Abstract
Continuous turnover of intracellular components by autophagy is necessary to preserve cellular homeostasis in all tissues. Alterations in macroautophagy, main responsible for bulk autophagic degradation, have been proposed to contribute to pathogenesis in Huntington’s disease (HD), a genetic neurodegenerative disorder caused by an expanded polyglutamine tract in huntingtin protein. However, the precise mechanism behind macroautophagy malfunctioning in HD is poorly understood. In this work, using cellular and mouse models of HD and cells from HD patients, we have identified a primary defect in the ability of autophagic vacuoles to recognize cytosolic cargo in HD cells. Autophagic vacuoles form at normal or even enhanced rates in HD cells and are adequately eliminated by lysosomes, but they fail to efficiently trap cytosolic cargo in their lumen. We propose that inefficient engulfment of cytosolic components by autophagosomes is responsible for their slower turnover, functional decay and accumulation inside HD cells.
All cells rely on quality control mechanisms that assure the maintenance of cellular homeostasis. Malfunction of the systems that contribute to cellular quality control – chaperones and proteolytic systems – leads to the accumulation of abnormal and damaged components inside cells, with subsequent functional decline and loss of cellular viability1. Altered homeostasis is particularly detrimental in post-mitotic cells, such as neurons, explaining thus the large number of pathologies resulting from poor quality control in these cells (i.e. neurodegenerative disorders)1. Huntington’s disease (HD) is a hereditary neurodegenerative disease in which mutation in a protein, in this case the ubiquitously expressed protein huntingtin (htt), converts it into a pathogenic protein product2. This pathogenic mutation produces an extension of the polyglutamine track located in the N–terminus region of htt. When mutant htt is not efficiently removed, it accumulates inside cells in the form of toxic oligomeric species and aggregates, eventually leading to cell death and onset of the disease2.
Reports from several groups indicate that removal of toxic forms of cytosolic mutant htt occurs preferentially in the lysosomal system (via autophagy)3,4, in part through htt acetylation5, whereas wild–type htt and mutant htt in the nucleus are normally degraded by the proteasome6. Macroautophagy, a type of autophagy in which whole regions of cytosol are sequestered in double membrane vesicles (known as autophagosomes (APH)) and delivered to lysosomes by vesicular fusion for degradation7, has been shown to contribute to the removal of both soluble and aggregate forms of mutant htt8. Indeed, protein aggregation and cell loss can be significantly slowed by activating macroautophagy in different HD models4,9,10. The observation that mutant htt accumulates in the affected cells suggests that a defect in macroautophagy could impair clearance of the mutant protein3. In support of this hypothesis, alterations in the endocytic and autophagic systems have been described in different experimental models of HD and in brains from affected patients. Early reports described an increased number of APHs in mouse striatal neurons expressing full–length or truncated human mutant htt11, in brains from HD patients and HD mouse models12,13, and in non–neuronal cells, such as lymphoblasts from HD patients14. This increased presence of endocytic and autophagic compartments was attributed to enhanced endocytosis and autophagy.
In contrast, recent studies have proposed a possible role for htt in the normal function of the autophagic/lysosomal system, and that a failure in the ability of mutant htt to perform that still unidentified function could explain the altered clearance of the toxic protein in HD15. The dynamic interaction of htt with the endoplasmic reticulum15, an organelle genetically linked to the formation of APHs16, the association of htt with late endosomes and autophagic vesicles15 and the interaction of htt with Rab5, involved in APH formation17, all indirectly support a relationship between htt and the autophagic system. However, the possible function of htt in macroautophagy and the step(s) of the autophagic process affected by mutant htt are poorly understood.
Here we have used biochemical, morphological and functional assays to directly analyze the status of the autophagic system in different cell types (primary neurons, striatal cell lines, fibroblasts and hepatocytes) from two HD mouse models and in lymphoblasts derived from HD patients. We found that the previously reported massive expansion of autophagic compartments in HD cells11 does not result in the predicted increase in proteolysis, but instead that the turnover of cytosolic components is impaired in these cells. Analysis of the different autophagic compartments revealed that failure to degrade cytosolic cargo via macroautophagy is not due to a defect in fusion of APHs with lysosomes or reduced proteolytic activity following this fusion, but rather originates for the most part from inefficient cargo loading. Our study identifies for the first time a defect in cargo recognition in HD that preferentially affects organelle sequestration by inducible macroautophagy and could explain the higher presence of lipid droplets and altered mitochondria observed in HD cells. The fact that the autophagic defect is, for the most part, detected in both neuronal and non–neuronal cells, whose alterations often precede and are independent of the neurological disorder, further support the importance of the autophagic failure to HD pathogenesis. These findings underline the importance of autophagic selectivity for specific organelles, as “random” formation of APHs per se is apparently not sufficient to guarantee adequate organelle clearance.
Results
Macroautophagic activity is reduced in HD
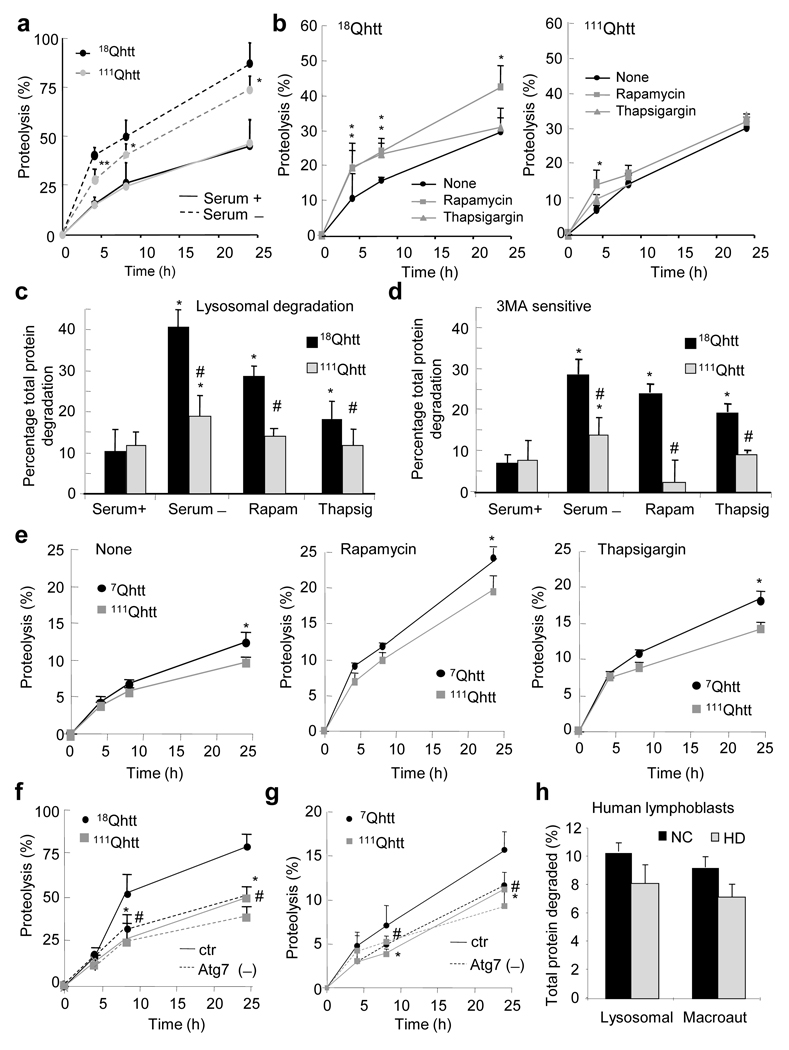
To determine the effect of expression of full size mutant htt on the activity of the autophagic system, we first compared total rates of intracellular protein degradation in embryonic fibroblasts (MEFs) from knock–in mice in which distinct CAG (glutamine) tracts (18 (control) or 111 (HD)) have been inserted in the exon–1 of the murine htt homolog to generate 18Qhtt and 111Qhtt mice, respectively18. The basal rate of degradation of long–lived proteins was comparable in both groups of cells, but the increase in protein degradation in response to serum removal, attributable for the most part to activation of the autophagic system, was significantly impaired in 111Qhtt MEFs (Fig. 1a). These differences were particularly marked during the first 4–6 hours of starvation, when macroautophagy is the prevalent form of active autophagy7 (Supplementary Fig. 1a). Rates of protein degradation were also lower in 111Qhtt MEFs in response to other autophagy stimuli such as rapamycin or ER stress induced by thapsigargin (Fig. 1b). In all these conditions, the observed changes in protein degradation mainly resulted from changes in lysosomal dependent proteolysis. The percentage of protein degradation mediated by lysosomes, determined as that sensitive to inhibition by NH4Cl (which collapses acidic pH in lysosomes)19, was comparable in both groups of cells under basal conditions, but there was a significant decrease (37.5+2.2% to 22.3+3.2%) in the amount of lysosomal protein degradation in the 111Qhtt MEFs upon activation of autophagy by different stimuli (Fig. 1c). We found that the decreased degradation under these conditions in 111Qhtt MEFs was due to their reduced ability to activate macroautophagy (determined as the percentage of lysosomal degradation inhibited by 3–methyladenine19) (Fig. 1d). Serum removal, rapamycin and thapsigargin increased macroautophagic protein degradation in control MEFs but it failed to do that in the 111Qhtt MEFs. The differences in protein degradation between 18Qhtt and 111Qhtt were mainly due to failure of macroautophagy. Rates of protein degradation were comparable in control and HD cells once macroautophagic activity was eliminated through knock–down of Atg7, an essential autophagy gene (Fig. 1f and Supplementary Fig. 1d). Overexpression of pathogenic human exon–1 polyQ htt (146Qhtt) in fibroblasts in culture reproduced this defect in macroautophagy (Supplementary Fig. 1b). Together, these results suggest that the macroautophagic impairment that occurs with htt mutation is due to a toxic or dominant negative effect of the mutant protein.
