Summary
Brain structure and size requires precise division of neural stem cells (NSCs), which self-renew and generate intermediate neural progenitors (INPs) and neurons. The factors that regulate NSCs remain poorly understood, as do mechanistic explanations of how aberrant NSC division causes reduced brain size as seen in microcephaly. Here we demonstrate that Magoh, a component of the exon junction complex (EJC) that binds RNA, controls mouse cerebral cortical size by regulating NSC division. Magoh haploinsufficiency causes microcephaly due to INP depletion and neuronal apoptosis. Defective mitosis underlies these phenotypes as depletion of EJC components disrupts mitotic spindle orientation and integrity, chromosome number, and genomic stability. In utero rescue experiments revealed that a key function of Magoh is to control levels of the microcephaly-associated protein, LIS1, during neurogenesis. This study uncovers new requirements for the EJC in brain development, NSC maintenance, and mitosis, thus implicating this complex in the pathogenesis of microcephaly.
In humans, mice and flies, size and structure of the adult brain depends upon precise control of neural stem cell (NSC) division during development. NSCs, composed initially of neuroepithelial cells and then radial glia, are located in the ventricular zone (VZ) bordering the ventricle of the neocortex1. NSCs undergo proliferative symmetric and asymmetric divisions to replenish themselves and to produce intermediate neural progenitors (INPs), respectively. INPs, which reside adjacent to the VZ in the sub-ventricular zone (SVZ), are thought to produce the majority of cortical neurons2,3. Neurons are also produced by NSCs undergoing neurogenic asymmetric divisions. Thus, a balance of symmetric and asymmetric NSC divisions regulates the number of NSCs, INPs and neurons produced, and is critical for defining the adult brain. This balance may be influenced by alterations in the NSC mitotic division plane as well as asymmetric inheritance of proteins that regulate cell fate4–6. However, the exact mechanisms defining NSC divisions remain poorly understood.
Defects in NSC division can cause microcephaly, a congenital neurodevelopmental disorder characterized by dramatically smaller brain size and cognitive deficiency of varying severity7. Microcephaly and microcephaly syndromes are associated with genes that regulate aspects of NSCs, including cell division and genomic integrity8–10. CENPJ, ASPM and CDK5RAP2, three genes associated with autosomal recessive microcephaly, encode proteins that regulate centrosomes, the microtubule organizing centers that anchor the mitotic spindle8,9,11. Increased and decreased levels of LIS1, a microtubule-associated protein essential for mitotic spindle integrity, is associated with human microcephaly syndromes12–14. In addition, microcephaly-associated syndromes, Seckel syndrome and microcephalic osteodysplastic primordial dwarfism (MOPD) type II, are caused by mutations in genes encoding centrosome and DNA damage signaling proteins15–17. Although studies of these genes highlight the fundamental role of mitosis in the etiology of microcephaly, very few genes are known to regulate both NSC mitosis and microcephaly in vivo. Thus the genetic mechanisms regulating these processes are poorly defined. In this study we utilized a new mouse mutant to explore how NSC aberrations cause microcephaly, and to uncover an essential regulator of NSC mitosis and brain size.
Results
Magoh haploinsufficiency causes microcephaly
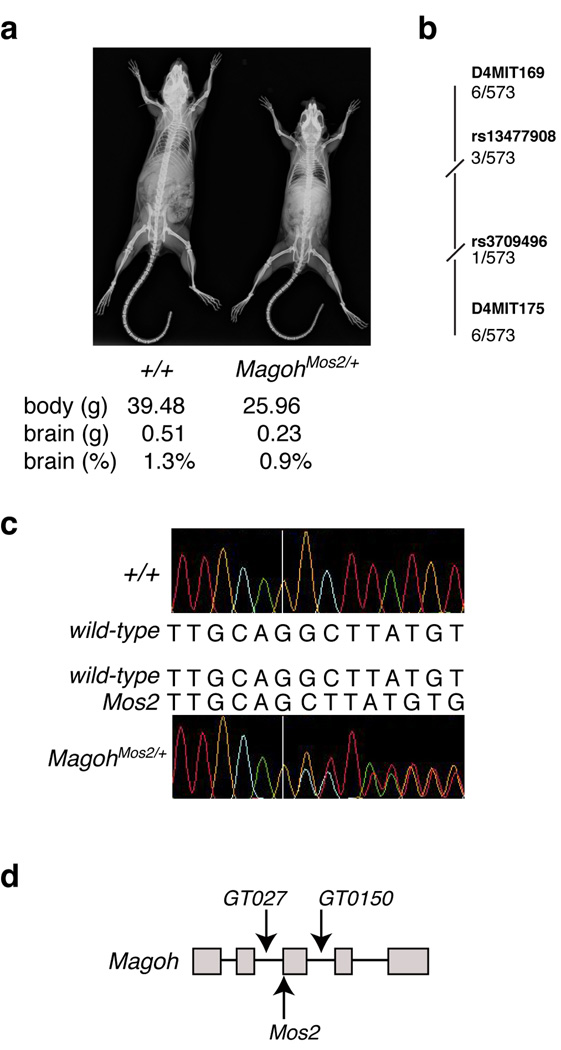
From an N-ethyl-N-nitrosourea (ENU) mutagenesis screen, we discovered Mos2 (Modifier of Sox10)18, a hypopigmented mouse mutant that is homozygous lethal prior to E9.5 and heterozygous lethal with incomplete penetrance (Table 1). Mos2+/− animals were 33% smaller than control littermates (P<0.05) with no significant differences in hematology, serum chemistries or histology of most organs (Fig. 1a, Supplementary Table 1, Supplementary Data). In contrast, Mos2+/− brains weighed significantly less than those of control littermates (compare 0.234 g to 0.509 g) (P<0.05) and were 28% smaller when normalized for overall reduced body size (compare 0.9% ± 0.1 to 1.3% ± 0.06) (Fig. 1a, Supplementary Table 1). These analyses demonstrate that Mos2+/− animals exhibit reduced body size and microcephaly in addition to hypopigmentation and lethality.
Table 1.
| Cross | Observed genotypes | |||
|---|---|---|---|---|
| Genotype X+/+ | +/+ | MagohMos2/+ | Tg-BAC; +/+ |
Tg-BAC; MagohMos2/+ |
| MagohMos2/+ | 90% (500) | 10% (57) ** | N/A | N/A |
| MagohMos2/+(E18.5) | 50% (14) | 50% (14) | N/A | N/A |
| MagohGT0150/+ | 89% (321) | 11% (40) ** | N/A | N/A |
| MagohGT027/+ | 93% (192) | 7% (15) ** | N/A | N/A |
| Tg-BAC1; MagohMos2/+ |
37% (65) | 0% | 33% (58) | 30% (51) ** |
| Tg-BAC2; | 31% (84) | 1% (3) | 33% (88) | 35% 96) ** |
| MagohMos2/+ | ||||
Percentage of viable offspring observed from mice of the indicated genotype crossed to wild-type (+/+). Offspring were genotyped at weaning except where indicated (E18.5). The number of animals analyzed is in parentheses. N/A, not applicable
P<0.001.
Figure 1. Mutation of Magoh causes microcephaly and reduced body size.
a, X-ray images of control and MagohMos2/+ adult mice. Average body size (g), brain size (g), and brain size as a percentage of total body weight is listed beneath each genotype. b, Representation of linkage analysis on chromosome 4 with the indicated SNPs and SSLPs (bold) and the number of recombinants/meioses evaluated. For all markers shown, P<0.0005. c, Sequence chromatogram and corresponding sequences of control (top) and MagohMos2/+(bottom) genomic DNA. The MagohMos2/+ DNA sequence contains a G deletion (right of the white line), and from this position onward two peaks are apparent, representing the control and Mos2 alleles. d, Schematic representation of Magoh gene, with exons (grey boxes), introns (lines, not drawn to scale), and locations of three alleles indicated.
Linkage analysis using 573 backcross animals segregating the Mos2 phenotypes defined a 2.4 Mb critical region containing 38 annotated genes (Fig. 1b). Sequence analysis of 12 genes within this interval revealed one mutation (198delG) in Magoh not present in parental strains (Fig. 1c). MAGOH encodes a component of the core exon junction complex (EJC) which also contains RBM8A, EIF4A3, and CASC3 and binds spliced mRNA upstream of exon-exon junctions19–23. Magoh (NM_010760) is highly conserved, showing 100% amino acid identity between mouse and human.
