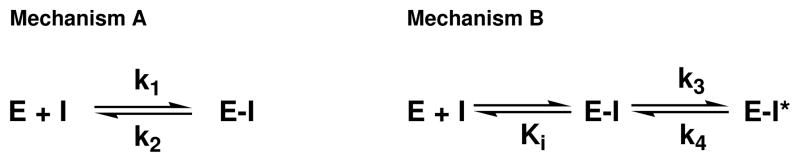Abstract
Inhibition of siderophore biosynthetic pathways in pathogenic bacteria represents a promising strategy for antibacterial drug development. E. coli synthesize and secrete the small molecule iron-chelator siderophore, enterobactin, in response to intracellular iron depletion. Here we describe a detailed kinetic analysis of EntE, one of six enzymes in the enterobactin synthetase gene cluster. EntE catalyzes the ATP-dependent condensation of 2, 3-dihydroxybenzoic acid (DHB) and phosphopantetheinylated EntB (holo-EntB) to form covalently arylated EntB, a product that is vital for the final assembly of enterobactin. Initial velocity studies show that EntE proceeds via a Bi Uni Uni Bi ping-pong kinetic mechanism with a kcat equal to 2.8 s−1 and Km values of 2.5, 430, 2.9 μM for DHB, ATP, and holo-EntB-ArCP, respectively. Inhibition and direct binding experiments suggest that, during the first half-reaction (adenylation), DHB binds first to the free enzyme, followed by ATP and the release of pyrophosphate to form the adenylate intermediate. During the second half-reaction (ligation), phosphopantetheinylated EntB binds to the enzyme followed by the release of products, AMP and arylated EntB. Two hydrolytically-stable adenylate analogues, 5′-O-[N-(salicyl)sulfamoyl]adenosine (Sal-AMS) and 5′-O-[N-(2, 3-dihydroxybenzoyl)sulfamoyl]adenosine (DHB-AMS), are shown to act as slow-onset tight-binding inhibitors of the enzyme with appKi values of 0.9 and 3.8 nM, respectively. Direct binding experiments, via isothermal titration calorimetry, reveal low picomolar dissociation constants for both analogues to EntE. The tight-binding of Sal-AMS and DHB-AMS to EntE suggests that these compounds may be developed further as effective antibiotics targeted to this enzyme.
Keywords: enterobactin, EntE, steady-state kinetics, antibacterial agents
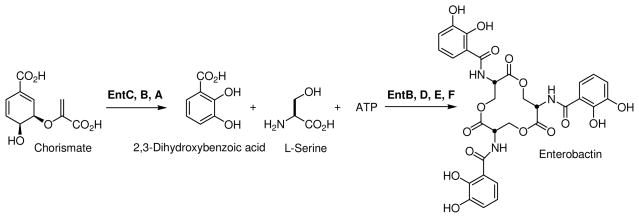
Iron is an essential element that is vital for the growth and virulence of most pathogenic bacteria (1). Key biological processes, such as oxygen transport, amino acid synthesis, respiration, and DNA biosynthesis, require micromolar concentrations of intracellular iron (2). However, in vertebrate hosts the concentration of free iron is too low to support bacterial growth (3). Consequently, many bacterial species have evolved mechanisms to capture ferric iron from their surroundings and deliver it to cells for metabolic processes via the biosynthesis of small molecule iron-chelators known as siderophores (4). The siderophore molecule enterobactin, a cyclic trimeric lactone of N-(2,3-dihydroxybenzoyl)-serine, is synthesized and secreted by Escherichia coli, as well as other species of enteric bacteria, in response to intracellular iron starvation (5). Enterobactin is the product of the enterobactin synthetase, which is a non-ribosomal peptide synthetase (NRPS) composed of six genes, entA-F (3).
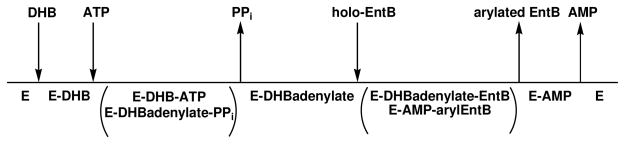
During the first phase of enterobactin biosynthesis, a central metabolite produced by the shikimate pathway, chorismate, is converted to 2, 3-dihydroxybenzoate (DHB) via the sequential catalytic activities of EntC, -B, and –A (3, 6). EntB, -D, -E, and –F are then required to catalyze the ATP-dependent assembly of enterobactin from three molecules each of DHB and L-serine (Scheme 1) (3). Briefly, EntD, a phosphopantetheinyl transferase, uses coenzyme A to phosphopantetheinylate S245 of the aryl carrier protein domain (ArCP) of EntB (3, 7). Next, EntE catalyzes the transfer of DHB onto the phosphopantetheinylated (holo) EntB to yield the covalently arylated EntB (5). Finally, arylated EntB, ATP, and L-serine are used as substrates for the reaction catalyzed by EntF to generate enterobactin (5, 8).
Scheme 1.
E. coli enterobactin biosynthesis occurs via a non-ribosomal peptide synthetase composed of six genes (entA-F).
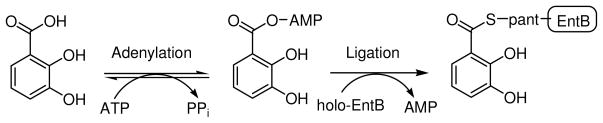
In addition to being essential for bacterial growth, enterobactin synthetase is absent in mammals and thus the enzyme components of this pathway represent promising targets for novel antibacterial agents (9). Moreover, previous studies have revealed that the inhibition of enzymes involved in siderophore assembly, including EntE homologues, can abolish siderophore biosynthesis and result in bacterial death (9–11). Belonging to the family of aryl acid adenylating enzymes (AAAE), EntE is characterized by a two-step adenylation/ligation reaction: first, the enzyme catalyzes the condensation of DHB and ATP to form an adenylate intermediate, followed by the ligation of DHB onto the phosphopantetheinylated cofactor that is bound to the ArCP of EntB (Scheme 2) (12). The product of the EntE reaction, arylated EntB, serves as the aryl donor for amide bond formation in the final assembly of enterobactin, and thus the EntE product is crucial for the completion of enterobactin biosynthesis (5).
Scheme 2.
Adenylation/Ligation reaction catalyzed by EntE.
In this study, we report the steady-state kinetic parameters and kinetic mechanism of the E. coli dihydroxybenzoate-AMP ligase, EntE. Furthermore, we show the inhibition of this enzyme by two hydrolytically-stable adenylate analogues that act as slow-onset tight-binding inhibitors. The mechanistic and inhibition studies provided herein reveal new details of the EntE reaction and thus may facilitate the development of novel antibacterial agents targeted to the enterobactin synthetase.
MATERIALS AND METHODS
Materials
All chemicals were purchased from Sigma-Aldrich Chemical Co. Enzymes used in molecular cloning were supplied by New England Biolabs. Plasmid pET-28a(+) and Escherichia coli strain BL21(DE3) were obtained from Novagen.
