonen L. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum Mol Genet. 2005 doi: 10.1093/hmg/ddi328. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Kaguni L. Baculovirus expression reconstitutes Drosophila mitochondrial DNA polymerase. The Journal of biological chemistry. 1999;274:28972–28977. doi: 10.1074/jbc.274.41.28972. [DOI] [PubMed] [Google Scholar]
- 30.Fan L, Sanschagrin PC, Kaguni LS, Kuhn LA. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc Natl Acad Sci U S A. 1999;96:9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernette C, Kaguni L. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster. Purification, subunit structure, and partial characterization. The Journal of biological chemistry. 1986;261:14764–14770. [PubMed] [Google Scholar]
- 32.Boulet L, Karpati G, Shoubridge EA. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF) American journal of human genetics. 1992;51:1187–1200. [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14:33–38, 27-38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. The EMBO journal. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]