The MagohMos2 allele is predicted to cause a frameshift resulting in a truncated protein. However, Western analyses of MagohMos2/+ cortical lysates showed reduced MAGOH levels, but normal protein size (Supplementary Fig. 1a). Expression profile analysis of E10.5 cortices revealed a two-fold reduction in Magoh mRNA levels in MagohMos2 versus control (P<0.005) (Supplementary Table 2). This result was confirmed by quantitative gene expression analysis of E10.5 and E12.5 control and MagohMos2 cortices (compare normalized values for E10.5 of 0.91 ± 0.14 to 0.45 ± 0.14 and for E12.5 of 0.81 ± 0.03 to 0.52 ± 0.15) (P<0.0005). Expression of the MagohMos2 mutant allele was not detectable by restriction digestion or sequence analysis of RT-PCR products (Supplementary Fig. 1b,c). Aberrantly sized RT-PCR products were also not observed, suggesting abnormal splicing of MagohMos2 mutant transcript does not occur at high frequency in the neocortex (Supplementary Fig. 1b). Because the MagohMos2 transcript contains a premature nonsense codon and was not detectable, we propose that the mutant allele undergoes nonsense mediated decay (NMD), a process dependent upon EJC function. However, we cannot distinguish whether NMD of Magoh occurs due to degradation of a properly spliced mutant transcript or degradation of mis-spliced mRNA containing a premature termination codon.
Two approaches confirmed that mutation of Magoh caused the Mos2 phenotypes. First, transgenic mice carrying either of two BACs expressing wild-type Magoh fully rescued the hypopigmentation, lethality, reduced body size and microcephaly phenotypes of MagohMos2/+ mice (Supplementary Fig. 2, and Table 1). Second, two independent Magoh mutant alleles, MagohGT0150/+ and MagohGT027/+, exhibited hypopigmentation, lethality, reduced body size and microcephaly, as seen in MagohMos2/+ (Fig. 1d, Supplementary Fig. 2, and Table 1). Taken together, the molecular and genetic evidence shows that MagohMos2 is a null allele and that Mos2/+ phenotypes result from Magoh haploinsufficiency.
Disorganized layers and fewer neurons in MagohMos2/+
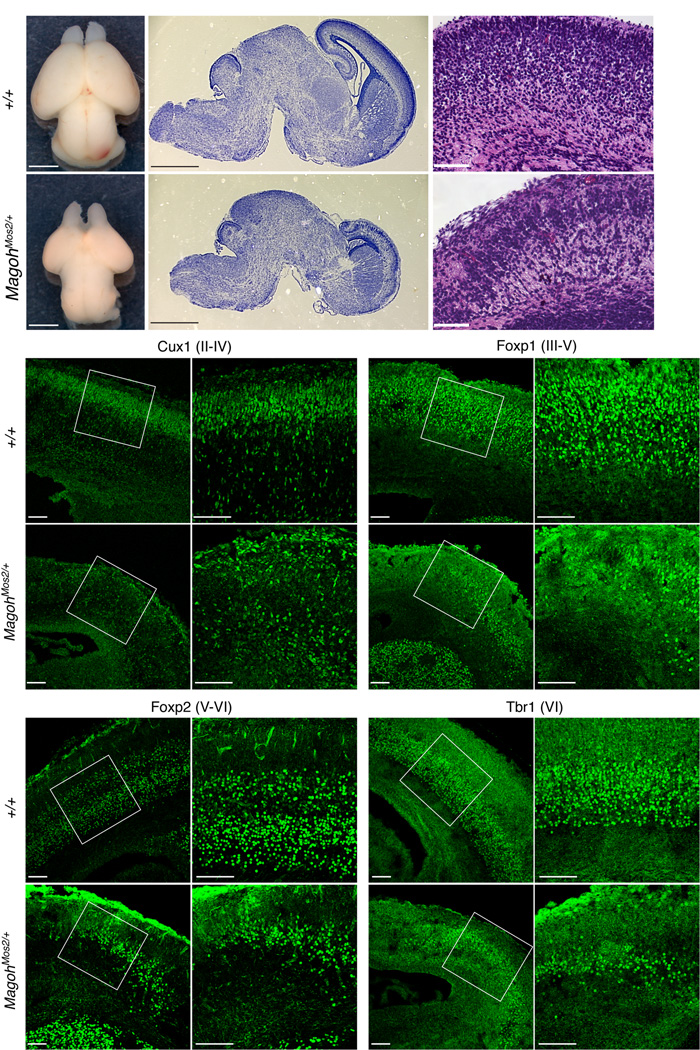
Histological analysis of adult MagohMos2/+ brains revealed hypoplasia and global reduction of the cerebral cortex, corpus callosum, and cerebellum (data not shown). The microcephaly was apparent prenatally, as brains of E18.5 MagohMos2/+ embryos were notably smaller, with the cerebral cortex disproportionately reduced in size (Fig. 2a,b,d,e). The cortical layers of E18.5 MagohMos2/+ brains were thinner and somewhat disorganized (Fig. 2c,f). Cux1-positive neurons, produced late in neurogenesis and residing in outer layers of the cerebral cortex (layers II-IV), did not form a cohesive layer in MagohMos2/+ (Fig. 2g,h,k,l). In contrast, older and deeper layers that are produced early in neurogenesis (layers V-VI), marked by Foxp1, Foxp2, and Tbr1, were present but markedly thinner in MagohMos2/+ brains relative to control (Fig. 2i,j,m–v, Supplementary Fig. 3a). MagohMos2/+ E18.5 brains also contained 60% fewer Foxp1-, Foxp2- and Tbr1-positive neurons (Supplementary Fig. 3b). Overall these analyses show that MagohMos2/+ brains contain fewer neurons and that multiple neuronal layers are dramatically affected.
Figure 2. E18.5 MagohMos2/+ brains contain disorganized layers and reduced neurons.
a,d, Whole mount brains, b,e, Nissl stained sagittal sections, and c,f, H & E stained coronal sections from indicated genotypes. g–v, Confocal micrographs of coronal sections from indicated genotypes stained with antibodies against Cux1 (g,h,k,l), Foxp1 (i,j,m,n), Foxp2 (o,p,s,r), and Tbr1 (q,r,u,v). The neuronal layers they mark are in parentheses. The boxes in g,i,k,m,o,q,s, and u indicate the regions shown in h,j,l,n,p,r,t, and v, respectively. Scale bars, 1 mm (a,b,d,e), 100 µm (c, f, g–v).
Evaluation of younger MagohMos2/+ embryos indicated that the onset of microcephaly occurs at approximately E12.5. E10.5 and E11.5 control and MagohMos2/+ cortices showed similar thickness and overall size, and were properly patterned as evidenced by in situ hybridization with Wnt1, Gli3, and Bmp4a (Supplementary Fig. 3c, data not shown). However, cortical thickness of MagohMos2/+ embryos was reduced 10% at E12.5, 25% at E13.5, and 50% at E14.5 relative to controls. Consistent with a role in neurogenesis, we detected MAGOH protein expression throughout the cortex from E11.5 to E16.5, spanning the onset and peak of neurogenesis (Supplementary Fig. 1d). At E14.5, Magoh mRNA expression is enriched in the VZ and SVZ of the cortex, regions composed of NSCs and INPs (Supplementary Fig. 1e).