Expression and Purification of EntE
The recombinant plasmid containing the entE gene from E. coli (a generous gift from Andrew Gulick) was transformed into competent E. coli BL21(DE3) cells (13). The transformed cells were used to inoculate 6 L of LB containing 50 μg/mL ampicillin. The culture was grown to mid-log phase (A600 ~ 0.8) at 37°C, then induced by the addition of 0.5 mM IPTG, and further incubated overnight at 18°C.
All purification procedures were performed at 4°C. The cells were harvested by centrifugation and suspended in buffer A [20 mM Tris, pH 8.0, 200 mM NaCl, and 20 mM imidazole] containing protease inhibitors and DNase I (0.1 μg/mL). The cells were then lysed by sonication, and cell debris was removed by centrifugation at 38000 × g for 45 min. The supernatant was loaded onto a Ni-NTA column pre-equilibrated with buffer A, and washed with 10 column volumes of the same buffer. The bound proteins were eluted with a linear imidazole gradient (from 20 to 250 mM) at a flow rate of 1 mL/min. Pure fractions, as determined by SDS-PAGE, were pooled and dialyzed overnight against a buffer containing 20 mM Tris, pH 8.0, 0.5 mM EDTA, 0.1 mM DTT, and 10% glycerol. The protein was concentrated by centrifugation through an Amicon concentrator with a 30 kDa cutoff membrane to a final concentration of 4 mg/mL.
Cloning, Expression, and Purification of EntB-ArCP
The aryl carrier protein domain (residues 188-285) of the entB gene from E.coli was sub-cloned from the recombinant plasmid containing the entB gene into a pET23a(+) vector (Novagen) using the PCR primers EntBf (5′-GATTCCATATGTCCCTGAAATATGTGGCCG-3′) and EntBr (5′-GAATTCCTCGAGTTTCACCTCGCGGGAGAG-3′) containing the underlined NdeI and XhoI restriction sites, respectively (13). The recombinant plasmid, harboring the ArCP domain of the entB gene, bearing an N-terminal His6 tag, was transformed into competent E. coli BL21(DE3) cells. The transformed cells were used to inoculate 6 L of LB containing 100 μg/mL ampicillin. The culture was grown to mid-log phase (A600 ~ 0.8) at 37°C, then induced by the addition of 1 mM IPTG, and further incubated overnight at 20°C.
EntB-ArCP was purified using Ni-NTA affinity as described above for EntE. Fractions containing EntB-ArCP, as determined by SDS-PAGE, were pooled and dialyzed overnight against buffer B [20 mM Tris, pH 8.0, 2 mM DTT, and 10% glycerol]. The protein was then concentrated to 4 mL and applied to a Superdex S-75 column, pre-equilibrated with buffer A (excluding imidazole). Pure fractions, as determined by SDS-PAGE, were pooled, dialyzed overnight against buffer B, and concentrated by ultrafiltration to a final concentration of 4.2 mg/mL in a total volume of 9 mL.
Phosphopantetheinylation of EntB-ArCP by Sfp
Sfp phosphopantetheinyl transferase was used to transfer the phosphopantetheinyl group from Coenzyme A to S245 of EntB-ArCP. The recombinant plasmid containing the Bacillus subtilis sfp gene (a generous gift from Jun Yin) was transformed into E. coli, expressed, and purified as described by Yin and coworkers (7), (14). To convert the apo form of EntB-ArCP to the phosphopantetheinylated form, i.e. holo-EntB-ArCP, the following reaction mixture was prepared: 200 μM apo-EntB ArCP, 200 μM Coenzyme A, 100 mM HEPES, pH 7.8, 10 mM MgCl2, and 4 μM Sfp. After incubation at 25°C for hour, the reaction resulted in phosphopantetheinylated EntB-ArCP, as confirmed by Fourier transform mass spectrometry, and was directly used in steady-state assays.
Enzyme Activity Assay
Initial velocities of the EntE reaction were assayed spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyruvate kinase, and lactate dehydrogenase as described previously (15). The decrease in absorbance of reduced nicotinamide (NADH) at 340 nm (ε340 = 6220 M−1s−1) was measured at 25°C using a UVIKON XL spectrophotometer. The standard reaction contains 100 mM HEPES at pH 7.8, 10 mM MgCl2, 250 mM NaCl, 1 mM PEP, 0.15 mM NADH, 18 units of myokinase, 18 units of pyruvate kinase, and 18 units of lactate dehydrogenase in addition to substrates in a final volume of 1 mL. After incubation for 5 min at 25°C, reactions were initiated by the addition of EntE, typically at a final concentration of 14 nM, and followed for ~ 2 min. EntE enzymatic activities were corrected for the background activity, i.e., the decrease in absorbance at 340 nm caused by ATP hydrolysis. The rate of arylated EntB-ArCP formation is proportional to the rate of NADH oxidation, where two molecules of NADH are oxidized for each molecule of arylated EntB formed.
Initial Velocity Experiments
Kinetic constants for ATP were determined at fixed, saturating concentrations of both holo-EntB-ArCP (20 μM) and DHB (80 μM) and at variable concentrations of ATP (0.1 – 5 mM). Kinetic constants for aryl acid substrates were determined at fixed, saturating concentrations of both holo-EntB-ArCP (20 μM) and ATP (5 mM). Kinetic constants for holo-EntB-ArCP and D-pantethine were determined at fixed, saturating concentrations of both ATP (5 mM) and DHB (80 μM). Initial velocities were determined with at least five different concentrations of the varied substrate. Individual substrate saturation kinetic data were fitted to eq 1 using Sigma Plot 11.0:
| (1) |
where V is the maximal velocity, A is the substrate concentration, and K is the Michaelis-Menton constant (Km). Initial velocity patterns were determined at various concentrations of one substrate in the presence of different fixed concentrations of a second substrate with the concentration of the third substrate kept saturating and constant. Initial velocity data were fit to eq 2 for a parallel pattern and to eq 3 for an intersecting pattern:
| (2) |
| (3) |
where Ka and Kb are the Michaelis constants for the varied substrates A and B, and Kia is the dissociation constant for substrate A. Dead-end inhibition studies with AMPCPP (a non-hydrolyzable analogue of ATP) were performed at various concentrations of one substrate, a fixed concentration of a second substrate, and the concentration of the third substrate kept saturating and constant while in the presence of different fixed concentrations of AMPCPP (0 – 0.5 mM). Inhibition data were fit to eq 4 for a competitive pattern and to eq 5 for an uncompetitive pattern:
| (4) |
| (5) |
where I represents the concentration of inhibitor, and Kis and Kii are the inhibition constants for the slope and intercept terms, respectively.