Figure 1. Autophagic activity is reduced in HD cells.
(a–d) Degradation of long–lived proteins in MEFs from wild type (18Qhtt) and mutant huntingtin knock–in mice (111Qhtt). (a–b) Rates of protein degradation after serum removal (a) or treatment with rapamycin or thapsigargin (b). (c) Lysosomal degradation calculated as degradation sensitive to NH4Cl. (d) Contribution of macroautophagy calculated when adding 3–methyladenine (3–MA). (e) Degradation of long–lived proteins in striatal cells from wild type (7Qhtt) and mutant huntingtin knock–in mice (111Qhtt) in response to different autophagic stimuli. (f–g) Rates of protein degradation in wild type and HD MEFs (f) and striatal cells (g) control or after RNAi for Atg7 (Atg7 (–)). (h) Contribution of macroautophagy to the degradation of long–lived proteins in lymphoblasts from normal control (NC) and from HD patients. All values are expressed in percentage and are mean+s.d. of values from 2–4 different individuals and 3–4 different experiments. *,# significant with untreated (*) or control (#) for p < 0.05.
We found a significant decrease in macroautophagic proteolysis also in neuronal cells. Rates of protein degradation in response to stressors known to stimulate macroautophagy in neuronal cells, such as rapamycin or ER–stressors, were significantly lower in the striatal neuronal cell line derived from the 111Qhtt knock–in mice than in that from 7Qhtt mice (Fig. 1e) 20. Differences between control and HD were already noticeable under basal conditions, suggesting that the compromise of the autophagic function could be more severe in neurons. As in MEFs, the lower rates of protein degradation in striatal neuron–derived cells resulted from changes in lysosomal degradation (sensitive to NH4Cl) (Supplementary Fig. 1c). Genetic blockage of macroautophagy also reduced the differences in protein degradation between 7Qhtt and 111Qhtt striatal–derived cells, further suggesting that macroautophagy was compromised in HD cells (Fig. 1g and Supplementary Fig. 1e). We observed a similar pattern for macroautophagy impairment in postnatally–derived primary striatal cultures from HD94 mice that express mutant human exon–1 htt with 94 CAG repeats (Supplementary Fig. 1f; both wild type and HD94 primary neurons were grown over a monolayer of rat cortex–derived wild type astrocytes to eliminate the contribution of changes in this monolayer to the observed changes in protein degradation). Although it did not reach significance, a similar trend was detected when we compared the contribution of macroautophagy to protein degradation in lymphoblasts from HD patients to that in age–matched healthy individuals (Fig. 1h and Supplementary Fig. 2). In summary, our findings suggest that reduced rates of intracellular degradation due to compromised macroautophagy is a common feature of many cell types in HD and HD models.
Autophagosome clearance is not altered in HD
To elucidate the step(s) in macroautophagy that may be disrupted by mutant htt, we first determined if the inability of HD to engage in productive autophagy was due to altered signaling in response to autophagic stimuli. We found that HD MEFs, like control MEFs, sense starvation as they respond to this stimulus by decreasing their proliferation rate (Supplementary Fig. 3a). Analysis of mTOR, a negative regulator of macroautophagy, revealed no significant differences between control and HD cells in the global phosphorylation state of this kinase and a comparable decrease in response to autophagy stimuli in phosphorylation of p70S6K, one of the best characterized mTOR substrates (Supplementary Fig. 3b,c). These results indicate that mTOR is inhibited normally in response to nutrient deprivation in both MEFs and striatal cells (see Supplementary Text 1).
We then evaluated whether the decrease in macroautophagy in HD cells could result from a primary defect in the lysosomal compartment. Comparison of the level of major lysosomal cathepsins by immunoblot of lysosomes isolated from MEFs (Supplementary Fig. 4a) revealed no differences between 18Qhtt and 111Qhtt MEFs. Processing of these enzymes was also preserved, supporting that lysosomal pH is not markedly altered in HD cells. Lastly, the analysis of the proteolytic activity of disrupted lysosomes against a pool of radiolabeled cytosolic proteins revealed that lysosomes from HD cells displayed higher proteolytic efficiency than those from control cells, arguing against a primary defect in mature lysosomes due to the pathogenic htt mutation (Supplementary Fig. 4b).
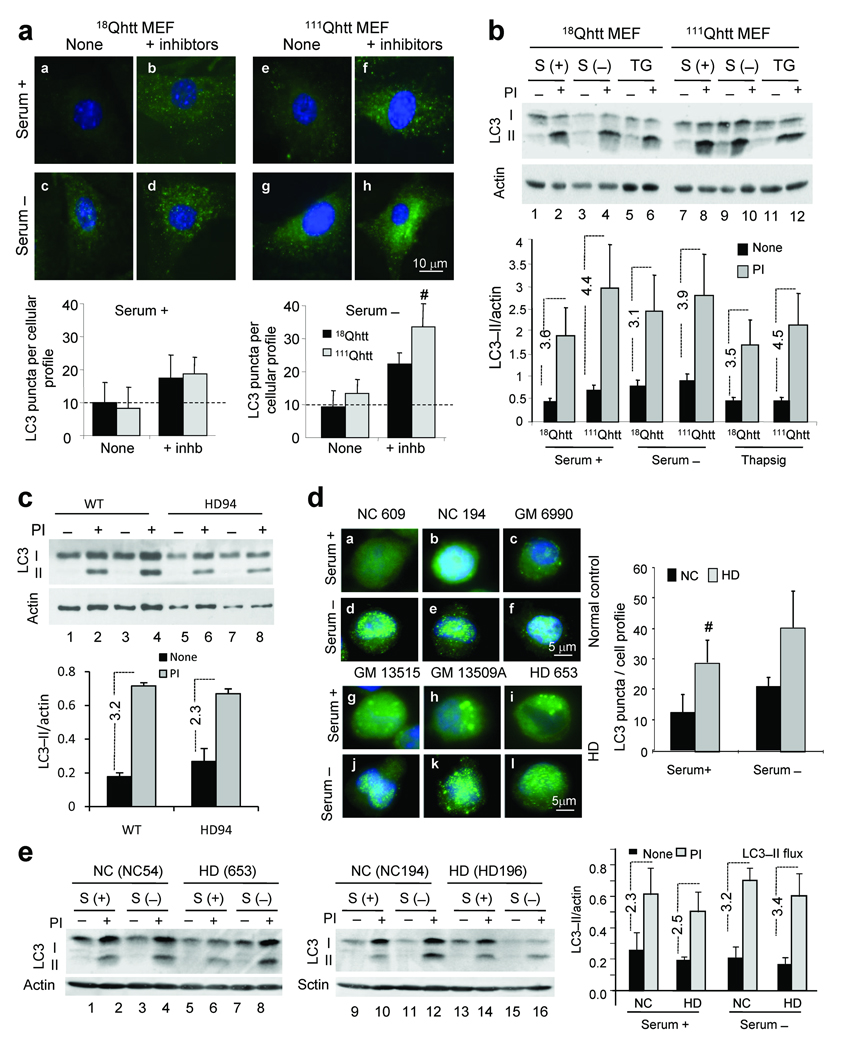
We then analyzed directly the cellular compartments involved in macroautophagy, namely the autophagosomes, and measured the cellular distribution and lysosomal turnover of LC321. Activation of macroautophagy results in the association of LC3 as LC3–II to both sides of the limiting membranes that form the APH21. The rate of degradation of autophagic vacuoles (AVs) following lysosomal fusion can be measured by comparing the effect of lysosomal proteolysis inhibitors on the levels of LC3–II19. Cytosolic LC3 shows a diffuse distribution by immunofluorescence staining whereas LC3–associated to APHs appears as bright fluorescent puncta. The number of APHs observed in 18Qhtt MEFs under basal conditions was low, but increased significantly when lysosomal degradation was inhibited, indicating basal autophagic activity (Fig. 2a). Removal of serum, treatment with rapamycin or induction of ER–stress further activated macroautophagy as reflected in the higher number of bright puncta, particularly visible once lysosomal degradation was inhibited (Fig. 2a and Supplementary Fig. 5a). Similar results were observed in the striatal neuron–derived cells upon activation of autophagy (Supplementary Fig. 5b). In clear contrast to the reduction in total rates of protein degradation observed in 111Qhtt MEFs and striatal–derived cells, the increase in number of LC3 positive puncta upon inhibition of lysosomal proteolysis in these cells was comparable or even higher than in control cells indicating a greater ongoing autophagic flux (Fig. 2a, bottom).