Magoh is required for proper numbers of INPs
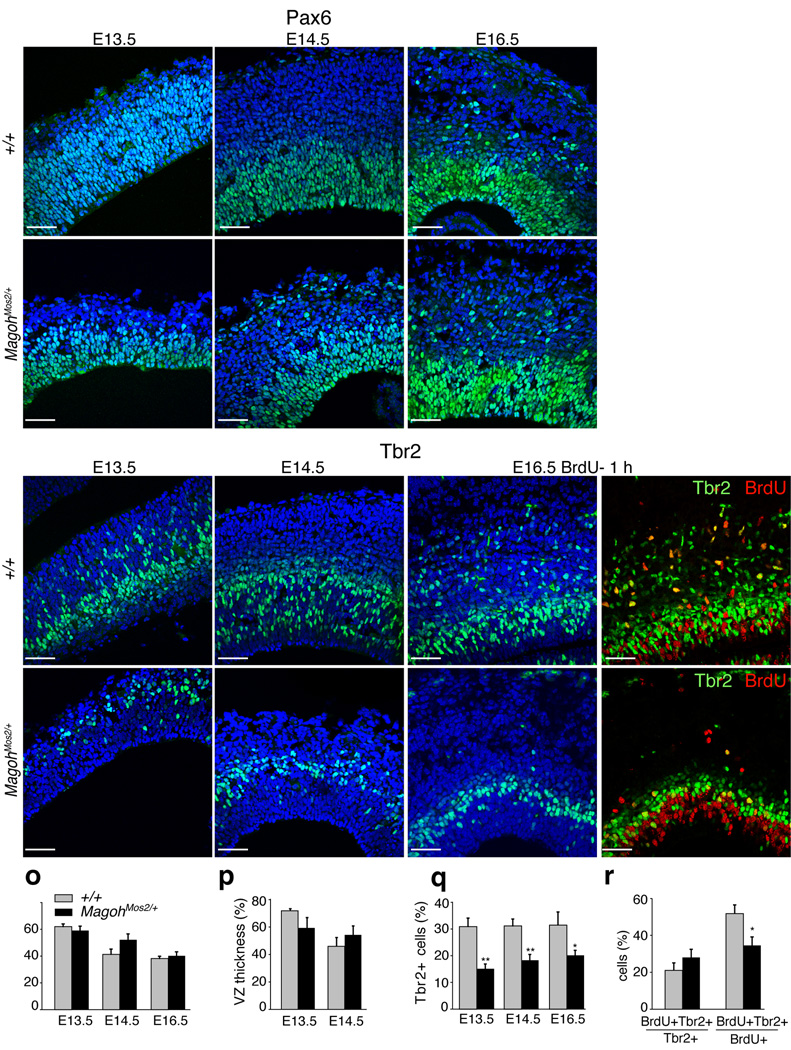
The Magoh expression pattern, combined with the neuronal depletion in MagohMos2/+ embryos suggested that the microcephaly reflects defects in NSCs and/or INPs. At E13.5, E14.5, and E16.5, the density of Pax6-positive NSCs and thickness of the VZ layer was similar in control and MagohMos2/+ (Fig. 3a–f,o,p). In contrast, Tbr2-positive cells of the SVZ and intermediate zone (IZ) layers showed a striking reduction in INPs in MagohMos2/+, apparent at E13.5, E14.5 and E16.5 (Fig. 3g–n,q). Consistent with this, MagohMos2/+ brains showed a four-fold reduction in the total number of BrdU-positive cells in the SVZ and IZ at E16.5 (P<0.005) (Supplementary Fig. 4a,b). The density of BrdU-positive cells remained constant, but the proportion of BrdU-positive cells expressing Tbr2 was significantly reduced in MagohMos2/+ (P<0.05) (Fig. 3i,j,m,n,r, Supplementary Fig. 4c). In support of these findings, both microarray and RT-PCR analyses showed that in MagohMos2/+ cortices, Pax6 mRNA levels were normal while Tbr2 mRNA levels were reduced (Supplementary Fig. 4d, Supplementary Table 2). Together these analyses indicate that Magoh is required for maintenance and/or generation of INPs. As INPs produce a large fraction of neocortical neurons, this phenotype is consistent with the reduction of neurons in E18.5 MagohMos2/+ brains.
Figure 3. Magoh is required for proper numbers of INPs but not NSCs.
a–n, Confocal micrographs of coronal sections from indicated genotypes and ages stained with DAPI (blue) and antibodies against Pax6 (green) (a–f), Tbr2 (green) (g–n), and BrdU (red) (j,n). o, Graph depicting the percentage of total DAPI cells that are Pax6+ from indicated genotypes at E13.5, E14.5, and E16.5. p, Graph depicting the thickness of the ventricular zone (VZ), as a percentage of total cortical thickness, from indicated genotypes at E13.5 and E14.5. q, Graph depicting the percentage of total DAPI cells that are Tbr2+ from indicated genotypes at E13.5, E14.5, and E16.5. r, Graph depicting the percentage of Tbr2+ cells that are BrdU+ (left) and the percentage of BrdU+ cells that are Tbr2+ (right). Pregnant females were dissected 1 hour after BrdU injection. For (o,p,q,r) the average values for all embryos (n=2–4 per genotype, 4–5 sections per embryo) is shown, quantitated within a 318 µm2 field. No asterisk indicates no significant differences were observed. *, P<0.05, **, P<0.005; Error bars, s.d. Scale bar (a) all images, 50µm.
Ectopic neuron differentiation and apoptosis in MagohMos2/+
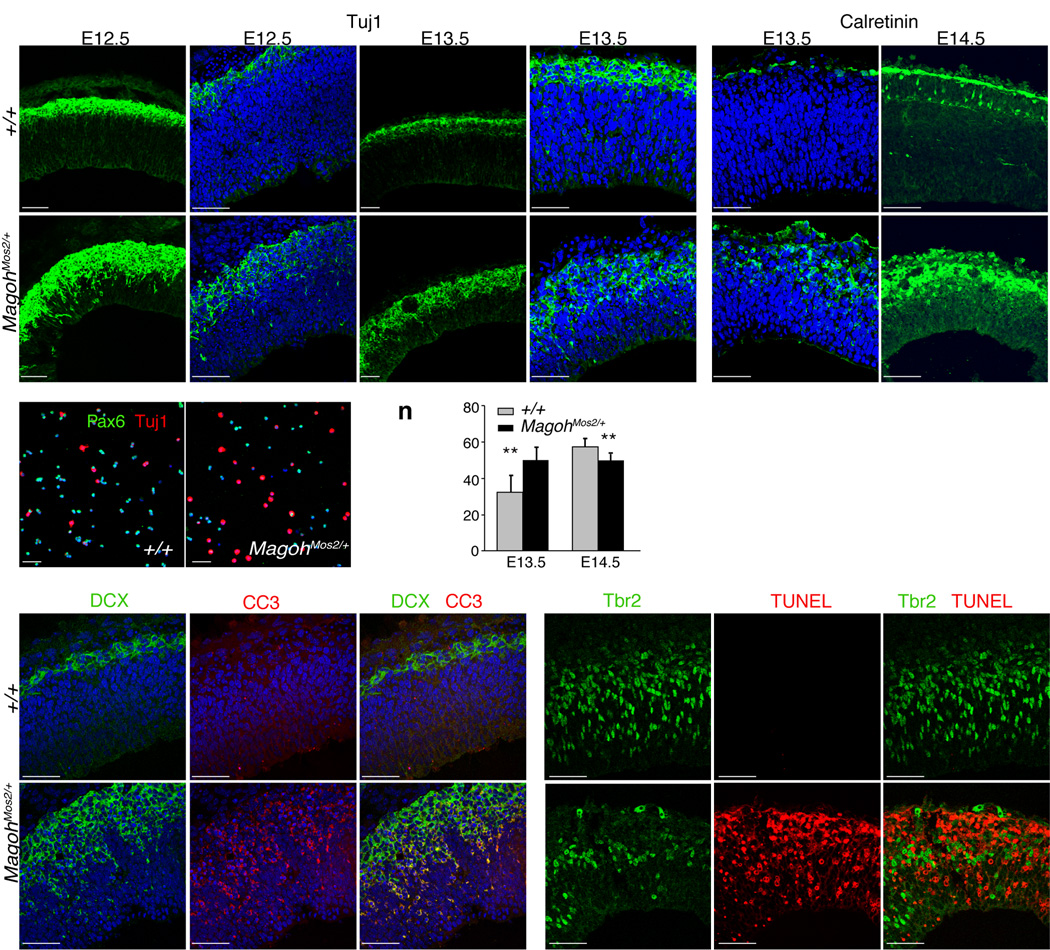
The INP depletion in MagohMos2/+ suggested that the balance of asymmetric divisions might be disrupted resulting in increased neuron production. To test this hypothesis, we evaluated new neuron production by monitoring Tuj1 and DCX expression. At E10.5 and E11.5, no neuronal defects were observed in MagohMos2/+ brains (data not shown, Supplementary Fig. 5a,b). However, beginning at E12.5 and continuing through E13.5, Tuj1 and DCX-positive cells were increased in MagohMos2/+ (Fig. 4a–d,g–j, Supplementary Fig. 5c–h). This finding was confirmed by quantitation of dissociated TuJ1-positive neurons of control and MagohMos2/+ cortices (compare 17.5% ± 3.3 to 25.8% ± 5.7) (P<0.0005) (Fig. 4m). Calretinin-positive cells were also expanded at E13.5 and E14.5, indicating that in MagohMos2/+, Cajal Retzius (CR) cells are ectopically produced (Fig. 4e,f, ,k,l). Consistent with these findings, in E13.5 MagohMos2/+ embryos the IZ and cortical plate (CP) layers were significantly thicker than in control embryos (P<0.005) (Fig. 4n). Together these results indicate that Magoh is required to prevent ectopic and precocious neurogenesis.