Bisubstrate Analogue Inhibition Experiments
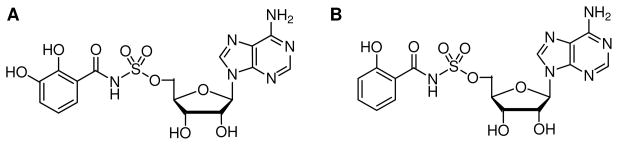
Two hydrolytically-stable adenylate analogues, 5′-O-[N-(salicyl)sulfamoyl]adenosine (Sal-AMS) and 5′-O-[N-(2, 3-dihydroxybenzoyl)sulfamoyl]adenosine (DHB-AMS) were tested as inhibitors of EntE (Scheme 3). Initial velocities of the EntE reaction were assayed spectrophotometrically using the coupled assay described using an Applied Photophysics Stopped-Flow spectrophotometer. Assays were performed at saturating DHB, saturating holo-EntB-ArCP, saturating ATP, and varied concentrations of either Sal-AMS or DHB-AMS. Reactions were performed by mixing 0.06 mL of the substrate-containing solution with 0.06 mL of 16 nM enzyme and the reaction was followed at 25°C for 4 min at 340 nm. The observed lag due to the coupling enzymes was corrected for by removing the first 20 seconds from all time-courses. Time-courses for each inhibitor concentration were determined five times and an average was calculated and used for subsequent data analysis. The time-courses were fit to eq 6 for slow-onset kinetics:
| (6) |
where [P]t is the concentration of product formed, vf is the final steady-state velocity, vi is the initial velocity, t is time, and kobs is the rate constant for conversion of the initial velocity to the final velocity (16). The dependence of kobs on inhibitor concentration was fit to eq 7 for the slow-association mechanism and eq 8 was used to calculate the appKi value:
| (7) |
| (8) |
where k2 and k1 are the rate constants shown in mechanism A, [I] is the concentration of the inhibitor, and appKi is apparent inhibition constant (Scheme 4) (16).
Scheme 3.
Structures of bisubstrate analogues. (A) 5′-O-[N-(2,3-dihydroxybenzoyl) sulfamoyl]adenosine (DHB-AMS) and (B) 5′-O-[N-(salicyl) sulfamoyl]adenosine (Sal-AMS).
Scheme 4.
Two possible kinetic mechanisms can explain non-linear kinetics: one-step slow-association (A) and two-step isomerization (B).
The intrinsic Ki for Sal-AMS was determined using the coupled assay as described previously for initial velocity experiments. Inhibition by Sal-AMS was performed at various concentrations of ATP (0.3 – 1.5 mM), different fixed concentrations of DHB (10, 50, and 80 μM), with the concentration of holo-EntB-ArCP kept saturating and constant and in the presence of different fixed concentrations of Sal-AMS (0 – 10 nM). Inhibition data were fit to eq 4 for a competitive pattern, yielding an appKi value for Sal-AMS at each concentration of DHB assayed. The linear dependence of appKi on DHB concentration was fit to eq 9 to determine the intrinsic Ki of Sal-AMS:
| (9) |
where Ki is the true inhibition constant for Sal-AMS, appKi is the apparent inhibition constant, A is the concentration of non-varied substrate (DHB), and Kia is the dissociation constant for substrate A.
Isothermal Titration Calorimetry Experiments
ITC titration experiments to investigate the binding affinity of Sal-AMS and DHB-AMS to EntE were performed using a Microcal VP-ITC microcalorimeter (Northhampton, MA). All measurements were carried out at 20°C in 30 mM Tris buffer pH 8.0 with 1 mM MgCl2. EntE was dialyzed (2 × 1L) against the above buffer, and all ligand solutions were prepared in the final dialysate. Protein and ligand concentrations of EntE, DHB-AMS, Sal-AMS, and salicylic acid were determined using experimentally derived values of ε280 = 58 790 M−1 cm−1, ε254 = 18 450 M−1cm−1, ε250 = 15 774 M−1 cm−1, and ε295 = 3785 M−1 cm−1, respectively. In individual titrations, injections of ligands were made into the enzyme solution. The quantity c = KAMt(0), where Mt(0) is the initial macromolecular concentration, is of importance in titration microcalorimetry (17). Experiments to determine ΔH (the binding enthalpy change in kcal mol−1) were performed with a c value in the range 6.7–39.9 × 104 while displacement ITC experiments were performed with a c value in the range 145–531. Ligand and protein concentration for the determination of ΔH were 70 μM Sal-AMS and DHB-AMS with 5 μM and 2.5 μM EntE respectively. The KA (the association constant in M−1) and n (the number of binding sites per monomer) values were determined in displacement ITC experiments with 200 mM salicylic acid added to both the enzyme and ligand solutions. Protein concentrations for the displacement ITC experiments, were increased to 5 μM for titration with DHB-AMS (70 μM) and 10 μM for titration with Sal-AMS (150 μM). All titrations were performed with a stirring speed of 307 rpm and a 300–600 s interval between 10 μL injections. The initial injection was not used for data fitting. Titrations were run past the point of enzyme saturation to determine and correct for heats of dilution. The experimental data for the stoichiometric titration of ligands into protein provided ΔH values. The experimental data for the competitive displacement ITC experiments were fitted to a theoretical titration curve using the Origin software package (version 7.0) provided with the instrument to afford values of KAapp and n (18). The KA for Sal-AMS and DHB-AMS were obtained from these KAapp values using eq 10:
| (10) |
where B is [salicylic acid] = 0.2 M and the KAB = 4.74 × 103 M−1 (12). The thermodynamic parameters ΔG and ΔS were calculated from eq 11:
| (11) |
where ΔG, ΔH, and ΔS are the changes in free energy, enthalpy, and entropy of binding. T is the absolute temperature, and R = 1.98 cal mol−1 K−1. The affinity of the ligand to protein is given as the dissociation constant (KD = 1/KA). Competitive and direct ITC were performed in three and two independent experiments respectively and analyzed independently, and the thermodynamic values obtained were averaged.
ITC titration experiments to determine the binding affinity of DHB to EntE were performed using a Microcal VP-ITC microcalorimeter. All measurements were performed at 25°C in 50 mM HEPES buffer pH 7.8 with 10 mM MgCl2. EntE was dialyzed (2 × 1L) against the above buffer, and all ligand solutions were prepared in the final dialysate. The protein concentration was determined using ε280 = 58 790 M−1 cm−1 and the concentration of DHB was determined by weight. The 1.46 mL sample cell was filled with a 50 μM protein solution and the 250 μL injection syringe with a 1 mM DHB solution which was injected at a rate of 5 μL every 230 s into the sample cell. The data were fit using the One Set of Sites Model provided in Origin 7.0 to determine KA, ΔH, and n.
RESULTS AND DISCUSSION
Cloning, Expression, and Purification of EntE
PCR amplification of the entE gene yielded a single fragment of the expected length (1611 bp). DNA sequencing of the cloned fragment confirmed the expected sequence and the absence of any mutations. Expression of the PCR product resulted in a soluble protein product with an apparent molecular mass, determined by SDS-PAGE, in agreement with the mass of 61 kDa deduced from the amino acid sequence. Approximately 13 mg of purified enzyme was obtained per liter of culture.
Cloning, Expression, and Purification of EntB-ArCP
PCR amplification of the EntB-ArCP gene yielded a single fragment of the expected length (288 bp). DNA sequencing of the cloned fragment confirmed the expected sequence and the absence of any mutations. Expression of the PCR product resulted in a soluble protein product with an apparent molecular mass, determined by SDS-PAGE, in agreement with the mass of 12 kDa deduced from the amino acid sequence. Approximately 6.2 mg of purified protein was obtained per liter of culture.