Figure 2. Formation and clearance of autophagic vacuoles is normal in HD cells.
(a) LC3 immunostaining of 18Qhtt and111Qhtt MEFs maintained in the presence (+) or absence (−) of serum and lysosomal proteolysis inhibitors. Bottom: mean number per cell (left) and average size (right) of LC3 positive vesicles. n = 4 (b) LC3 immunoblot in the same cells after serum removal or thapsigargin (TG) treatment. PI: protease inhibitors. Bottom: LC3–II levels and LC3–II flux (dotted lines). N = 4. (c) LC3–II values and LC3–II flux (dotted lines) in neuronal cultures from wild type (WT) or HD94 mice (HD) grown over wild type rat astrocyte monolayers analyzed as in b. n = 4. (d) LC3 staining of lymphoblasts from 3 normal control (NC) or HD patients maintained in the presence or absence of serum. Right: mean number of LC3 positive vesicles. Extended study in Supplementary Fig. 8b,c. (e) LC3–II levels in NC and HD lymphoblasts treated or not with protease inhibitors. Right: LC3–II values and LC3–II flux (dotted lines). Extended study in supplementary Fig. 8a. Values are all expressed as mean+s.d. Differences with control, where significant, are indicated with # for p < 0.05. Full–length blots are presented in Supplementary Fig. 20.
We confirmed the preservation of autophagic flux in mutant cells using immunoblot. Similar to the results with LC3 puncta, the increase in the amount of LC3–II after inhibition of lysosomal proteolysis wash higher in 111Qhtt MEFs under basal conditions and also upon induction of macroautophagy (Fig. 2b). Changes in levels of p62, a protein used as a marker of autophagic degradation19, were comparable in both cell types (Supplementary Fig. 6) confirming that clearance of APHs by lysosomes was normal or even moderately increased in the 111Qhtt MEFs. We did not find significant differences in the levels of the well characterized autophagy effectors (Atg7, Atg5/12 and Atg4) between 18Qhtt and 111Qhtt MEFs or 7Qhtt and 111Qhtt striatal cells, suggesting that abnormal levels of these factors do not explain the inhibition of macroautophagy–dependent degradation (Supplementary Fig. 7). In addition, levels of beclin–1, a major component of the autophagy activation complex, were significantly higher in HD cells (Supplementary Fig. 7a). Increased beclin–1 content could be a compensatory response to the lower rates of protein degradation in HD cells. Overall these results support normal macroautophagy induction and APH formation in these htt mutant cells.
Similarly, analysis of LC3–II flux in primary striatal neurons from the HD94 mice grown over a wild type rat astrocyte monolayer revealed no statistical significant difference from wild–type (Fig. 2c). Lymphoblasts obtained from six different HD patients show a higher presence of APHs, detected as LC3–positive puncta by immunofluorescence (Fig. 2d, compare a–c to g–i) and a comparable increase by immunoblot in the number of these organelles in response to serum (Fig. 2e and Supplementary Fig. 8a and 8b). There was thus an apparent paradox in our findings: how could the reduced macroautophagic proteolysis in mutant cells (Fig. 1) be consistent with their normal or even increased macroautophagic flux (Fig. 2)?
Reduced ability of autophagosomes to recognize cargo in HD
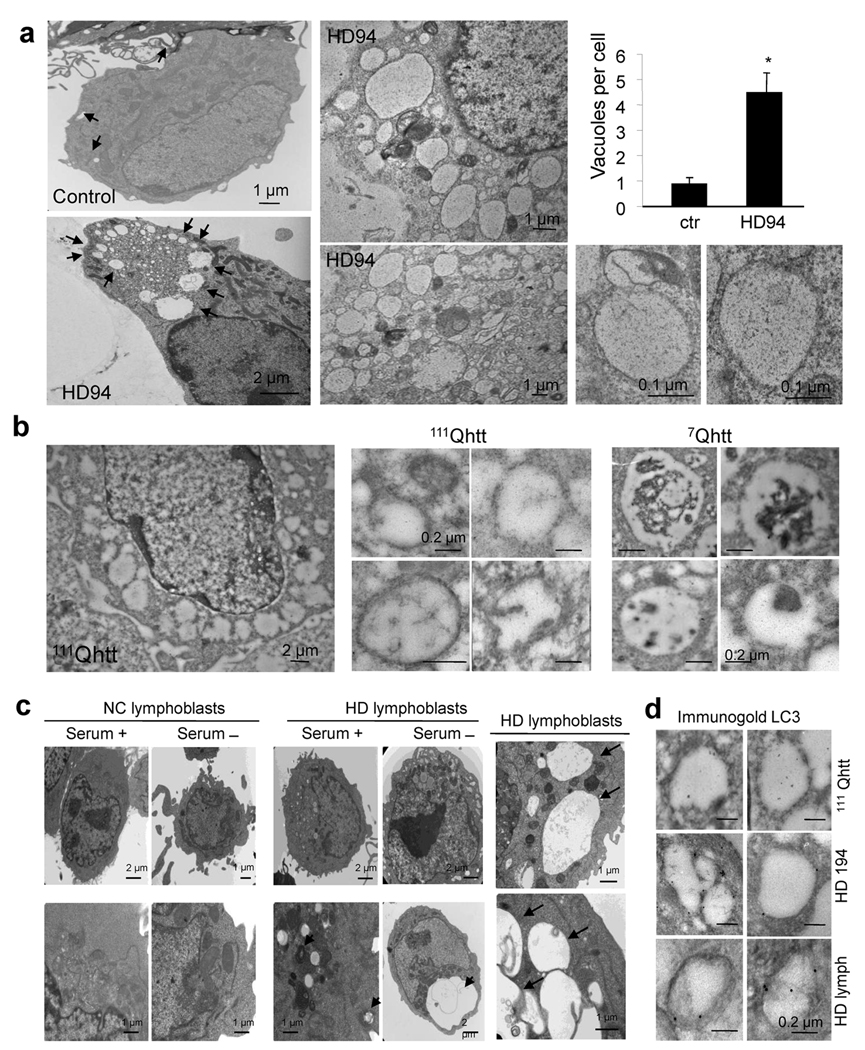
To explore this apparent conflict between macroautophagic proteolysis and LC3 flux, we examined AVs by electron microscopy in the different HD models. Analysis of neurons from HD94 mice (Fig. 3a), striatal cells from 111Qhtt mice (Fig. 3b) and lymphoblasts from some (4 out of 6) of the HD patients (Fig. 3c and Supplementary Fig. 9) revealed a markedly higher presence of vacuolated structures compatible with AVs than in their control. Morphometric analysis supported a higher number and size of AVs in HD cells (Fig. 3a and Supplementary Fig. 9c–e). The enhanced presence of AVs in HD model neurons and lymphoblasts is consistent with prior studies12,13.
Figure 3. Autophagic vacuoles in different HD cell types present abnormal characteristics.
(a) Electron micrographs of striatal neurons from control and HD94 mice grown over a wild type rat astrocyte monolayer. Higher magnification fields show the double membrane and clear content of the cytosolic vesicles. Right: number of vacuoles per cell profile (13–17 cell profiles in triplicate experiments). (b) Electron micrographs of striatal cells from wild type (7Qhtt) and mutant huntingtin knock–in mice (111Qhtt). Right: Higher magnification fields. (c) Electron micrographs of lymphoblasts from normal control (NC) and HD patients maintained in the presence or absence of serum. Right: higher magnifications areas containing enlarged electron–clear vesicles (arrows). (d) Immunogold for LC3 in striatal cells from mutant huntingtin knock–in mice (111Qhtt), striatal neurons from HD194 mice and lymphoblasts from HD patients. Full fields and more details of autophagic vacuoles are shown in Supplementary Fig. 10 and 11. * p < 0.05.