Figure 4. Magoh is required to prevent premature neuronal differentiation and apoptosis.
a–l, Low magnification (a,c,g,i) and high magnification (b,d,e,f,h,j,k,l) confocal micrographs of coronal sections from indicated genotypes and ages, stained for Tuj1 (green) and DAPI (blue) (a–d,g–j), and Calretinin (green) and DAPI (blue) (e,f,k,l). m, Confocal micrographs of dissociated E12.5 cortical cells from indicated genotypes stained for Pax6 (green), Tuj1 (red), and DAPI (blue). n, Graph depicting the thickness of the cortical plate (CP) and intermediate zone (IZ), as a percentage of total cortical thickness. The average value for all embryos (n=3–4 per genotype, 4–5 sections per embryo) is shown, quantitated within a 318 µm2 field. o–q, u–w, Confocal micrographs of E12.5 coronal sections of indicated genotypes stained for DCX (green) (o,u), CC3 (red) (p,v), DCX (green), CC3 (red) (q,w). r–t, x–z, Confocal micrographs of E14.5 coronal sections of indicated genotypes stained for Tbr2 (green) (r,x), TUNEL (red) (s,y), Tbr2 (green), TUNEL (red) (t,z). **, P<0.005; Error bars, s.d. Scale bars, 50 µm.
Although E12.5 and E13.5 MagohMos2/+ embryos contained ectopic neurons, E18.5 MagohMos2/+ embryos had fewer neurons relative to control littermates, suggesting that the ectopic neurons did not survive. In MagohMos2/+ but not control brains, we observed increased apoptotic cells (cleaved-caspase3 (CC3) and TUNEL-positive) between the SVZ and CP in a pattern similar to migratory neurons (Fig. 4o–z, Supplementary Fig. 5a–f). Co-localization of CC3 with Tuj1 and DCX confirmed that a majority of dying cells were new neurons (Fig. 4v,w, Supplementary Fig. 5). In contrast, Tbr2-positive cells were not TUNEL-positive, indicating that INPs did not undergo cell death (Fig. 4r–t,x–z). These results show that in MagohMos2/+, the majority of ectopically produced neurons undergo cell death. In summary, we conclude that the microcephaly in MagohMos2/+ embryos is due to both increased neuronal apoptosis and depletion of the neuron-producing INP population.
Magoh and core EJC components regulate the mitotic spindle
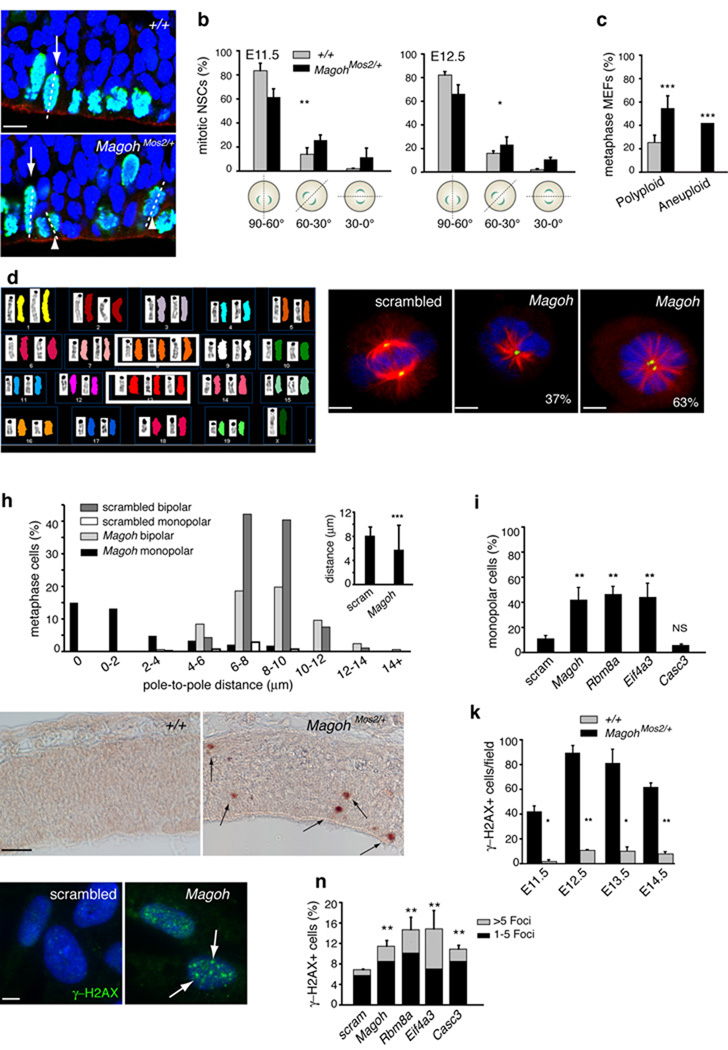
In microcephaly mutants such as Lis1hc/ko, NSC and INP depletion, ectopic neurogenesis, and neuronal apoptosis are associated with abnormal orientation of the NSC mitotic division plane24–26. In E10.5 MagohMos2/+ embryos, evaluation of dividing cells using phospho-histone H3 (PH3) showed no defects in division or PH3 number (Supplementary Fig. 6a,b). However, at E11.5 and E12.5, intermediate and horizontal divisions were significantly increased in MagohMos2/+ embryos (P<0.005, P<0.05, respectively) (Fig. 5a,b). The onset of this spindle defect at E11.5, prior to massive apoptosis or premature differentiation, suggests the spindle defect is not an artifact of general tissue disorganization (see Supplementary Fig. 5i–l). Thus, we conclude that Magoh is required for proper orientation of the NSC mitotic division plane.
Figure 5. Magoh and core EJC components regulate the mitotic spindle, ploidy, mitosis, and genomic stability.
a, Confocal micrographs of E11.5 coronal sections stained with DAPI (blue), rhodamine phalloidin (red) and for PH3 (green), with metaphase cells dividing vertically (arrows), and with intermediate orientation (arrowheads) indicated. b,c, Graphs depicting percentage of NSCs exhibiting indicated mitotic cleavage planes at E11.5 (left) and E12.5 (right) (b) and MEFs exhibiting polyploidy and aneuploidy (c). d, Representative SKY image from MagohMos2/+ MEF depicting aneuploidy (3 copies of chromosomes 8, 13). e–g, Confocal micrographs of HeLa cells treated with scrambled (e) and Magoh(f,g) siRNA, and stained with DAPI (blue), for α-Tubulin (green) and γ-Tubulin (red). Percentages indicate Magoh siRNA-treated monopolar cells containing one (f) or two (g) centrosomes. h, Graphs depicting percentages of siRNA-treated metaphase cells exhibiting indicated pole-to-pole distances, for bipolar and monopolar cells independently (left) and together (inset). i, Graph depicting percentages of siRNA-treated cells that are monopolar. j, Images of E11.5 coronal sections stained for γ-H2AX (brown, arrows). k, Graph depicting γ-H2AX+ cells at indicated ages. l,m, Images of siRNA-treated RPE cells stained with DAPI (blue) and for γ-H2AX+ foci (green, arrows). n, Graph depicting percentages of siRNA-treated RPE cells exhibiting >5 γ-H2AX+ (grey) and 1–5 γ-H2AX+ (black) foci. Graphs indicate average values for 2–4 embryos per genotype (b,c,k), for all siRNA-treated cells (h), and for 3–6 independent experiments (i,n). *, P<0.05, **, P<0.005, ***, P<0.0005; Error bars, s.d.; Scale bars, 5 µm (e, f, g), 10 µm (a,m), 50 µm (l).
The apoptosis exhibited by MagohMos2/+ neurons could result from aberrant chromosome numbers due to defective NSC division. Consistent with this, metaphase analysis indicated that MagohMos2/+ mouse embryonic fibroblasts (MEFs) had a two-fold increase in polyploid cells compared to control (P<0.0005) (Fig. 5c). Spectral Karyotyping (SKY) analysis of metaphase spreads also uncovered increased chromosomal aneuploidy in MagohMos2/+ MEFs compared to control (P<0.0005) (Fig. 5d). Aneuploidy was random relative to the number and type of chromosomes affected. These results demonstrate that Magoh is required to maintain chromosome number during mitosis.