Expression and Purification of Sfp
Expression of the sfp gene resulted in a soluble protein product with an apparent molecular mass, determined by SDS-PAGE, in agreement with the mass of 26 kDa deduced from the amino acid sequence. Approximately 9.6 mg of purified enzyme was obtained per liter of culture.
Initial Velocity Experiments
Kinetic parameters were determined for the three substrates of the EntE reaction revealing Km values of 2.5 ± 0.3, 430 ± 30, 2.9 ± 0.6 μM for DHB, ATP, and holo-EntB-ArcP, respectively, with a kcat equal to 2.8 ± 0.1 s−1 (Table 1). It was previously reported that EntB is a bifunctional protein: its N-terminus is an isochorismate lyase domain required for production of DHB and its C-terminus serves as an aryl carrier protein during the assembly of enterobactin (3). In addition, Gehring and colleagues have shown that the N-terminal 187 amino acids of EntB are not needed for the reaction of EntB with either EntD or EntE, and therefore we used only the ArCP domain (residues 188-285) for our kinetic analysis (3). Moreover, S245 of the EntB ArCP domain is phosphopantetheinylated by EntD, a phosphopantetheinyl transferase, resulting in the formation of phosphopantetheinylated EntB, which is the holo-form required for EntE catalysis (3). As described in Material and Methods, we used Sfp phosphopantetheinyl transferase to covalently transfer the 4′-phosphopantetheinyl group from coenzyme A onto S245 of apo-EntB-ArCP (14). The stoichiometric conversion of apo to holo-EntB-ArCP using this method was confirmed by Fourier transform mass spectral analysis (data not shown).
Table 1.
Kinetic Parameters of EntE1
| substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|
| ATP | 430 ± 30 | 2.8 ± 0.1 | 3.9 × 105 |
| holo-EntB-ArCP | 2.9 ± 0.6 | 2.8 ± 0.2 | 9.8 × 105 |
| D-pantetheine | 34200 ± 2000 | 0.9 ± 0.1 | 2.6 × 101 |
| DHB | 2.5 ± 0.3 | 2.8 ± 0.1 | 8.8 × 105 |
| salicylic acid | 70 ± 4 | 0.8 ± 0.1 | 1.1 × 104 |
| 4-aminosalicylic acid | 3100 ± 300 | 4.4 ± 0.2 | 1.5 × 104 |
| 3-hydroxybenzoic acid | 70 ± 8 | 0.30 ± 0.01 | 4.6 × 103 |
| 4-aminobenzoic acid | N/A | N/A | N/A |
Initial velocities of the EntE reaction were assayed spectrophotometrically using the coupled assay as described in the text. All assays were performed at 25°C and at pH 7.8.
Our present kinetic data reveal that the apo-form of EntB-ArCP is not a substrate for the EntE reaction, thus revealing the necessity of the phosphopantetheinyl modification of EntB for EntE substrate specificity. When D-pantetheine was tested as a substrate in place of holo-EntB-ArCP, the catalytic efficiency of EntE dropped four orders of magnitude due mostly to its decreased binding affinity to the enzyme. (Table 1). This significant decrease suggests that, in addition to the phosphopantetheine group, crucial interactions between EntB and EntE are necessary for EntE activity. In addition to our present kinetic data, there is an increasing amount of evidence for a direct interaction between EntB and EntE, thus highlighting the requirement of protein-protein interactions in the enterobactin biosynthetic machinery (13, 19).
Although an EntE crystal structure is not yet available, the three-dimensional structure of DhbE, a homologous B. subtilis protein involved in the biosynthesis of bacillibactin, may provide insight into the substrate binding in the EntE active site (20). To aid this endeavor and to elucidate structural components necessary for binding and/or catalysis, we tested additional aryl acids as substrates for EntE (Table 1). EntE has been shown to catalyze the adenylation of salicylic acid and we show here that EntE can also catalyze the arylation of holo-EntB-ArCP using salicylic acid, albeit with a catalytic efficiency decreased by one order of magnitude compared to that with DHB (21). 3-hydroxybenzoic acid was also tested as a substrate and it binds to EntE with a Km equal to that of salicylic acid, though the kcat is decreased by a factor of 2. We also tested 4-aminosalicylic acid as a possible EntE substrate and it too is a suitable substrate, but with a binding affinity decreased by three orders of magnitude when compared to that of DHB. However, when 4-aminobenzoic acid was tested as a substrate for EntE, no activity was observed, thus showing the necessity at least one hydroxyl group, either in the 2 or 3 position, for aryl acid binding to EntE.
In the two substrate-bound structures of DhbE (i.e. DHB and AMP bound and AHB-adenylate bound), the side-chain of the conserved N235 hydrogen bonds with the 2-hydroxyl group of DHB and the 3-hydroxyl group hydrogen bonds to the side chain hydroxyl of S240 (20). Alignment of DhbE and EntE sequences reveals that both N235 and S240 are conserved between EntE and DhbE (Figure S1). The ability of EntE to use both salicylic acid and 4-aminosalicylic acid as substrates, two molecules lacking the 3-hydroxyl group, demonstrates that the formation of a hydrogen bond at the conserved active site S240 is important, but not vital for binding and/or catalysis. Likewise, the ability of the enzyme to use 3-hydroxybenzoic acid as a substrate reveals that formation of a hydrogen bond between the conserved N235 and 2-hydroxyl group is preferred, but not essential for enzyme turnover. Conversely, the inability of EntE to use 4-aminobenzoic acid as a productive substrate suggests that the formation of at least one hydrogen bond, either between the conserved N235 and the 2-hydroxyl group and/or the conserved S240 and the 3-hydroxyl group, is imperative for substrate binding and subsequently, enzyme turnover. Therefore, EntE achieves maximal catalytic efficiency when the aryl acid substrate contains hydroxyl groups in both the 2 and 3 positions, i.e. DHB, its natural substrate. Adenylation domains have been suggested to act as gatekeepers of the NRPS assembly line, thus these enzymes are often selective for substrate recognition and activation (8, 20, 22). As suggested for DhbE, our data support that the conserved active site residues Asn and Ser also serve as the major determinates for the substrate specificity of EntE (20).
The kinetic mechanism of EntE was determined by initial velocity experiments using the coupled assay. The parallel lines observed in double-reciprocal plots of the initial velocity using either ATP or DHB as the varied substrate and holo-EntB-ArCP as the fixed, variable substrate, suggest a ping-pong kinetic mechanism for EntE (Figure 1). The intersecting lines observed in double-reciprocal plots of the initial velocity by varying concentrations of ATP and DHB at a fixed concentration of holo-EntB-ArCP are indicative of the formation of a ternary E-DHB-ATP complex (Figure 1). These data were fit to eq 2 or 3, yielding Km values for ATP, DHB, and holo-EntB-ArCP equivalent to those determined by individual substrate saturation kinetic data (Table 1).