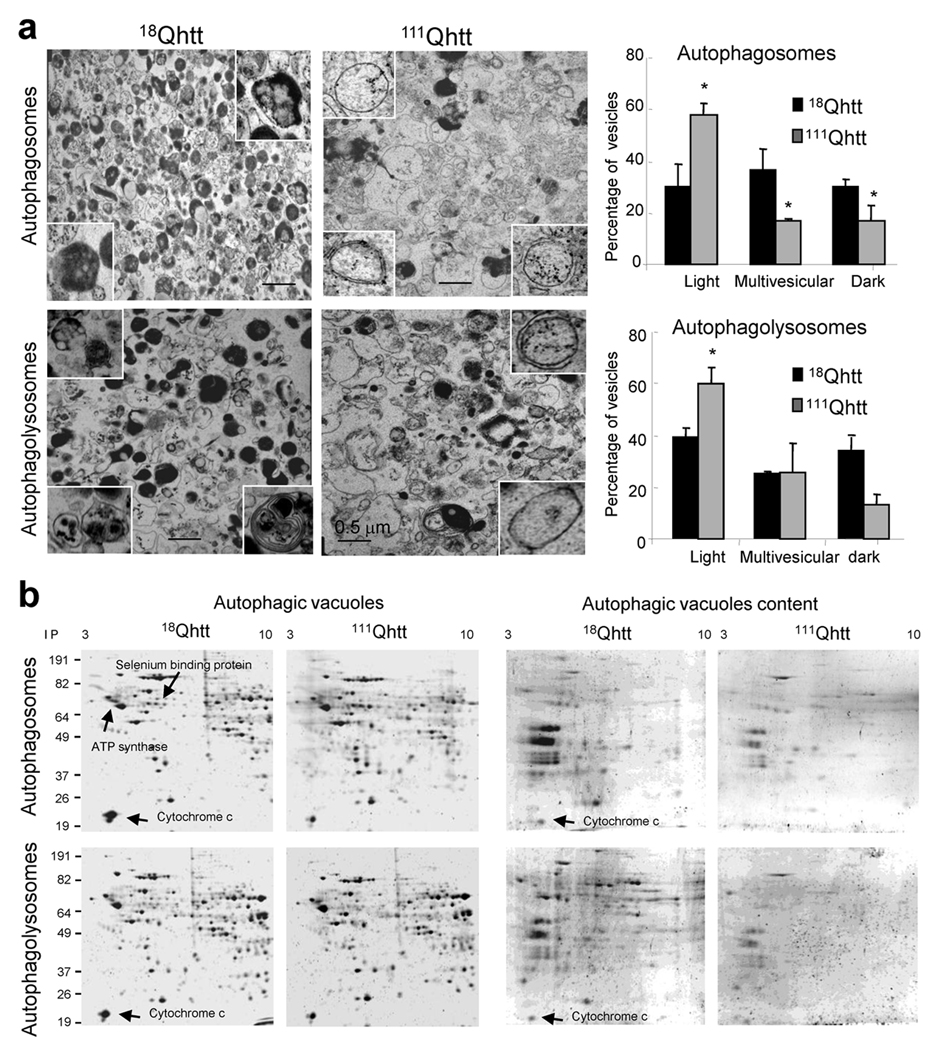
Another striking feature apparent in the electron micrographs of all the models was a lower electron density of the AVs. Rather than the typical double membrane vesicles with identifiable cytosolic content inside (Fig. 3b, right), the most abundant type of vesicles occupying the cytosol of HD cells were still double membraned, but appeared to be “empty” (high magnification panels Fig. 3). While for most cells the size of the “empty” structures was comparable to that of classic AVs (0.1–0.5 µm of diameter), in some cells much larger structures occupied extensive areas of the cytosol (Fig.3a, b). Immunogold for LC3 in the different samples revealed that most of the vesicles observed in the different HD models were positive for LC3, thus supporting the autophagic origin of these compartments (Fig. 3d and supplementary Fig. 10 and 11). We also found these “empty” vesicles in livers of 111Qhtt mice from where we can isolate fractions enriched in APHs or immature AVs, autophagolysosomes or mature AVs and secondary lysosomes22. Electron microscopic analysis of these isolated vesicles confirmed that in contrast to the presence of recognizable cytosolic structures inside the double membrane vesicles in the APH–enriched fraction from the 18Qhtt mice, the vesicles from the 111Qhtt mice were still double membraned (see insets Fig. 4a, top) but possessed an amorphous electrotranslucent content in their lumen (Fig. 4a). Thus, ultrastructural analysis in whole cells and in isolated organelles indicates that AVs in HD are relatively devoid of discernable organelle cargo.
Figure 4. Altered composition of autophagic–related compartments in HD cells.
(a) Electron micrographs of fractions enriched in autophagosomes and autophagolysosomes isolated from liver of 18Qhtt and 111Qhtt mice. Insets: higher magnification images of single vesicles. Right: Percentage of vesicles with an electron–clear (light), vesicular (multivesicular) or electron–dense content (dark). Mean+s.e.m of 3 different isolations (estimated > 1,000 AVs). (b) Bidimensional electrophoresis and SyproRuby staining of the same fractions. Samples on the right are the content of the same fractions isolated in the supernatant after hypotonic shock and high speed centrifugation. * p < 0.05.
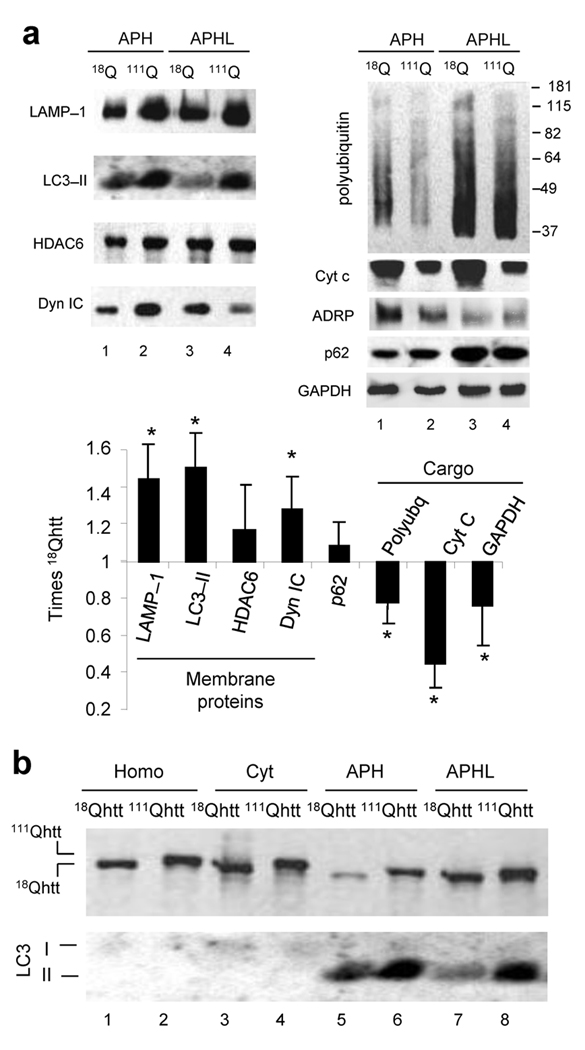
To confirm whether AVs in mutant cells were deficient in normal luminal cargo, we compared their total protein bidimensional electrophoretic pattern (Fig. 4b). Upon separation of membrane and cargo by hypotonic shock (markers shown in Supplementary Fig. 12), the content of AVs isolated from the 111Qhtt mice was markedly less enriched in proteins, with many luminal proteins detected in the 18Qhtt mice vesicles reduced or even absent from 111Qhtt mice fractions. Mass spectrometry analysis of some of the protein spots present in lower amounts in the APHs from 111Qhtt mice confirmed them as possible cargo (mitochondria proteins or cytosolic soluble proteins). In fact, when we analyzed the abundance of proteins associated with the AV membrane (Fig. 5a, left) and cargo proteins (Fig. 5a, right) in the isolated fractions, AV membrane–associated proteins such as the lysosome–associated membrane protein type 1 (LAMP–1; detected also in AVs), LC3 and dynein and HDAC6 (both required for AV trafficking) were significantly higher in AVs isolated from 111Qhtt mice (Fig. 5a). In contrast, common AV cargo such as polyubiquitinated proteins, mitochondrial makers (cytochrome C), lipid droplet structural proteins (ADRP) and soluble cytosolic proteins (GAPDH) were significantly reduced in the AVs isolated from 111Qhtt mice (Fig. 5a).
Figure 5. Altered properties of autophagic–related compartments in HD cells.
(a) Immunoblot for the indicated proteins in fractions enriched in autophagosomes (APH) and autophagolysosomes (APHL) isolated from liver of 18Qhtt and 111Qhtt mice. Bottom: Changes (folds increase or decrease) in the levels of each protein. Mean+s.d. n = 4. (b) Homogenates (Homo), cytosol (Cyt) and fractions enriched in autophagosomes (APH) and in autophagolysosomes (APHL) isolated from wild type (18Qhtt) and mutant huntingtin knock–in mice (111Qhtt) livers were subjected to immunoblot for htt. A representative immunoblot of 4 experiments with duplicated samples is shown. * p < 0.05. Full–length blots are presented in Supplementary Fig. 20.