To further evaluate the requirement of Magoh for cell division, siRNA knockdown in HeLa cells was used to deplete MAGOH levels (Supplementary Fig. 6c). Analysis of the mitotic spindle using α-Tubulin revealed that 42% of dividing Magoh knockdown cells failed to form a bipolar spindle and instead displayed an apparent monopolar spindle (P<0.005) (Fig. 5e–g,h,i). As spindle defects are frequently caused by dysfunctional centrosomes, we assessed centrosome number and integrity using γ-Tubulin, a pericentriolar marker of centrosomes. In Magoh knockdown cells, the average centrosome distance was reduced (5.78 µm versus 8.1 µm in control, P<0.0005) (Fig. 5h). Of cells exhibiting monopolar spindles, 37% contained a single centrosome and spindle, indicating these were bona fide monopolar spindles (Fig. 5f,h). 63% of cells with monopolar spindles contained two adjacently located centrosomes, with the majority exhibiting distances less than 4 µm apart (Fig. 5g,h, Supplementary Fig. 6d). Knockdown of the other core EJC components Rbm8a and Eif4a3, but not Casc3, caused spindle defects similar to Magoh knockdown (P<0.005, P<0.005, P=0.065, respectively) (Fig. 5i). Using FACS analysis, we observed an average two-fold increase in the proportion of cells in G2/M phases, confirming the disruption of mitosis (Supplementary Fig. 7a). Together these data show that EJC components are required for cells to proceed from prophase into metaphase, for proper centrosome number and separation, and for integrity of the bipolar mitotic spindle.
Given the widespread apoptosis in MagohMos2/+ embryos, and involvement of microcephaly-associated genes in DNA damage repair10,15, we evaluated if Magoh was required for genome stability using the double-stranded break marker γ-H2AX. In contrast to control brains, in MagohMos2/+, the number of γ-H2AX-positive cells was significantly increased beginning at E11.5 (Fig. 5j,k, Supplementary Fig. 7b). We extended this analysis to assess the role of additional EJC components in genome stability using siRNA depletion in RPE cells. Reduced expression of Magoh, Eif4a3, Rbm8a and Casc3 each resulted in significantly increased DNA damage foci (Fig. 5l,m,n, Supplementary Fig. 7c). Taken together these analyses show that EJC components are required for mitosis and DNA integrity, both processes disrupted in microcephaly.
Magoh acts upstream of LIS1 to regulate neurogenesis
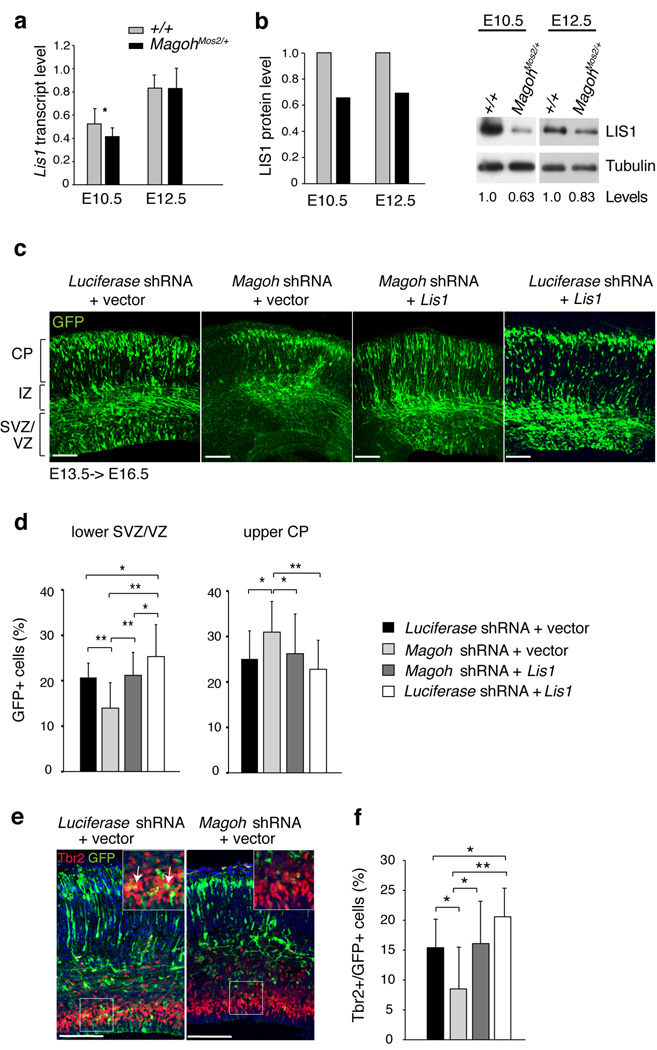
Similar to MagohMos2/+, Lis1−/− mutants exhibit microcephaly, altered NSC mitotic cleavage planes, precocious neurogenesis, increased apoptosis, and reduced neural progenitors24–26. Given the EJC’s role in transcript regulation, and similarities between Magoh and Lis1 mutant phenotypes, we hypothesized that Magoh regulates brain size in part by controlling LIS1 expression. Inspection of Lis1 mRNA levels in MagohMos2/+ cortices showed minimal changes at E10.5 (22% reduced) and no changes at E12.5 (Fig. 6a). In contrast, Western analyses showed that LIS1 protein levels were reduced in both MagohMos2/+ E10.5 and E12.5 cortices (by 34% and 30%, respectively) (Fig. 6b, Supplementary Fig. 8a). A significant reduction in LIS1 levels was also independently detected by quantitative 2D gel analysis of mutant E12.5 cortices (20% reduction) (P<0.00005) and by Western analyses of Magoh knockdown HeLa cells (50% reduction) (data not shown, Supplementary Fig. 8b). These changes in LIS1 expression are not reflective of global alterations in steady state mRNA and protein levels in E10.5 MagohMos2/+ cortices, as expression profile analysis identified only 147 transcripts (0.8% of genes on the microarray) with altered levels (P<0.05) (Supplementary Table 2). The vast majority of these changes were less than two-fold higher or lower indicating that the observed expression changes were not dramatic. In addition, 2D gel analyses uncovered only 8 significantly altered proteins (0.4%) (P<0.05). Consistent with our quantitative RT-PCR analyses, the mutant/wild-type ratio for Lis1 by microarray was 0.85 (P=0.1166). Thus, Magoh is required for proper LIS1 expression, suggesting that altered LIS1 levels may account for some phenotypes observed with Magoh haploinsufficiency.
Figure 6. Magoh acts upstream of the microcephaly-associated protein LIS1 to regulate neurogenesis.
a,b, Graphs representing Lis1 gene expression measured by quantitative PCR (a) and LIS1 protein expression measured from Western blot analyses (b). b, Representative cropped Western blots of cortical lysates from indicated ages and genotypes, probed with antibodies against LIS1 (46 kDa) and α-Tubulin (55 kDa) as a loading control. Below the lanes are densitometry values normalized for loading, with control normalized to 1.0. The full-length blots are presented in Supplementary Fig. 8e. c, Confocal micrographs of coronal sections from E16.5 brains in utero co-electroporated at E13.5 with pCAG-GFP (green) and the following: Luciferase shRNA + vector, Magoh shRNA + vector, Magoh shRNA + Lis1 Luciferase shRNA + Lis1. Note that in Magoh shRNA + vector brains there are fewer GFP+ cells in the SVZ/VZ layers. d, Graph depicting the percentage of GFP+ cells in lower SVZ/VZ and upper CP layers of the brain for indicated in utero electroporations. e, Confocal micrographs of brains from indicated in utero electroporations showing GFP (green) and stained for Tbr2 (red) and DAPI (blue). The inset shows a higher magnification view of the boxed region, depicting co-localization (yellow) of Tbr2 (red) and GFP (green). f, Graph depicting the percentage of GFP+ cells that express Tbr2 for indicated in utero electroporations. Graphs in (d,f) show averages of all sections. *, P<0.05, **, P<0.005; Error bars, s.d.; Scale bars, 100 µm.
We tested this hypothesis by asking whether Lis1 addition is sufficient to rescue phenotypes associated with Magoh loss of function. We utilized in utero electroporation to introduce GFP along with either Magoh shRNA or Luciferase shRNA (control) into E13.5 cortices, and then analyzed the distribution of GFP-positive cells at E16.5. GFP-positive cells in control electroporated brains were relatively evenly distributed throughout the CP, IZ, and SVZ/VZ layers (Fig. 6c,d, Supplementary Fig. 8c). However, in Magoh shRNA electroporated brains, GFP-positive cells were significantly increased in the upper CP layer and decreased in the lower SVZ/VZ layer (P<0.05 and P<0.005, respectively) (Fig. 6c,d), consistent with the neurogenesis defects observed in MagohMos2/+ brains. We observed similar distribution defects upon knockdown with two additional shRNA constructs against Magoh and upon knockdown with different dosages of Magoh shRNA, further evidencing the specificity of these phenotypes (data not shown and Supplementary Fig. 8d). The proportion of Magoh shRNA electroporated cells that were Tbr2-positive was also significantly reduced relative to the control (P<0.05) (Fig. 6e,f). This indicates that in utero knockdown of Magoh recapitulates the depletion of INPs in MagohMos2/+ brains.