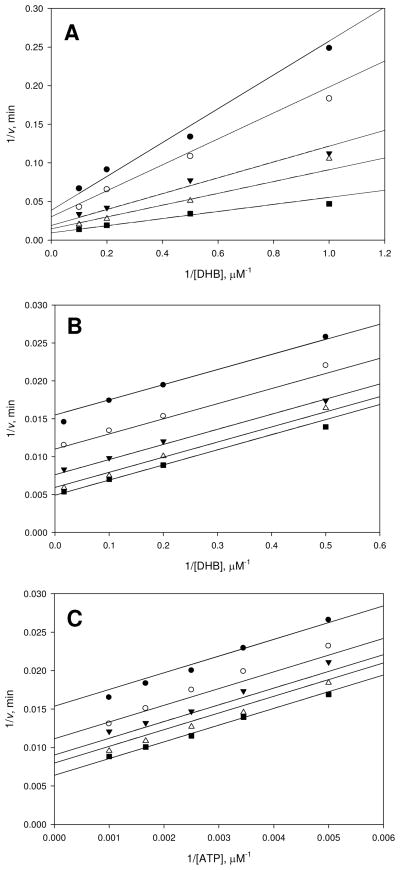
Figure 1.
Kinetic mechanism of EntE. (A) Assays were performed at varying concentrations of DHB (1–10 μM), saturating holo-EntB-ArCP (20 μM), and fixed concentrations of ATP: 150 (●), 200 (○), 350 (▼), 500 (△), and 1000 μM (■). (B) Assays were completed at varying concentrations of DHB (2–60 μM), saturating ATP (5 mM), and varying concentrations of holo-EntB-ArCP: 3 (●), 5 (○), 10 (▼), 20 (△), and 50 μM (■). (C) Assays were performed at varying concentrations of ATP (200–1000 μM), saturating DHB (80 μM), and varying concentrations of holo-EntB-ArCP: 3 (●), 5 (○), 7.5 (▼), 10 (△), and 20 μM (■). Initial velocities were measured spectrophotometrically in 100 mM HEPES pH 7.8 and 10 mM MgCl2 using the coupled assay as detailed in Materials and Methods. Points are experimental and the lines are global fits of the data to eq 3 for A, and eq 2 for B and C.
Pyrophosphate (PPi) was used to further probe the nature of the ping-pong kinetic mechanism. To verify that PPi is released after both DHB and ATP bind to the enzyme, 100 μM PPi was added to the reaction mix and the initial velocity pattern was determined using DHB as the variable substrate and holo-EntB-ArCP as the fixed, variable substrate. The addition of PPi changes the initial velocity pattern from parallel to intersecting and confirms that PPi is released after both DHB and ATP have bound to the enzyme, but before holo-EntB-ArCP binds (Figure 2).
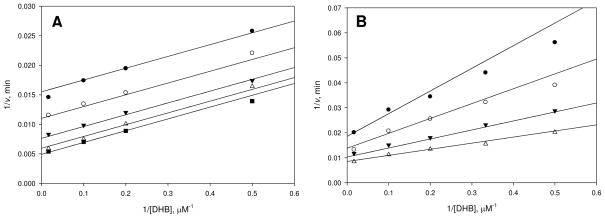
Figure 2.
Pyrophosphate as a probe of the ping-pong nature of the reaction. (A) Assays were performed at varying concentrations of DHB (2–60 μM), saturating ATP (5 mM), and varying concentrations of holo-EntB-ArCP: 3 (●), 5 (○), 10 (▼), 20 (△), and 50 μM (■). (B) Assays were performed at varying concentrations of DHB (2–60 μM), saturating ATP (5 mM), and varying concentrations of holo-EntB-ArCP: 3 (●), 5 (○), 10 (▼), 20 μM (△) in the presence of 100 μM PPi. Initial velocities were measured spectrophotometrically in 100 mM HEPES pH 7.8 and 10 mM MgCl2 using the coupled assay as described in the text. Points are experimental and lines are global fits of the data to eqs 2 and 3 for A and B, respectively.
In order to determine the order of addition of ATP and DHB, dead-end inhibition studies were performed using AMPCPP, a non-hydrolyzable analogue of ATP. AMPCPP exhibited competitive inhibition versus ATP and uncompetitive inhibition versus DHB (Figure 3). These data suggest that DHB binds to the enzyme prior to ATP during the first half-reaction (adenylation). Moreover, direct binding experiments using isothermal titration calorimetry were completed in order to verify the order of substrate binding in the first half-reaction. Consistent with our kinetic data, DHB can bind directly to the free enzyme with a Kd equal to 8.7 μM (Figure 4). In addition to our present data, the three-dimensional structure of DhbE also suggests that productive ATP binding can only occur after DHB has bound to the enzyme as the adenine ring of ATP would likely block access to the DHB binding pocket (20).
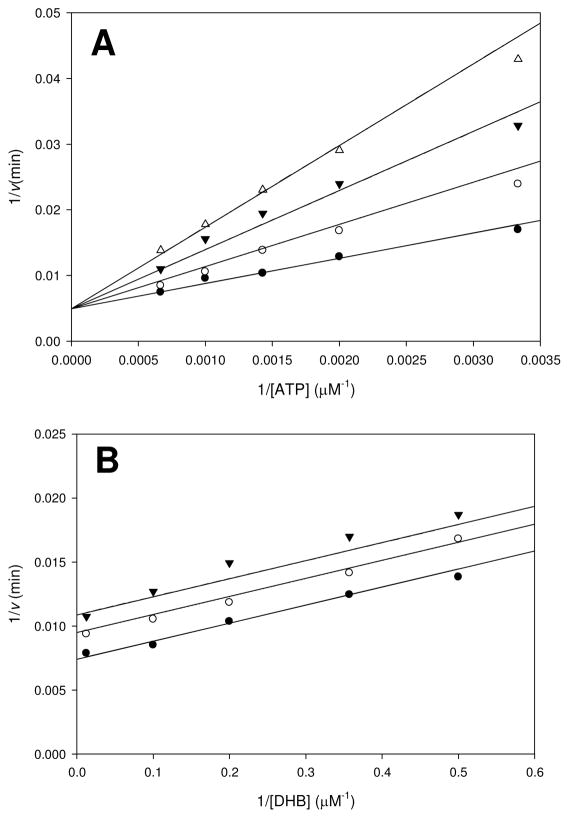
Figure 3.
DHB binds EntE before ATP. (A) Assays were performed at varying concentrations of ATP (0.3 – 1.5 mM), saturating DHB (80 μM), fixed holo-EntB-ArCP (4 μM), and varying concentrations of AMPCPP: 0 (●), 150 (○), 300 (▼), and 500 μM (△). (B) Assays were performed at varying concentrations of DHB (2 – 80 μM), saturating ATP (5 mM), fixed holo-EntB-ArCP (4 μM), and varying concentrations of AMPCPP: 0 (●), 30 (○), and 50 μM (▼). Initial velocities were measured spectrophotometrically in 100 mM HEPES pH 7.8 and 10 mM MgCl2 using the coupled assay as described in the Materials and Methods. Points are experimental and lines are global fits of the data to eqs 4 and 5 for A and B, respectively.