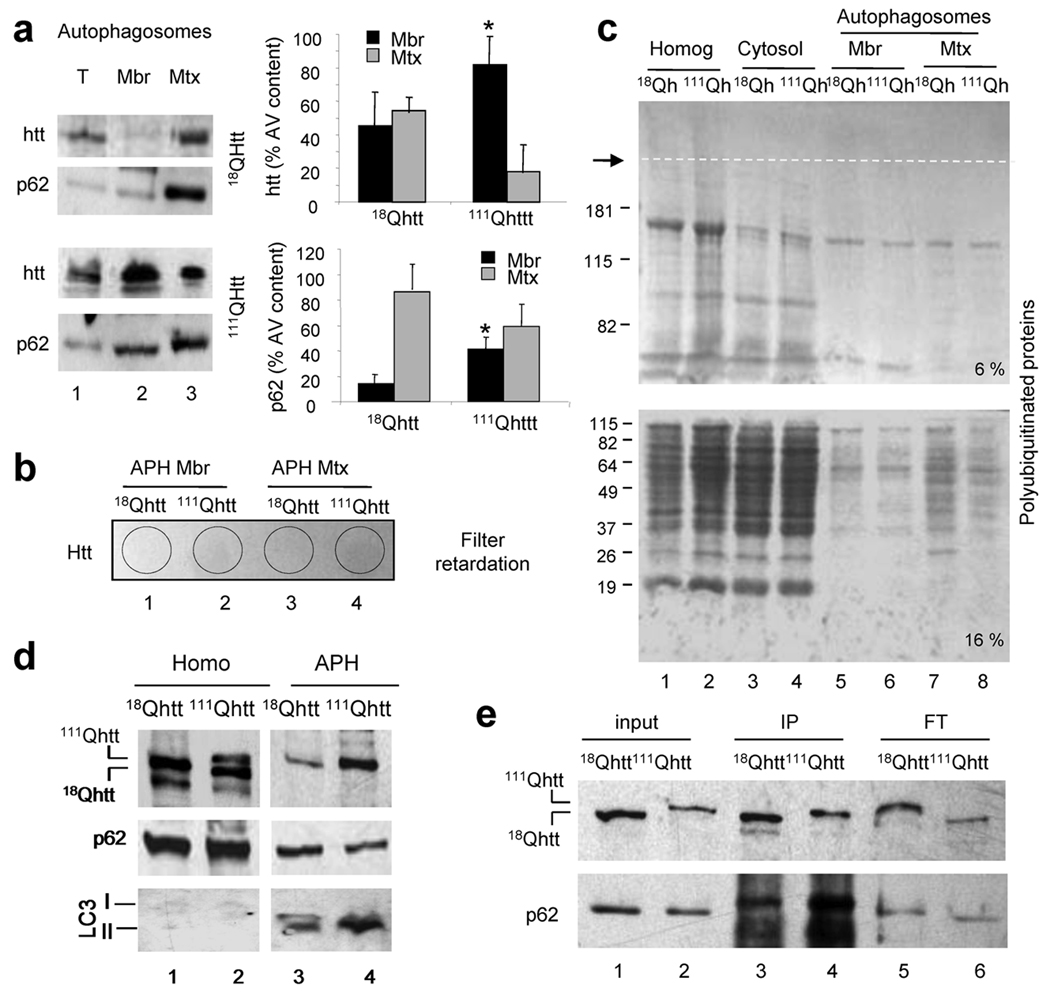
Both autophagy–related compartments also contained htt as described in other experimental systems5, but levels of htt in AVs from 111Qhtt mice were remarkably higher, resembling the trend observed for membrane associated proteins (Fig. 5b). Whereas wild–type htt distributed almost evenly between both fractions, mutant htt was more abundant at the APH membrane (Fig. 6a). Filter retardation assays and immunoblot for htt (Fig. 6b) and analysis of the gel stacking of immunoblots for polyubiquitinated proteins (common components of aggregates) (Fig. 6c) did not revealed aggregation of mutant httt in any of the fractions; indeed, htt aggregation under these conditions is negligible both by immunoblot and immunofluorescence in these cells (Supplementary Fig. 13). It is still possible that reversible changes in the oligomeric state of htt when associated to the APHs could contribute to the inefficient sequestration/engulfment of cytosolic cargo in HD. Interestingly, levels of p62 were comparable in 18Qhtt and 111Qhtt mice AVs (Fig. 5a), but similar to htt, we detected enhanced p62 binding to AV membranes from 111Qhtt mice (Fig. 6a). Interaction of p62 and htt has been recently described in total cell lysates5 but we could coimmunoprecipitate both proteins directly from AVs isolated from 111Qhtt mice brain, where as in liver, levels of htt in AVs were remarkably higher than in control (Fig. 6d). When normalized for htt, the levels of p62 associated to 111Qhtt were markedly higher (Fig. 6e). As p62 participates in the recognition by AVs of polyubiquitinated protein aggregates and organelles23, it is possible that this abnormal association between 111Qhtt and p62 causes the impairment of cargo recognition in HD cells.
Figure 6. Distribution of htt and p62 in autophagic vacuoles.
(a, d) Immunoblot for htt or p62 of total autophagosomes (T) and their corresponding membranes (Mbr) and matrices (Mtx) isolated from wild type (18Qhtt) and mutant huntingtin knock–in mice (111Qhtt) livers. Left: Representative immunoblots. Right: Distribution of htt and p62 between Mbr and Mtx calculated by densitometric quantification in six different immunoblots as the ones shown here. Values are mean+s.d. * p<0.05 compared to wild type values. (b) Filter retardation analysis of the same fractions as in a, and blotted for htt. Negative signal indicated absence of aggregates retained in the filter. (c) Immunoblot for ubiquitin in homogenates (homog), cytosol and the autophagic fractions described in a, was performed in a 6% (top) and a 16% (bottom) gel. Dotted line indicates separation between the stacking and running part of the gel. (d) Immunoblot for LC3 and p62 of homogenates (Homo) and autophagosomes (APH) isolated from wild type (18Qhtt) and mutant huntingtin knock–in mice (111Qhtt) brains. (e) Membranes of autophagic vacuoles shown in d were subjected to immunoprecipitation for htt in mild co–immunoprecipitation buffer. Note that input for 111Qhtt AVs was 1/3 of the input used for 18Qhtt to avoid problems with antibody saturation. Levels of htt (top) and p62 (bottom) in the input, immunoprecipitate (IP) and flow through (FT) are shown. Full–length blots are presented in Supplementary Fig. 21.
We then directly measured the fusion between AVs and lysosomes using an in vitro fusion assay with isolated organelles. The number of fusion events between LC3–labeled APHs (green) and LAMP–2B labeled lysosomes (red) (identified as colocalization between both fluorophores) was similar in fractions from 18Qhtt or 111Qhtt mice (Supplementary Fig. 14a,b), consistent with comparable LC3 flux in control and HD cells. Interestingly the homotypic fusion ability of APHs (APH with APH) from 111Qhtt mice was higher (33%) than in 18Qhtt mice, perhaps explaining the formation of the abnormally large AVs observed in some of the HD cells (Fig. 3a,b,c). Surprisingly, lysosomes from 111Qhtt mice fused with APHs from 18Qhtt mice at rates nearly twice those when both compartments were from the same genotype (Supplementary Fig. 14c), probably to compensate the increased homotypic fusion of the APHs in these cells. We also found an enhanced interaction between APHs from 111Qhtt mice and endocytic compartments to form amphisomes as levels of the endocytic markers mannose–6–phopsphate receptor and Rab 5, were higher in AVs from 111Qhtt mice (Supplementary Fig.14 d). Enhanced amphisome formation could explain recent reports revealing that functional multivesicular endosomes are necessary for mutant htt clearance24.
In summary, the morphological and biochemical analysis provided an explanation for the lower rates of inducible macroautophagy–dependent degradation in these cells despite similar or higher rates of APH formation and clearance. Although APHs form and fuse normally with lysosomes in HD cells, the lower amount of cytosolic cargo in their lumen leads to lower net protein degradation.