Next, we evaluated if Lis1 addition could rescue defects caused by Magoh knockdown. Brains co-electroporated with Lis1 expression vector and Magoh shRNA showed GFP-positive cells distributed evenly throughout the cortex, similar to control brains (Fig. 6c,d, Supplementary Fig. 8c). The distribution in the upper CP and lower SVZ/VZ layers was significantly different from that of Magoh shRNA alone (P<0.05 and P<0.005, respectively), indicating that Lis1 expression rescued the altered distribution caused by Magoh knockdown (Fig. 6c,d). Also consistent with a genetic relationship, an increase in GFP-positive cells in the SVZ/VZ seen with Lis1 expression alone12, was abrogated in the co-electroporated brains (Fig. 6c,d, Supplementary Fig. 8c). Lis1 and Magoh shRNA co-electroporation also rescued the loss of Tbr2-positive cells (Fig. 6e,f). Together these results demonstrate that: Lis1 is sufficient to restore Magoh-depleted cells to their proper distribution and fate, Magoh is a critical regulator of LIS1 levels, and LIS1 is one of the key downstream targets of Magoh that regulates brain development.
Discussion
Despite the fundamental importance of the core EJC in regulating RNA metabolism, the requirement of EJC components in vertebrate development has not been evaluated. Here we utilized mouse genetics to uncover a new cellular requirement for the EJC in mitosis and to show that mutation of Magoh disrupts brain size as a result of defective NSC division, INP depletion and neuronal apoptosis. Our study indicates that Magoh is an essential regulator of stem cell maintenance and division and thus has implications not only for microcephaly syndromes, but also for chromosome integrity and stem cell disorders, such as cancer.
We have demonstrated that core EJC components regulate mitosis by modulating mitotic spindle integrity, in part by regulating centrosome separation and duplication. However, EJC components may also control mitotic spindle integrity by regulating microtubule organization, such as in Drosophila oogenesis where the Magoh ortholog, mago, is required for microtubule polarization27. Together this indicates that core EJC components have a fundamental, conserved, but largely unexplored role in microtubule organization and cell division.
The requirement of the EJC in mitosis is manifested as altered chromosome number and loss of genome integrity. The observed polyploidy and aneuploidy are consistent with defects in centrosomes and mitotic progression. Delays in mitotic progression also likely explain the increased frequency of spontaneous DNA damage. Of note, depletion of the NMD component UPF1 also causes increased DNA damage, but does so by disrupting DNA replication28. While we cannot rule out a similar role for the EJC in replication, there is no evidence of the core EJC associating with DNA or of an increase in S phase cells upon loss of EJC components, as measured by FACS analysis or by BrdU incorporation in E16.5 brains. Thus, our working model is that defects in mitosis precede and result in accumulated DNA damage.
We propose that in MagohMos2/+ mice, defective mitosis alters cell fates and causes microcephaly. Either misoriented or dysfunctional mitotic spindles may interfere with production and/or maintenance of INPs and neurons by altering distribution of fate determinants4,6. Hence we predict that neuronal apoptosis is caused by inheritance of aberrant chromosome number and/or damaged DNA. The observed alterations in cell fate are also consistent with an overall shift in the balance of asymmetric, proliferative and neurogenic NSC divisions (Supplementary Fig. 9). Interestingly, this function is conserved across kingdoms, as knockdown of mago in spermatogenesis of the plant, Marsilea, skews the balance of symmetric and asymmetric cell divisions29.
A key regulator of mitosis is LIS1, and our analyses indicate that an essential function of Magoh in brain development is to regulate LIS1 levels. The microarray, quantitative real time PCR and Western analyses together showed that in Magoh mutants, LIS1 protein levels are more significantly reduced than Lis1 mRNA levels. This suggests Magoh may regulate LIS1 protein expression, an observation consistent with the established role for Magoh and the EJC in regulating protein translation19,22. Hypomorphic or hypermorphic Lis1 levels disrupt neurogenesis, leading to microcephaly12,25,30. We show that restoring normal LIS1 levels is sufficient to rescue neurogenesis phenotypes of cellular distribution and progenitor number caused by Magoh knockdown. Consistent with a genetic relationship, MagohMos2/+ and Lis1−/− mutants have similar phenotypes24,25. A few differences exist, such as the onset of spindle orientation defects, which in Lis1−/− is evident at E9.5, but in MagohMos2/+ is first evident at E11.526. However, the Mos2 mutation is heterozygous and causes LIS1 reduction, but not loss, so the difference in mutant phenotypes may be due to LIS1 dosage.
It is important to note that Magoh mutants also exhibit cellular phenotypes not previously associated with Lis1 function, including increased Cajal-Rejtius cells, DNA damage and ploidy. This may indicate potentially new roles for LIS1, particularly in genomic stability. Alternatively, these Magoh mutant phenotypes may be due to dysregulation of other critical targets of Magoh. Through our electroporation experiment, we have uncovered LIS1 as a critical target of Magoh in generating progenitors between E13.5 and E16.5. However it is unlikely that LIS1 is the only critical target of Magoh, and that the complex process of neurogenesis that initiates prior to E13.5 is dependent upon additional downstream genes. Future studies will reveal for example, whether any of the genes altered in the microarray or proteomics experiments are also essential targets of Magoh.
Our findings implicate core EJC components in the pathogenesis of microcephaly syndromes. Of note, MAGOH is found within a 55-gene deletion on 1p32.3 associated with mental retardation and brain size abnormalities31. In addition, RBM8A is one of 15 genes in a 0.4 Mb microdeletion on 1q21.1 associated with microcephaly32,33. Therefore, the core EJC functions we have uncovered open up a fruitful area of research for the role of this complex in brain development and disease, stem cell maintenance, and chromosome anomaly disorders such as cancers.
METHODS
Mapping and sequencing of mutations
Initial mapping of Mos2 was performed as previously described using hypopigmentation as criteria for affected animals18. For fine mapping, both affected and unaffected animals were genotyped using SSLP markers and SNPs polymorphic between BALB/cJ and C57BL/6J strains. P values were calculated using Chi-square analyses. PCR and sequencing of genomic DNA was performed by Harvard Partners Healthcare Center for Genetics and Genomics, Harvard Medical School.
Quantitation
The thickness of the cortex, VZ (Pax6-positive layer), IZ and CP (TuJ1-positive layer), and E18.5 neuronal layers was measured from confocal micrographs using Zeiss AIM software. Measurements of E18.5 length and width were made from micrographs. Quantitation of BrdU, Tuj1, Tbr2, and Pax6-positive cells was performed from confocal micrographs using ImagePro6.3 and ImageJ software. For analysis of E12.5 cortical cultures, only cells that were TuJ1 positive and Pax6 negative were included in measurements. For this analysis, the average value was measured for n=3 embryos each genotype, n=3345 cells, n=2457 cells for control and MagohMos2/+, respectively. Measurements of co-localization in cells (Tbr2, BrdU and Tbr2, GFP) were made using Adobe Photoshop CS3. Mitotic orientation of dividing NSCs was calculated on micrographs by measuring the angle of chromosomes relative to the ventricle as detected using DAPI and anti-PH3 staining to mark prophase and metaphase cells and either a cytoplasmic marker or membrane marker to visualize the ventricular surface. Centrosome distances were calculated from confocal Z stacks (0.44 µm slices) using ImagePro6.3 software. Centrosome distances were calculated on the following: scrambled bipolar (n=268), scrambled monopolar (n=12), Magoh bipolar (n=201), and Magoh monopolar (n=143). For measurement of average centrosome distance of all siRNA treated cells, scrambled (n=280) and Magoh (n=345) were analyzed. Centrosome number was calculated from visual inspection of confocal Z stacks. For analysis of monopolar spindles the following number of cells were evaluated: scrambled (n=281 cells), Magoh (n=629 cells), Rbm8a (n=340 cells), Eif4a3 (n=335 cells), and Casc3 (n=235 cells). Analysis of γ-H2AX+ cells in sections was calculated within a 350 µm2 field. For all siRNA analyses significance was calculated relative to the scrambled control. For in utero electroporation experiments, GFP-positive cell location was quantified from confocal micrographs by dividing the cortex into 5 equal bins, using Adobe Photoshop CS3. For each genotype, all stainings were repeated on several sections each from at least three independent embryos.