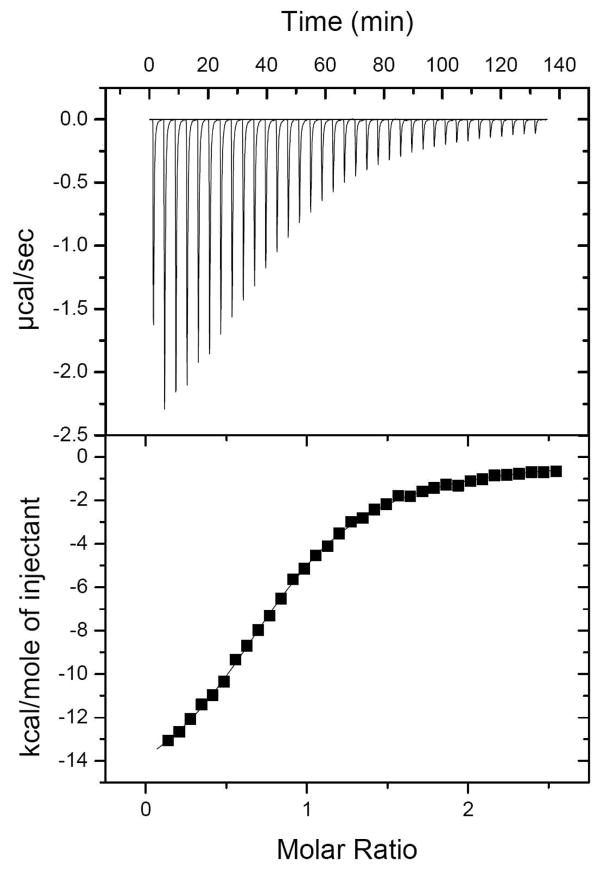
Figure 4.
Isothermal calorimetry profile of EntE (50 μM) with DHB (1 mM). Experiments were performed as described in Materials and Methods. Top: data obtained from automatic injections of 5 μl of DHB. Bottom: the integrated curve showing experimental points (■) and the best fit (−). A fit of the data to a one-set of sites model produced the following values for the binding of DHB to EntE: n = 0.824 ± 0.00512, ΔH = −16500 ± 140 cal/mol, ΔS = −32.2 cal mol−1 K −1, and KA = 115000 ± 2800 M−1.
The results from initial velocity and dead-end inhibition studies of EntE, as well those from direct binding experiments, are fully consistent with a Bi Uni Uni Bi ping-pong kinetic mechanism for EntE, as illustrated in Scheme 5. In this proposed mechanism, DHB binds first to the free enzyme followed by the binding of ATP to form a ternary E-DHB-ATP complex. Next, the enzyme releases pyrophosphate with the formation of DHB-adenylate bound in the enzyme active site. In the absence of holo-EntB, ATP can slowly react with the enzyme-bound DHB-adenylate to form P1, P3-diadensoine-5′-tetraphosphate (Ap4A) (23). When present, holo-EntB binds last to the enzyme and a thioester bond is formed between DHB and the reactive terminal thiol group of the phosphopantetheine moiety of EntB. The resulting products, arylated EntB and AMP, are then released from the enzyme active site. The order of product release for arylated EntB and AMP was not defined in this study because product inhibition by AMP versus ATP could not be determined due to the coupling enzyme assay used. However, we suggest that the enzyme releases arylated EntB prior to AMP based on previous kinetic studies with other ATP-dependent enzymes that catalyze thioester-forming reactions (24). Moreover, the proposed Bi Uni Uni Bi ping-pong mechanism has been well-documented for other adenylate-forming enzymes, including 4-chlorobenzoate: coenzyme A ligase and acetyl-CoA synthetase, as well as for EntE homologues, such as VibE from Vibrio cholerae (25, 26).
Scheme 5.
Proposed Bi Uni Uni Bi Ping-Pong Kinetic Mechanism of EntE.
Bisubstrate Analogue Inhibition Experiments
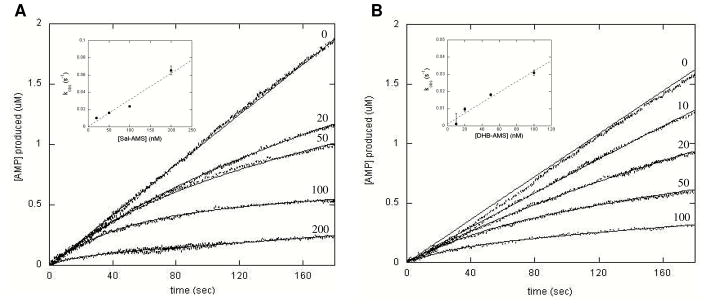
Two hydrolytically-stable adenylate analogues, 5′-O-[N-(salicyl)sulfamoyl]adenosine (Sal-AMS) and 5′-O-[N-(2, 3-dihydroxybenzoyl)sulfamoyl]adenosine (DHB-AMS), were synthesized as described previously (27) and examined as inhibitors of EntE (Scheme 4). During the initial analysis of these small molecules, it became apparent that the time-courses of product formation displayed non-linear kinetics and exhibited a time-dependent inhibition suggestive of slow-onset inhibition (Figure 5) (16, 28). Slow-onset inhibition is characterized by time-courses that display a rapid initial rate followed by a slower steady-state rate (16, 29). Moreover, this type of inhibition has been observed in several enzymatic systems when bisubstrate analogues are used as enzyme inhibitors (30, 31).
Figure 5.
Bisubstrate analogues act as slow-onset inhibitors of EntE. (A) Assays were performed at saturating concentrations of all three substrates: DHB (80 μM), holo-EntB-ArCP (20 μM), and ATP (5 mM), varying, fixed concentrations of Sal-AMS (as labeled in figure, in nM), and 8 nM EntE. (B) Assays were performed as in A, but with varying, fixed concentrations of DHB-AMS (as labeled in figure, in nM). Initial velocities were measured spectrophotometrically in 100 mM HEPES pH 7.8 and 10 mM MgCl2 using the coupled assay as described in Materials and Methods. Dashed lines represent an average of five traces at a given inhibitor concentration and fits of the data to eq 6 are shown as solid lines. Inset: Plot of kobs values obtained from eq 6 versus concentration of Sal-AMS (A) or DHB-AMS (B).
Two possible kinetic mechanisms, as described by Walsh and Morrison, can explain non-linear kinetics (Scheme 5) (16). Mechanism A depicts a single-step mechanism where the association of inhibitor with enzyme is slow. Conversely, mechanism B depicts a two-step mechanism where the association of inhibitor with enzyme is rapid, followed by a second step which is the slow equilibration to a more tightly-bound complex (EI*) (16, 32). For both Sal-AMS and DHB-AMS, a plot of kobs (obtained from eq 6) versus inhibitor concentration yields a linear dependence and this pattern is a hallmark of the one-step slow-onset mechanism (Figure 5, inset) (16). The dependence of kobs on inhibitor concentration can be fit to eq 7 to obtain the rate constants shown in mechanism A and this allows for the determination of the appKi for each inhibitor. This analysis results in appKi values of 0.9 and 3.8 nM for Sal-AMS and DHB-AMS, respectively, at saturating concentrations of all three substrates.