Cellular consequences of impaired cargo recognition in HD
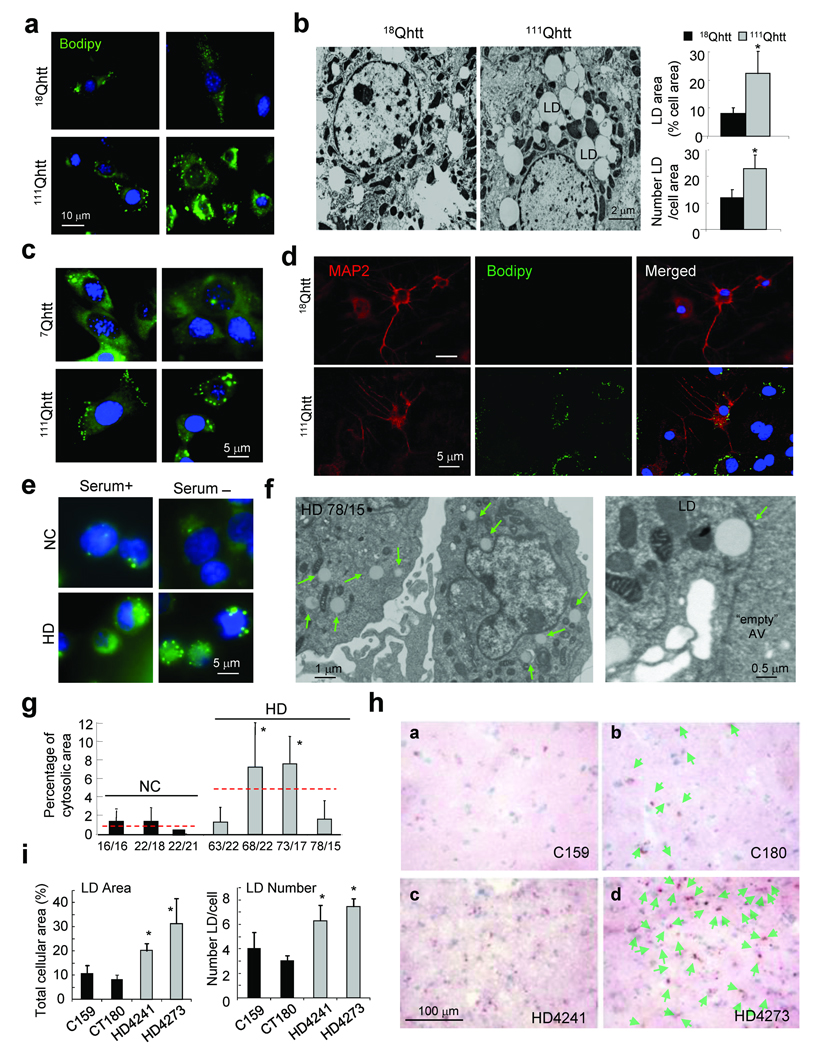
The reduced rates of macroautophagy–dependent protein degradation observed in HD cells did not result in higher levels of soluble cytosolic proteins such as GAPDH. However, markers of two organelles, lipid droplets and mitochondria, were particularly reduced inside AVs from 111Qhtt mice (Fig. 5a). Selective degradation of lipid droplets by macrolipophagy was recently described by our group25. Consistently with the reduced ability to recognize cargo, both electron microscopy analysis and fluorescent label revealed a dramatic increase in the content of lipid droplets in MEFs (Fig. 7a), livers of 111Qhtt mice (Fig. 7b), striatal cell lines (Fig. 7c), primary striatal neurons and glia from the 111Qhtt knock–in mice (Fig. 7d), and lymphoblasts from the HD patients (Fig.7e–g and supplementary Fig.15b). The content of lipid droplets in HD cells did not further increase upon blockage of the lysosomal degradation or when fusion of autophagosomes to lysosomes is prevented (with vinblastine), as it is normally observed in control cells, further supporting that the observed increase in lipid droplet content in HD cells is due to problems in their degradation and not in their formation (Supplementary Fig. 15a). In all HD cell types the number of lipid droplets per cell and area occupied by the droplets was higher, although the average size of the lipid droplets remained unchanged (Fig. 7 and Supplementary Fig. 15). Lastly, we performed Oil Red O staining of striatal tissue from two advanced stage HD and two age–matched control patients (49–54 yr) and found an increase in the density of Oil Red O puncta in each patient (Fig. 7h, panels c,d, and 7i) when compared with control subjects (Fig. 7h, panels a,b and 7i). Interestingly, the density of nuclei in HD striatum was 22% higher than in controls, consistent with the loss of neuropil associated with the disease. However, even when we calculated the number of Oil Red puncta per cell, HD patients displayed more puncta per cell.
Figure 7. Consequences of altered recognition of autophagic cargo in HD on cellular lipid content.
(a,c–e) Neutral lipids in MEFs from 18Qhtt and 111Qhtt mice (a), striatal cells from 7Qhtt and 111Qhtt mice (c), primary striatal neurons from 18Qhtt and 111Qhtt mice grown in a monolayer of their own astrocytes (d) and lymphoblasts from a control and HD patient (e) were stained with Bodipy 493/503. MAP2 staining highlights neurons. Extend study is shown in Supplementary Fig. 15b. (b, f) Electron micrographs of livers from 18Qhtt and 111Qhtt mice (b) and lymphoblast from a HD patient (with 78/15 polyQ repeats) (f). LD: lipid droplets (green arrows). Right (in b): Number of LD, mean area of LDs and percentage of cellular area occupied by LD. Mean+s.d. n = 3. (g) Fraction of cellular cytosol occupied by LD quantified in 3 NC and 4 HD. Numbers of polyQ repeats are shown at the bottom. Mean+s.d. of >100 cell profiles. Dotted red lines: mean value of all NC and HD patients. * p < 0.05. (h) Oil Red O staining of striatal tissue from brain of normal control (top) and two HD patients (bottom). Nuclei are highlighted with hematoxylin. Lipid droplets are indicated in the right panels with green arrows. (i) The percentage of total cellular area occupied by lipid droplets (left) and the average number of lipid droplets per cell was calculated for each of the samples by quantification of 8–9 different fields. *p < 0.001, ANOVA, p < 0.01 for each HD patient vs. each control patient, neither control nor HD patients were different from each other, Tukey post–hoc test.
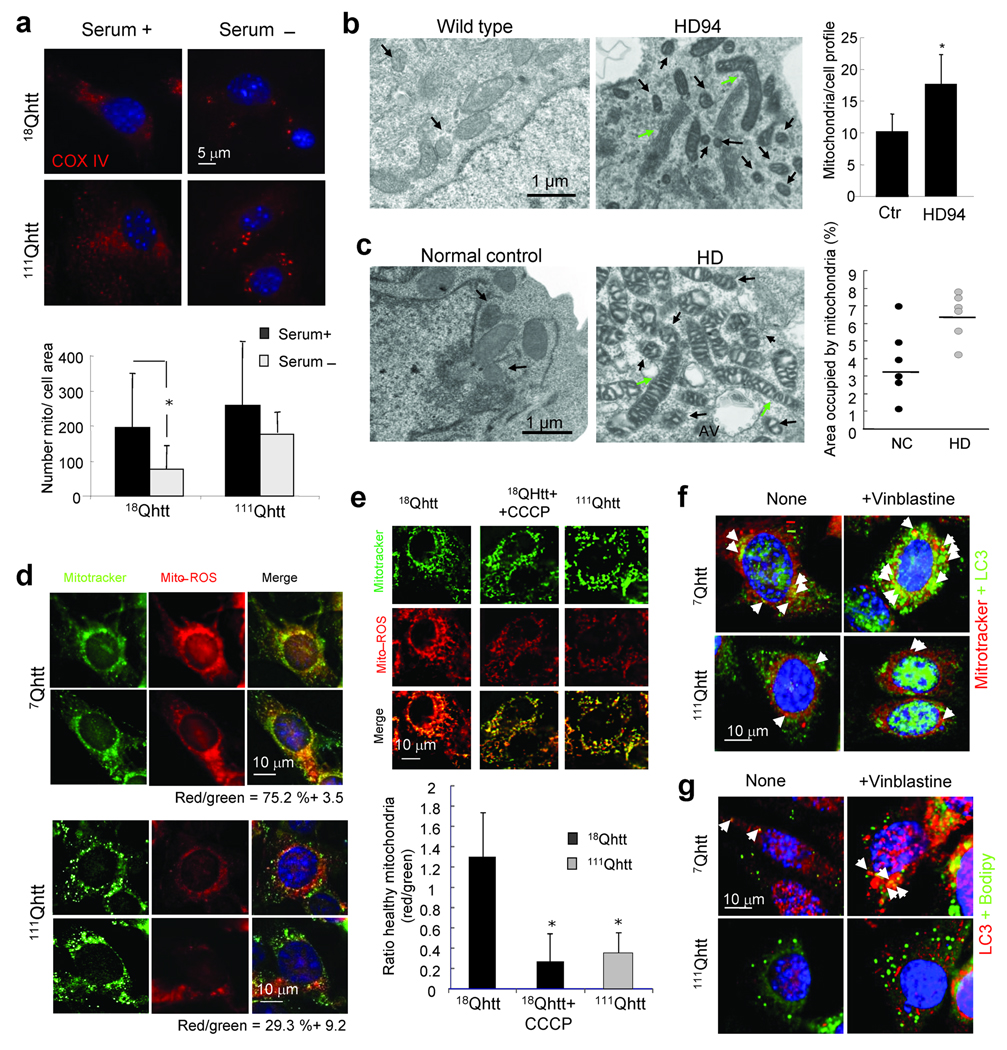
Similarly, in contrast to 18Qhtt MEFs, where starvation–induced macroautophagy decreased total number of mitochondria, induction of macroautophagy did not significantly change the pool of mitochondria in MEFs from 111Qhtt mice (Fig. 8a). Morphometric analysis of electron micrographs of neurons from HD94 mice also revealed a significantly higher number of total mitochondria per cell profile in these animals (Fig. 8b). Lastly, although with variations between individuals, the portion of cellular area occupied by mitochondria was higher in lymphoblasts from HD patients than in control (Fig. 8c); these differences could be due to the disease stage, age, or other factors related to HD pathology.
Figure 8. Altered mitochondria turnover in HD cells.