Mouse Genetics and husbandry
The MagohMos2/+ line was generated as previously described, and maintained on a C57BL/6 background18. MagohGT0150/+ and MagohGT027/+ mice were generated from ES cells containing gene trap insertions from Sanger. Briefly, chimeras were made by injecting 4–6 ES cells into 8-cell stage C57BL/6 embryos, and blastocysts were injected into pseudo-pregnant C57BL/6 females. Chimeras with high ES cell contribution (evaluated using coat color) were crossed to C57BL/6 mice and evaluated for germline transmission. BAC constructs were purchased from CHORI (Children’s Hospital Oakland Research Institute) and CsCl purified (Lofstrand Labs, Gaithersburg, MD). Tg-BAC1 and Tg-BAC2 lines were made by injecting RP24-a175K18 and RP23-274G4 BAC constructs, respectively, into C57BL/6 blastocysts. For this study, C57BL/6 inbred animals (purchased from The Jackson Laboratory) were used as a control and outcrossed MagohMos2/+ mice were used (>10 generations on C57BL/6J). BrdU injections were performed intraperitoneally (50 µg/g body weight). For embryo collection, 0.5 days (E0.5) gestation was designated upon identification of a plug in a female. All mice described were bred and housed in an NHGRI animal facility according to NIH guidelines. Animal care was in accordance with NHGRI institutional standards and was approved by the IACUC, NHGRI, NIH Institutional Review Board.
Genotyping
MagohMos2/+ mice were genotyped using a real-time PCR Taqman™ SNP genotyping assay on an ABIPrism7000. For this assay, a region containing the point mutation is amplified by PCR. Two fluorescently labeled single-stranded oligonucleotides (probes), one complementary to the wild-type product, and one complementary to the mutant product are multiplexed in the assay. Allelic discrimination was performed to detect the relative amount of each allele at the conclusion of the PCR reaction. The following cycling conditions were used: 1. 50°C for 2 minutes, 2. 95°C for 10 minutes, 3. 92°C for 30 seconds, 4. 60°C for 60 seconds (3–4, 40 cycles). The following primers and probes were used: GGCCCCTTTAGGCTCATACATTTTA (forward primer), ATCAATAATTCTCTTTAACTCTTCCATCACACT (reverse primer), CACATAAGCCTGCAATC (VIC probe, C57BL/6 allele), and CACATAAGCTGCAATC (FAM probe, MagohMos2 allele) (designed using Applied Biosystems software, Foster City, CA). BAC animals were genotyped by traditional PCR to detect the presence of the BAC construct in genomic DNA using the following cycling conditions: 1. 94°C for 3 minutes, 2. 93°C for 30 seconds, 3. 58°C for 30 seconds, 4. 72°C for 30 seconds, (steps 2–4, 35 cycles) 5. 72°C for 7 minutes. The following primers were used: Forward: CGCTGCAAAACGCTGACGGAACAGTAG, and reverse: CTCAGCGTATGGTTGTCGCCGGATGTAT.
RT-PCR and Sequencing
RNA from E10.5 or E12.5 cortex was used for first-strand cDNA synthesis using a High Capacity cDNA Reverse Transcription Kit (ABI, Foster City, CA) according to manufacturers instructions. PCR amplification was subsequently done using primers within exon 1 and 4 with the following conditions: 1. 94°C for 3 minutes, 2. 94°C for 30 seconds, 3. 57°C for 90 seconds, 4. 72°C for 30 seconds, (steps 2–4, 35 cycles) 5. 72°C for 10 minutes. The following primers were used: Forward: CGTTACTACGTGGGCCACAA, and Reverse: GGTTGACATCAATAAGGGAACC. RT-PCR products were digested with AluI (New England Biolabs) before gel electrophoresis. Site-directed mutagenesis was used to generate a mutant Magoh cDNA construct containing the point nucleotide deletion (198delG) found in Mos2 mutants (New England Biolabs). RT-PCR products from wild-type and mutant cortices were commercially sequenced (ACGT, Wheeling, IL) using Big Dye terminator cycle sequencing reagents (ABI). For both the AluI digest and for sequencing, a mock heterozygote cDNA sample was generated as a control by combining wild-type and mutant cDNA plasmid DNA in a 1:1 mix before PCR amplification.
Quantitative gene Expression
Gene expression was measured in E10.5 and E12.5 cortical RNA using the following Taqman™ real-time PCR gene expression assays: Magoh (Mm00487546_m1); Lis1/Pafah1b1 (Mm00443070_m1); Pax6 (Mm00443081_m1); Tbr2/Eomes (Mm01351984_m1). Expression was measured relative to a standard curve diluted from wild-type E12.5 embryonic head control cDNA. Expression for each sample was normalized to Gapdh expression measured using the same standard curve and wild-type levels were set to 1.0. 20 µl reactions were carried out in 1X Fast Universal PCR Master Mix (ABI) with the following cycle conditions: 1. 95°C for 20 seconds, 2. 95°C for 3 seconds, 3. 60°C for 30 seconds, (steps 2–3, 40 cycles). For all expression data, graphs were averaged from 3–4 replicate reactions for each of 4–6 control and MagohMos2/+ cortices.
Metaphase and SKY analyses
Metaphase preparations were performed by standard air-drying technique34. Fluorescent In Situ Hybridization was performed with Spectrum Orange (Vysis, Downers Grove, IL)-labeled DNA by nick translation technique, essentially as described35,36. SKY analysis was carried out as described37,38.
Western Analyses and Quantitation
E10.5 and E12.5 cortices were dissected and frozen at −80°C. MagohMos2/+ and control embryos from the same litter were pooled and lysed in RIPA lysis buffer containing protease inhibitors (Pierce, Rockford, IL). Cortical extracts of mutant and wild-type samples from multiple litters were run on either 16% or 4–12% SDS-Polyacrylamide gels (Invitrogen) and analyzed by Western blotting using the following primary antibodies: mouse anti-Magoh (1:100, Santa Cruz), rabbit anti-Magoh (1:100, Abcam), mouse anti-LIS1 (1:1000, Sigma), and mouse anti-α-Tubulin at 1:1000 (Sigma); and secondary antibodies at 1:20,000: anti-rabbit HRP and anti-mouse HRP (Amersham, Piscataway, NJ). Western blots were quantitated by densitometry (Image J). Values for each lane were first normalized to α-Tubulin and the average ratio of mutant to wild-type was calculated for each blot. For LIS1 measurements, data was averaged for E10.5 from 3 blots including 8 litters (8 wild-type, 8 mutant samples), and for E12.5 from 7 blots including 13 litters (12 wild-type, 11 mutant samples). Samples were run in replicates of 1–3 blots.
Cell culture analysis and Immunofluorescence
HeLa cells were grown to approximately 55% confluency and transfected with 50 nM siRNAs (or 12 nM each for 4 siRNAs and 6 nM each for 8 siRNAs) (Qiagen, Valencia, CA) using siLentFect (Bio-Rad, Hercules, CA). Cells were grown for 48 hours, and then harvested for either FACS cell cycle analysis, Western analysis, or plated onto chamber slides for an additional 12 hours. Expression of both Magoh and Magoh2 (a homolog with 99% amino acid identity) was targeted, as HeLa cells express both genes. The following siRNAs were used: Control: AllStars Negative siRNA; MAGOH1: Hs_Magoh_1, 2, 5, 6; MAGOH2: Hs_FLJ10292_6,7,8,9; RBM8A: Hs_Rbm8a_1, 5, 6, 7; EIF4A3: Hs_Eif4a3_2, 3, 5, 6; CASC3: Hs_Casc3_5, 6, 7, 8. RPE cells were transfected with 100 nM siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were treated at 0 and 24 hours. RNA was collected at 48 hours and cells were fixed and stained at 72 hours. Dissociated cortical cultures were prepared from E12.5 embryos and stained as previously described39. HeLa cells were plated onto coverslips and sub-confluent cells were fixed in methanol at −20°C for 20 minutes, and stained as previously described40. The following mouse antibodies were used: mouse anti-α-tubulin (1:500, Sigma, Saint Louis, MO), mouse anti-TuJ1 (1:1000), rabbit anti-γ-tubulin (1:500, Sigma), anti-γ-H2AX (1:2000, Millipore); and rabbit anti-Pax6 (1:2000, Millipore). Analysis was performed using microscopes described below.