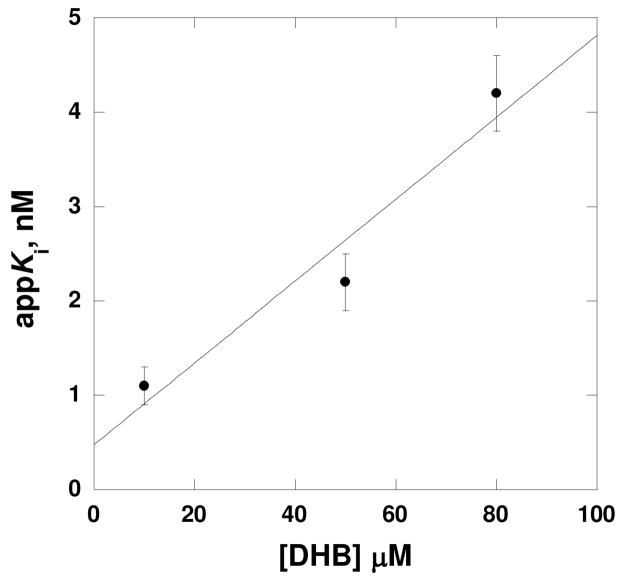
In addition to the slow-onset analysis of both bisubstrate analogues, we also used steady-state kinetics to determine the intrinsic Ki for Sal-AMS. When the appKi value of Sal-AMS versus ATP was determined at several concentrations of DHB, the appKi value decreased as the DHB concentration decreased, as expected for an inhibitor that binds to both substrate-binding sites (Figure 6) (33). Extrapolation to 0 μM DHB allows for the determination of the intrinsic Ki value for the binding of the bisubstrate to the free enzyme, yielding a value of 470 pM. As shown below, our ITC analysis yields a KD value for Sal-AMS that is comparable to that obtained kinetically.
Figure 6.
Linear dependence of appKi values at various concentrations of DHB. Symbols are experimentally determined Ki values determined from assays performed at various concentrations of ATP (0.3 – 1.5 mM), different fixed concentrations of DHB (10, 50, and 80 μM), and saturating holo-EntB-ArCP (20 μM) while in the presence of different fixed concentrations of Sal-AMS (0 – 10 nM) and 14 nM EntE. Eq 4 was used to calculate the appKi value at each concentration of DHB assayed and the line is a fit of the data to eq 9, yielding an intrinsic Ki of 0.47 nM for Sal-AMS.
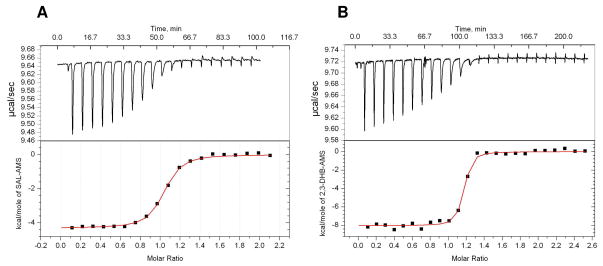
We employed ITC studies to further investigate the binding affinities of Sal-AMS and DHB-AMS to EntE (Figure 7). The binding enthalpies were obtained by stoichiometric titration of EntE with ligands at high c values. Due to the extremely high affinity of both Sal-AMS and DHB-AMS for EntE, displacement ITC experiments were required using salicylic acid as the competitive ligand in order to obtain values for KA and n (18). The thermodynamic binding parameters are presented in Table 2. The titration of Sal-AMS resulted in an experimental KD of 77.5 pM, −ΔH value of 12.7 kcal mol−1, TΔS value of 0.9 kcal mol−1, and n value of 1.01 ± 0.01. The titration of DHB-AMS resulted in a 10-fold lower KD of 7.5 pM with a −ΔH value of 15.4 kcal mol−1, TΔS value of −0.4 kcal mol−1, and n value of 1.12 ± 0.03. Furthermore, the ITC demonstrated that there is one substrate-binding site per EntE monomer.
Figure 7.
Representative ITC profiles of (A) EntE (10 μM) with Sal-AMS (150 μM) and salicylic acid (200 mM) and (B) EntE (5 μM) with DHB-AMS (70 μM) and salicylic acid (200 mM). Top: data obtained for automatic injections of 10 μL of Sal-AMS (A) and DHB-AMS (B). Bottom: the integrated curve showing experimental points (■) and the best fit (−).
Table 2.
Thermodynamic Parameters Obtained from Calorimetric Titration of EntEa
| ligand | KA (× 109 M−1) | −ΔG (kcal mol−1) | −ΔH (kcal mol−1) | TΔS (kcal mol−1) |
|---|---|---|---|---|
| SAL-AMS | 13.3 | 13.6 | 12.7 | 0.9 |
| 2,3-DHB-AMS | 159.5 | 15.0 | 15.4 | −0.4 |
Determined at 293 K. Relative error values: KASAL-AMS 19.4%; KA2,3-DHB-AMS 56.5%; ΔH 1.3–1.6%
Here we reveal low picomolar inhibition and dissociation constants for both Sal-AMS and DHB-AMS to EntE. These small molecules are rationally-designed bisubstrate analogues that are hydrolytically stable structural isoteres of the adenylate intermediate and designed to simultaneously occupy both substrate binding sites at the enzyme active site (34). As exemplified by the kinetic and ITC data presented here, bisubstrate analogues have the potential to bind to enzymes with extremely high affinity and specificity, thus addressing the two critical aspects of any effective therapeutic agent (35). The tight-binding of Sal-AMS and DHB-AMS to EntE suggests that these compounds have great promise to be used as novel antibacterial agents targeted to siderophore biosynthesis. Moreover, because this pathway is absent in mammals, it is likely that this class of antibiotics will be specific and selective to bacteria, thus causing little or no effect to the human host (9).
Although we have focused on enterobactin, many pathogenic bacteria utilize related aryl-capped siderophores for iron acquisition and it is expected that these bisubstrate analogues and other derivatives may be effective against a broad array of clinically relevant pathogens (34). Furthermore, other adenylate analogues based on the prototypical Sal-AMS and DHB-AMS used here, have also been shown to inhibit EntE homologues, such as DhbE, YbtE, and MbtA from B. subtilis, Y. pestis, and M. tuberculosis, respectively (9–11, 27). These adenylate analogue inhibitors have been shown in vitro to abolish mycobactin biosynthesis in M. tuberculosis and exhibit potent growth inhibition (27). MbtA inhibition by Sal-AMS and related derivatives occur at MIC99 values that are equal to that of one of the most powerful anti-tuberculosis drugs, isoniazid (MIC99 = 0.18 μM) (27). Moreover, the same study revealed that none of the tested inhibitors displayed toxicity when evaluated against a murine leukemia cell line at inhibitor concentrations up to 400 times that of the MIC99 value (27). The use of these bisubstrate analogues as inhibitors of adenylate-forming enzymes, such as MbtA and EntE, in siderophore biosynthetic pathways, is proving a promising approach for the development of novel antibacterial therapeutics.
Supplementary Material
Acknowledgments
We would like to thank Dr. Brenda A. Frankel for cloning EntB-ArCP and also for purifying the Sfp protein. In addition, we would like to thank Dr. Rong Guan for assistance with the ITC experiments used to determine the KD of DHB.