(a) COX IV immunofluorescence in 18Qhtt and 111Qhtt MEFs maintained in the presence or absence of serum. Bottom: Number of mitochondria per cell. Mean+s.d. of 10–20 cells in 3 different experiments. (b) Electron micrographs of striatal neurons from wild type and HD94 mice grown over a wild type rat astrocyte monolayer. Arrows: Abnormally short (black) or abnormally long (green) mitochondria. Right: Number of mitochondria per cell profile. Mean+s.d. of 10 cells per group in triplicates. (c) Left: Electron micrographs of lymphoblasts from normal control or HD patients. Arrows are as in b. Right: Percentage of cellular area occupied by mitochondria in different individuals. Line indicates the mean value of the population. (d, e) Striatal cells from 7Qhtt and 111Qhtt mice (d) and MEFs from 18Qhtt and 111Qhtt mice (e) co–stained with mitotracker and mito–ROS. Right: Merged images. Percentage of colocalization is indicated at the bottom in d and is displayed in the graph at the bottom in e. CCCP was added to control cells in e as a positive control of depolarization. Differences with control are significant for * p < 0.05. (f,g) Striatal cells from 7Qhtt and 111Qhtt mice untreated or treated with vinblastine were co–stained for LC3 and mitotracker (f) or Bodipy 493/503 (g). Arrows point to colocalization events. Extended study in Supplementary Fig. 16.
Inefficient macroautophagy of intracellular components will slow their normal turnover, increasing their probability to undergo undesirable changes and become damaged. To determine the status of the mitochondria in the different HD models used in this study we analyzed the percentage of “healthy” polarized mitochondria (labeled with MitoRos) compared to the total pool of mitochondria (labeled with Mitotracker, which highlights both polarized and depolarized mitochondria). We found that both HD neuronal (striatal–derived cells shown in Fig. 8d) and non–neuronal (MEFs shown in Fig. 8e) cells display a higher percentage of depolarized mitochondria than their corresponding control cells (the mitochondria depolarizing agent CCCP is used as a positive control in Fig. 8e). The increased content of cytosolic organelles (lipid droplets and mitochondria) observed in HD cells could be, at least in part, due to their reduced recognition and degradation by macroautophagy. Indeed, we found a significant decrease in the colocalization of both organelles with the autophagic marker LC3 in HD cells, even when autophagosome/lysosome fusion was inhibited to increase the time that cytosolic cargo remains in autophagosomes (Fig. 8f and g and Supplementary Fig. 16).
Abnormal intracellular lipid stores and the persistence of altered mitochondria in HD cells could contribute to the increased production of reactive oxygen species and elevated oxidative stress extensively reported in HD26. In fact, despite absence of overt pathology in the 111Qhtt mice, the total content of carbonyl groups, indicative of protein oxidation, in the brain of these animals was markedly higher than in 18Qhtt mice (Supplementary Fig. 17). Accumulation of damaged intracellular components is a common feature of different organ–specific mouse models deficient for essential autophagy genes, and we find that even a relatively acute blockage of macroautophagy in wild type MEFs is sufficient to increase the amount of intracellular LD and oxidized proteins (Supplementary Fig. 18), suggesting that these features observed in HD cells could be, at least in part, direct consequence of the altered macroautophagic function reported here.
In conclusion, our findings support that macroautophagy activity is compromised in HD cells, mainly due to a defect in the ability of autophagic vacuoles to sequester different cellular components, and that this inefficient removal of cytosolic organelles could contribute to cellular toxicity in HD.
Discussion
In this study, we have identified a functional decline in the activity of the autophagic/lysosomal system that is common to different cellular and mouse models of HD and is also evident in cells and striatal tissue from HD patients. Autophagosomes form and fuse efficiently with lysosomes in HD cells, and structural components of the autophagosome membrane, such as LC3, undergo complete proteolysis upon lysosomal delivery via macroautophagy. However, morphological and biochemical analysis of the HD autophagosomes reveal a marked reduction of their content on cytosolic cargo, particularly evident for cytoplasmic organelles. We propose that this decreased efficiency in the sequestration of cellular components by the autophagic system could contribute to the increased levels of protein aggregates, lipid stores and dysfunctional mitochondria characteristic of HD cells (Supplementary Fig. 19).
Abnormal expansion of the autophagic/lysosomal system has become a common feature of different neurodegenerative disorders and protein conformational diseases in general. Thus, besides HD, increased numbers of autophagosomes have been described in the affected cells in Alzheimer’s disease, Parkinson’s disease, prion disorders, methamphetamine and other neurotoxins and stroke, among others27–29. However, higher levels of autophagic vacuoles are often associated with increased protein degradation, such as in neuronal cells expressing mutant forms of Tau30 or in models expressing pathogenic forms of α–synuclein31,32. In other neurodegenerative disorders, such as AD, the massive accumulation of autophagic vacuoles does not associate with increased degradation, but the autophagic vacuoles contain visible cytosolic cargo33. Similarly, cargo was also visible in the autophagosomes induced in neuronal cells by treatment with pro–aggregating compounds such as L–DOPA34 or methamphetamine27, which collapses lysosomal pH gradients to inhibit hydrolases. Consequently, the reasons behind the expansion of the autophagic compartments seem to differ from one disease to another, ranging from upregulation of autophagosome formation to impaired autophagosome/lysosome fusion or inefficient degradation of the cytosolic cargo once delivered to lysosomes. In this study, we have identified in HD a defect in yet a different autophagic step – cargo sequestration – that, to the best of our understanding, has not been reported in any of these other disorders. This could be, however, a defect shared also by other pathologies, but could have been overlooked if only the dynamics of the autophagic vacuoles, but not their cargo, have been tracked (see Supplementary Text 2).
The observation that lipid droplets and mitochondria are both excluded from HD cell AVs suggests that there is a general defect in the autophagic system rather than a primary defect in mitochondria or lipid droplets. Our results support that the defect is mainly in cargo sequestration/engulfment rather than a problem with autophagosome formation or degradation in the lysosomal compartment because: 1) we did not find accumulation of undegraded products inside lysosomes or autophagolysosomes (instead we saw “empty” vacuoles; 2) we did not find differences in levels of major lysosomal proteases; 3) cathepsins seem to be normally processed in their active forms, supporting that the lysosomal pH is not significantly altered and 4) the proteolytic activity of isolated lysosomes (when the membrane is disrupted) is comparable in control and HD. While the reasons for the decreased ability to recognize cargo of APHs in HD and HD models remain elusive, we propose that the mutant htt protein, possibly through its abnormal association with p62, is responsible for this failure to engulf cargo. Htt associates with various organelle membranes15 and the presence of mutant htt on these membranes could prevent their recognition by the APH. This could also explain the failure to recognize polyubiquitinated proteins, as mutant htt also binds to polyubiquitinated aggregates8. While the cargo–recognition hypothesis may not explain the reduction in cytosolic soluble proteins in APH it is possible that some of these proteins could be part of large oligomeric protein complexes or that they associate to the surface of the organelles that fail to be recognized in HD. Alternatively, given the extensive network of proteins interacting with htt, it is plausible that their interaction with mutant htt instead prevents those proteins from being engulfed. For example, mutant htt can form a complex with GAPDH 35, one of the cytosolic proteins less abundant in the lumen of AVs from HD cells (Fig. 5a). The mutant htt present in the luminal side of the AV membrane (Fig. 6a) could also contribute to poor cargo recognition. Future advances in our understanding of the mechanisms that mediate cargo recognition in macroautophagy, should help test this hypothesis.
Failure to properly turnover organelles could explain some of the alterations in cellular homeostasis described in HD cells. Impairment of autophagy in non–neuronal cells in HD may become evident under the conditions that induce macroautophagy, such as during the period in between meals in organs as in liver, or in response to stressors such as ER–stress, as demonstrated by our studies in MEFs. The almost daily frequency with which macroautophagy is induced in these cells could contribute to explain the chronic nature of the disease. In contrast, our studies in striatal neuronal cell lines and primary neuronal cultures reveal that in this case the autophagic defect is not limited to inducible autophagy but it is already apparent under basal conditions. The reasons behind the higher susceptibility of the neuronal autophagic system to the pathogenic protein remain unknown, but might provide a basis for the greater severity of the pathology in neurons as the disease progresses. The negative consequences of impaired organelle degradation arise both from their loss of function but also from toxic effects due to their accumulation. Indeed, the autophagic system itself could be among the targets of this toxicity. Thus, we have recently described that impaired autophagy of lipid droplets leads to their intracellular accumulation and this eventually exerts an inhibitory effect on macroautophagic activity, perpetuating a vicious cycle25. It is thus plausible that the massive accumulation of lipid droplets observed in HD patients could eventually contribute to further failure of the autophagic system in these cells. Alterations in mitochondria have been extensively reported in different models of HD36, and overexpression of mutant htt is sufficient to alter mitochondrial dynamics, reduce mitochondria fusion and increase mitochondria fragmentation37. Recent studies have shown that alterations in mitochondrial fusion and fission induce macroautophagy to favor their removal 38. However, the reduced ability to sequester cytosolic cargo in HD cells would make any upregulation of macroautophagy futile.
In summary, we have identified a defect in the recognition of cargo by macroautophagy common to models of HD, lymphoblasts from HD patients, with consequences validated in the striatum of HD patients. Our findings could have important therapeutic implications because upregulation of macroautophagy has been successfully used to slow progression of HD pathology in animal models8. Further beneficial effects could be expected by developing means to improve recognition of cargo in HD cells.