Staining of mouse Sections
Embryos (E12.5 and younger) or brains (E13.5 and older) were dissected and fixed overnight in 4% paraformaldehyde, washed in 1X PBS, soaked sequentially in 10 and 20% sucrose overnight, and frozen in NEG-50 medium (Richard-Allan Scientific, Kalamazoo, IL). Coronal sections (12–19 µm thick) were prepared using a Leica CM3050S cryostat, and staining was performed using primary antibodies (either 2 hours at room temperature or overnight at 4°C) and secondary antibodies (15 minutes at room temperature) diluted and first blocked in 10% normal goat serum. For staining of nuclear antigens, sections were first permeabilized with 0.25% triton-X 100 for 10 minutes. For BrdU staining, sections were first heated at 55°C in 1N HCl for 15 minutes or for double immunofluorescence, were boiled in Citric Acid Antigen Unmasking solution for 30 minutes (Vector Laboratories). Stained sections were mounted using Vectashield with DAPI or with Propidium Iodide (Vector Laboratories). Stained sections were analyzed primarily with a Zeiss510LSM NLO Confocal Microscope with Meta Detector and also with a Zeiss Fluorescence Axioskop2 microscope with IPLab imaging software. Images were cropped as necessary using Adobe Photoshop CS3. The following antibodies were used: Rabbit: anti-cleaved Caspase-3 (1:200, Cell Signaling, Danvers, MA), anti-phospho-Histone H3 (PH3) (1:200, Upstate Biotechnology, Charlottesville, VA), anti-Magoh (1:200, Proteintech, Chicago, IL), anti-Cux1 (1:100, Santa Cruz, Santa Cruz, CA), anti-Foxp1 (1:200, Abcam, Cambridge, MA), anti-Foxp2 (1:100, Abcam), anti-Tbr1 (1:500, Abcam), anti-Tbr2 (1:1000, Abcam), anti-Calretinin (1:200, Millipore, Billerica, MA), anti-Pax6 (1:1000, Millipore), Mouse: anti-TuJ1 (1:400, Covance, Berkeley, CA), Goat: anti-doublecortin (1:50, Santa Cruz); anti-γ-H2AX (1:2000, Millipore); Rat: rat anti-BrdU (1:200, Abcam). Secondary antibodies: Alexafluor 488, 568, and 594 (1:200, Invitrogen, Eugene, OR), mouse anti-HRP (Roche). TUNEL staining was performed according to manufacturer’s directions using Apoptag Fluorescein in situ apoptosis kit (Millipore). Nissl and H & E staining of paraffin sections was performed according to standard procedures (Histoserv, Gaithersburg, MD). In situ hybridization images were taken from genepaint.org 41.
In Utero electroporation
Gene transfer into embryonic CD1 mice was performed using in utero electroporation (IUE) as previously described with the Nepagene CUY21EDIT electroporator42. E13.5 embryos were electroporated with 3.7 µg DNA in a total 1 µl volume (2 µg of either pSM2-Luciferase-shRNA (#RHS1705) or pSM2c-Magoh-shRNA (#RMM1766–984609047) accompanied by 0.7 µg of pCAGGS-GFP and as noted, 1 µg each of pCAGGS-Lis1, and either pCAGGS-empty or pCLE-IRES-Plap). For empty vectors either pCAG or pCLE were used, with similar results. E16.5 embryonic brains were prepared as described above. ShRNAs were purchased from Open Biosystems. Lis1 was subcloned into pCAGGS vector from a pcDNA3-Lis1 construct (Addgene). Graphs include analysis of the following: Luciferase shRNA + vector (n=13 sections, 5 brains), Magoh shRNA + vector (n=13 sections, 4 brains), Magoh shRNA + Lis1 (n=22 sections, 8 brains), Luciferase shRNA + Lis1 (n=22 sections, 9 brains). For quantitation of Tbr2+GFP+ cells, analysis included n=13–22 sections, 4–9 brains, each electroporation.
Microarray and Proteomic analyses
Microarray was performed as previously described using 5 biological replicates each of RNA prepared from E10.5 MagohMos2/+ and control cortices 43. To ensure reproducible dissections, the olfactory epithelium was included in cortical dissections. Proteomic analyses were performed using 3 biological replicates each of protein prepared from MagohMos2/+ and control cortices. Proteomic analyses and statistical analyses of data were performed by Applied Biomics, CA.
Statistical analysis
Supplementary Table 3 lists all statistical tests used for data analysis and actual P values calculated.
Supplementary Material
Acknowledgements
We thank Pavan laboratory members for helpful advice including Laura Baxter for reading of the manuscript. For technical assistance we thank the following: Art Incao (mouse husbandry), Gene Elliot and Amy Chen (mouse transgenics), Amalia Dutra, Eugenia Pak, and Stephen Witchovitch (metaphase, SKY, microscopy assistance), Stacie Anderson and Martha (FACS analysis), Bhavesh Bhorate (microarray statistics), Mark Bryant (pathology analysis), Harvard Partners Center for Genetics and Genomics (candidate gene sequencing), Julia Fekecs and Darryl Leja (Figure assistance). This research was funded in part by an NIGMS PRAT fellowship and K99/R00 Pathway to Independence Award (to D.L.S.), by the Intramural Research program of NIH/ NHGRI (to W.J.P., K.M.), by NIH/NHGRI (to N.G.). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession numbers. The data obtained in our microarray experiments were deposited at the GEO website under accession number GSE 19168.
Note: Supplementary Information is available on the Nature Neuroscience website.
Author Contributions D.L.S. designed the study, performed all experiments except as noted, and wrote the paper. D.E.W. performed all mRNA analyses and assisted with quantitative analyses. K.C.S. and T.J.P. performed all in utero electroporations and dissections of electroporated brains. D.M.L. performed staining of E18.5 markers. A.J.P. assisted with HeLa cell analyses, and quantitation of NSCs and INPs. H.L. performed RPE cell analyses. D.L.S., W.J.P., C.A.W., N.G., and K.M. were involved in research design and all authors were involved in data analysis. D.L.S. and W.J.P. prepared the manuscript. All authors have agreed to the content in the manuscript, including the data as presented.
Reprints and permissions information are available at www.nature.com/reprints.
REFERENCES
- 1.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 2.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;Vol. 30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 4.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 5.Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 7.Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 9.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AP, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.do Carmo Avides M, Glover DM. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- 12.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faulkner NE, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;Vol. 2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 14.Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 15.Griffith E, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 17.Rauch A, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 18.Matera I, et al. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 2008;17:2118–2131. doi: 10.1093/hmg/ddn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. Embo J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 24.Pawlisz AS, et al. Lis1-Nde1-dependent neuronal fate control determines cerebral cortical size and lamination. Hum Mol Genet. 2008;17:2441–2455. doi: 10.1093/hmg/ddn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gambello MJ, et al. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yingling J, et al. Neuroepithelial stem cell proliferation requires Lis1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micklem DR, et al. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- 28.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 29.van der Weele CM, Tsai CW, Wolniak SM. Mago nashi is essential for spermatogenesis in Marsilea. Mol Biol Cell. 2007;18:3711–3722. doi: 10.1091/mbc.E06-11-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada M, et al. Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly. Nat Med. 2009;15:1202–1207. doi: 10.1038/nm.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulatinho M, Llerena J, Leren TP, Rao PN, Quintero-Rivera F. Deletion (1)(p32.2-p32.3) detected by array-CGH in a patient with developmental delay/mental retardation, dysmorphic features and low cholesterol: A new microdeletion syndrome? Am J Med Genet A. 2008;146A:2284–2290. doi: 10.1002/ajmg.a.32454. [DOI] [PubMed] [Google Scholar]
- 32.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundsteen C, Lind AM. A test of a climate room for preparation of chromosome slides. Clin Genet. 1985;28:260–262. doi: 10.1111/j.1399-0004.1985.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 35.Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 36.Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liyanage M, et al. Multicolour spectral karyotyping of mouse chromosomes. Nat Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 38.Schrock E, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 39.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 40.Kittler R, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 41.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- 43.Buac K, et al. NRG1 / ERBB3 signaling in melanocyte development and melanoma: inhibition of differentiation and promotion of proliferation. Pigment Cell Melanoma Res. 2009;22:773–784. doi: 10.1111/j.1755-148X.2009.00616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.