Abbreviations
- AMP
adenosine monophosphate
- Ap4A
P1, P3-diadenosine-5′-tetraphospate
- ArCP
aryl carrier protein
- AMPCPP
α, β-methyleneadenosine 5′-triphosphate
- ATP
adenosine triphosphate
- bp
base pairs
- CoA
coenzyme A
- DHB
2, 3-dihydroxybenzoic acid
- DHB-AMS
5′-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl]adenosine
- DNA
deoxyribonucleic acid
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
isopropyl β-D-thiogalactopyranoside
- LB
Luria broth
- ITC
isothermal titration calorimetry
- NADH
nicotinamide adenine dinucleotide
- Ni-NTA
nickel nitriloacetic acid
- NRPS
non-ribosomal peptide synthetase
- PCR
polymerase chain reaction
- PEP
phosphoenolpyruvate
- PPi
inorganic pyrophosphate
- Sal-AMS
5′-O-[N-(salicyl)sulfamoyl]adenosine
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
This work was supported by NIH grant AI60899 (to J.S.B.), T32 GM08572 (to A.L.S.), and AI070219 (to C.C.A.)
Supporting Information Available
One figure showing the sequence alignment of adenylate-forming enzymes EntE and homologues. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandy M, Butler A. Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev. 2009;109:4580–4595. doi: 10.1021/cr9002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 4.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gehring AM, Mori I, Walsh CT. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 6.Sakaitani M, Rusnak F, Quinn NR, Tu C, Frigo TB, Berchtold GA, Walsh CT. Mechanistic studies on trans-2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (Ent A) in the biosynthesis of the iron chelator enterobactin. Biochemistry. 1990;29:6789–6798. doi: 10.1021/bi00481a006. [DOI] [PubMed] [Google Scholar]
- 7.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 8.Ehmann DE, Shaw-Reid CA, Losey HC, Walsh CT. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc Natl Acad Sci U S A. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miethke M, Bisseret P, Beckering CL, Vignard D, Eustache J, Marahiel MA. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. Febs J. 2006;273:409–419. doi: 10.1111/j.1742-4658.2005.05077.x. [DOI] [PubMed] [Google Scholar]
- 10.Gupte A, Boshoff HI, Wilson DJ, Neres J, Labello NP, Somu RV, Xing C, Barry CE, Aldrich CC. Inhibition of siderophore biosynthesis by 2-triazole substituted analogues of 5′-O-[N-(salicyl)sulfamoyl]adenosine: antibacterial nucleosides effective against Mycobacterium tuberculosis. J Med Chem. 2008;51:7495–7507. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreras JA, Ryu JS, Di Lello F, Tan DS, Quadri LE. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat Chem Biol. 2005;1:29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 12.Neres J, Wilson DJ, Celia L, Beck BJ, Aldrich CC. Aryl acid adenylating enzymes involved in siderophore biosynthesis: fluorescence polarization assay, ligand specificity, and discovery of non-nucleoside inhibitors via high-throughput screening. Biochemistry. 2008;47:11735–11749. doi: 10.1021/bi801625b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 15.Zheng R, Blanchard JS. Steady-state and pre-steady-state kinetic analysis of Mycobacterium tuberculosis pantothenate synthetase. Biochemistry. 2001;40:12904–12912. doi: 10.1021/bi011522+. [DOI] [PubMed] [Google Scholar]
- 16.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 17.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 18.Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat Protoc. 2006;1:186–191. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 19.Khalil S, Pawelek PD. Ligand-induced conformational rearrangements promote interaction between the Escherichia coli enterobactin biosynthetic proteins EntE and EntB. J Mol Biol. 2009;393:658–671. doi: 10.1016/j.jmb.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 20.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusnak F, Faraci WS, Walsh CT. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 22.Keating TA, Suo Z, Ehmann DE, Walsh CT. Selectivity of the yersiniabactin synthetase adenylation domain in the two-step process of amino acid activation and transfer to a holo-carrier protein domain. Biochemistry. 2000;39:2297–2306. doi: 10.1021/bi992341z. [DOI] [PubMed] [Google Scholar]
- 23.Sikora AL, Cahill SM, Blanchard JS. Enterobactin synthetase-catalyzed formation of P(1),P(3)-diadenosine-5′-tetraphosphate. Biochemistry. 2009;48:10827–10829. doi: 10.1021/bi901680m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YS, Kang SW. Steady-state kinetics of malonyl-CoA synthetase from Bradyrhizobium japonicum and evidence for malonyl-AMP formation in the reaction. Biochem J. 1994;297(Pt 2):327–333. doi: 10.1042/bj2970327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry. 2008;47:8026–8039. doi: 10.1021/bi800698m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reger AS, Carney JM, Gulick AM. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry. 2007;46:6536–6546. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, 3rd, Aldrich CC. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J Med Chem. 2006;49:31–34. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 28.de Carvalho LP, Argyrou A, Blanchard JS. Slow-onset feedback inhibition: inhibition of Mycobacterium tuberculosis alpha-isopropylmalate synthase by L-leucine. J Am Chem Soc. 2005;127:10004–10005. doi: 10.1021/ja052513h. [DOI] [PubMed] [Google Scholar]
- 29.Sculley MJ, Morrison JF, Cleland WW. Slow-binding inhibition: the general case. Biochim Biophys Acta. 1996;1298:78–86. doi: 10.1016/s0167-4838(96)00118-5. [DOI] [PubMed] [Google Scholar]
- 30.Khalil EM, De Angelis J, Ishii M, Cole PA. Mechanism-based inhibition of the melatonin rhythm enzyme: pharmacologic exploitation of active site functional plasticity. Proc Natl Acad Sci U S A. 1999;96:12418–12423. doi: 10.1073/pnas.96.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faller B, Farley D, Nick H. Finasteride: a slow-binding 5 alpha-reductase inhibitor. Biochemistry. 1993;32:5705–5710. doi: 10.1021/bi00072a028. [DOI] [PubMed] [Google Scholar]
- 32.Lightcap ES, Silverman RB. Slow-binding inhibition of gamma-aminobutyric acid aminotransferase by hydrazine analogues. J Med Chem. 1996;39:686–694. doi: 10.1021/jm950437v. [DOI] [PubMed] [Google Scholar]
- 33.Yu M, Magalhaes ML, Cook PF, Blanchard JS. Bisubstrate inhibition: Theory and application to N-acetyltransferases. Biochemistry. 2006;45:14788–14794. doi: 10.1021/bi061621t. [DOI] [PubMed] [Google Scholar]
- 34.Neres J, Labello NP, Somu RV, Boshoff HI, Wilson DJ, Vannada J, Chen L, Barry CE, 3rd, Bennett EM, Aldrich CC. Inhibition of siderophore biosynthesis in Mycobacterium tuberculosis with nucleoside bisubstrate analogues: structure-activity relationships of the nucleobase domain of 5′-O-[N-(salicyl)sulfamoyl]adenosine. J Med Chem. 2008;51:5349–53470. doi: 10.1021/jm800567v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broom AD. Rational design of enzyme inhibitors: multisubstrate analogue inhibitors. J Med Chem. 1989;32:2–7. doi: 10.1021/jm00121a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.