METHODS
Animals, Cells and human brain tissue
18Qhtt and 111Qhtt knock–in mice were from The Jackson Laboratory and were maintained and used for our study under an animal protocol approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine. HD94 mice were generated as described39. MEFs were prepared as described40. Striatal cell lines were from Coriell Repository. Mouse fibroblasts expressing 146Qhtt–exon 1 and human lymphoblasts from normal controls and HD patients were gifts from N. Wexler, M. Andresen and J. Gusella. Striatal neuron cultures from postnatal HD94, 18Qhtt and 111Qhtt mice were prepared as described41. Fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma) in the presence of 10 % fetal bovine serum (FBS). Striatal cells were maintained in the same medium supplemented with high glucose and 2 mM L-glutamine in addition of FBS. Lymphoblasts were cultured in Roswell Park Memorial Institute medium (RPMI 1640) (Gibco) containing 15% FBS and 2 mM L-glutamine. Striatum neuronal cultures from day 0–2 postnatal HD94 mice (generated crossing CaMKIIα-tTA and HD94htt-BiTetO-lacz mice) were prepared and maintained as described41. The origin of the monolayer of astrocytes for the culture of primary neurons is indicated in each experiment. Frozen brain tissue from the caudate was obtained from the New York Brain Bank upon written consent from 4 patients: Control 1 (F, 54yr), Control 2 (F, 52 yr), HD patient 1 (F, 49 yr), HD patient 2 (M, 53 yr).
Chemicals
Sources of chemicals and antibodies were as described25,42. The antibodies against LC3, cytochrome C, mTOR and mTOR substrates were from Cell Signaling Technology. The antibodies against mannose–6–phosphate receptor and Rab 5 were a gift from A. Wolkoff. The antibody against huntigtin (MAB2166, clone 1HU–4C8) was from Chemicon Int., against GAPDH and actin from Abcam, against, p62 from Biomol. against Atgs, beclin–1, from Novus Biologicals, against COX IV from MitoSciences, against HDAC6, cathepsin B and D from Santa Cruz Biotechnology, and against polyubiquitin from Invitrogen. Mitotracker and Mitotracker CMXRos were from Invitrogen.
Intracellular protein turnover
To measure degradation of long-lived proteins in cultured fibroblasts, lymphoblasts and neurons, confluent cells were labeled with [3H]leucine (2 µCi/ml) for 48 h at 37°C and then extensively washed, incubated in radioactivity free media for 1 hour (to assure degradation of short-lived proteins) and then maintained in complete (10 % FBS) or serum-deprived medium containing an excess of unlabeled leucine (to prevent reutilization of the radiolabeled amino acid released into the media from the break-down of intracellular proteins)43. Aliquots of the medium taken at different times were precipitated with TCA to separate the radioactivity incorporate into intact proteins that could have been secreted into the media, from the radioactivity in free amino acids released into the media after proteolysis of intracellular proteins. Proteolysis was measured as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at each time point43. Total radioactivity incorporated into cellular proteins was determined as the amount of acid-precipitable radioactivity in labeled cells immediately after washing. Lysosomal-dependent degradation was inhibited by addition of 20 mM NH4Cl and macroautophagy-dependent degradation by addition of 10 mM 3-methyl-adenine in the incubation media. In all studies, cell viability was monitored in parallel wells to discard possible increase in cell death as a result of the different treatments. All of the studies were performed with confluent cells and proper monitoring was carried out to assure negligible cell loss during the experiments.
Isolation of subcellular fractions
Autophagosomes and autophagolysosomes were isolated from mouse liver after 12 h starvation by differential centrifugation and floatation in metrizamide gradients as previously described22. Fractions enriched in the different autophagic compartments were recovered from the interfaces of the metrizamide gradient and washed by centrifugation in 0.25 M sucrose. Membranes of the organelles in the different fractions were separated for the luminal fraction by high speed centrifugation (100,000 g for 30 min) after hypotonic shock44.
Fluorescence and immunocytochemical staining
Cells grown on coverslips were fixed with a 3 % formaldehyde solution, blocked, and then incubated with the primary and corresponding fluoresce-conjugated secondary antibodies42. Mounting medium contained DAPI (4',6-diamidino-2-phenylindole) to highlight the cellular nucleus. Neutral lipids were stained with Bodipy 493/503 as described25. Images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss Ltd., Thornwood, NY), with a ×63 objective and 1.4 numerical aperture, subjected to deconvolution with the manufacturer’s software and prepared using Adobe Photoshop 6.0 software (Adobe Systems Inc., Mountain View, CA). Quantification was performed in individual frames after deconvolution and on images with maximum projection of all Z-stack sections using ImageJ software (NIH). Colocalization was calculated with the JACoP plugin of the Image J software in single Z-stack section of deconvoluted images. For each cell, the mean value of colocalization in three different Z-stack sections was calculated and a minimum of 20 cells per slide were quantified. Particle number was quantified with the “analyze particles” function in thresholded single sections with size (pixel2) settings from 0.1–10 and circularity 0–1.
Electron microscopy, immunogold and morphometric analysis
Neuronal cells in culture, lymphoblasts, mouse livers sectioned in 1mm3 cubes or isolated autophagic compartments pellet after centrifugation were fixed in 2.5 % gluteraldehyde in SC (100 mM sodium cacodylate, pH 7·43) at room temperature for 45 min25. The pellet was then rinsed in SC, post-fixed in 1 % osmium tetroxide in SC followed by 1 % uranyl acetate, dehydrated through a graded series of ethanols and embedded in LX112 resin (LADD Research Industries). Ultrathin sections were cut on a Reichert Ultracut E, stained with uranyl acetate followed by lead citrate and viewed on a JEOL 1200EX transmission electron microscope at 80 kV. Immunogold labeling was performed in cell pellets fixed in 4 % paraformaldehyde/ 0.1 % glutaraldehyde in 0.1 M cacodylate buffer. Briefly, samples were fixed for 1 h at room temperature, dehydrated, embedded in Lowicryl and cut in ultrathin sections as described above. Each grid was washed in 50 mM glycine in phosphate buffered saline, blocked and preincubated in the antibody incubation buffer for 1 h. Blocked grids were incubated with the antibody against LC3 for 2 h, extensively washed and incubated with the gold-conjugated secondary antibody (1:100) for another 2 h. Control grids were incubated with either an irrelevant IgG and the secondary antibody under the same conditions or only with the secondary antibody. After extensive washing, samples were fixed a second time for 5 min in 2 % glutaraldehyde washed and negatively stained with 1% uranyl acetate for 15 min. Morphometric measurements were conducted in digital micrographs using Image J software. For the HD and normal control lymphoblasts, all analysis was conducted by an individual blind to all of the treatments and cell types. The relevant organelles and cell edges were traced, and density determined. All of the samples were processed and images acquired and processed identically.
Histochemical detection of Lipid droplets in human brain
Frozen brain tissue from the caudate was obtained from the New York Brain Bank from 4 patients: Control 1 (F, 54 yr), Control 2 (F, 52 yr), HD patient 1 (F, 49 yr), HD patient 2 (M, 53 yr). Brains were chilled between 2–4 h post-mortem and frozen 6–30 h postmortem. The two HD patients were rated at a grade of neuropathological severity of 4/4. Tissue blocks of region SBB6 (encompassing caudate, putamen, and nucleus accumbens), were cryosectioned and Oil Red O and hematoxylin staining was performed by the Histology Service of the Experimental Molecular Pathology facility at Columbia University Medical Campus. Standard eosin staining was omitted, as its pink color would interfere with identification of Oil Red O label.
Gray matter regions in the sections were selected randomly and fields of 300 × 380 µm were photographed with differential interference contrast optics. The micrographs were processed identically and analyzed for the number of hematoxylin-labeled nuclei, and number and area of Oil Red O-labeled objects. Analysis of 3–9 micrographs per patient were performed by three observers, two of whom were blind to the conditions, and all observers reported nearly identical determinations.
Statistical analysis
Results are shown as the mean+s.d. Student’s t test for unpaired data was used for statistical analysis and a one–way analysis of variance (ANOVA) was used for multiple comparisons. A value of p<0.05 was considered statistically significant.
General methods
Protein was determined by the Lowry method and lysosomal enzymatic activities were measured as reported45. After SDS-PAGE and immunoblotting, the proteins recognized by the specific antibodies were visualized by chemiluminescence methods (Renaissance, NEN-Life Science Products). Membranes were exposed to BioMax Light Kodak films (Kodak Scientific Films) for increasing periods of time ranging from 5 seconds to 10 minutes. Densitometric quantification of the immunoblotted membranes was performed with an Image Analyzer System (Inotech S-100, Sunnyvale, CA). The intensity of the bands was quantified using the square “spot denso” routine of the Image Analyzer System. Isoelectric focusing (IEF) was done using the Protean IEF Cell (Bio-Rad) at 20 °C with rapid ramping to voltage 10000 V at a current limit of 50 µA using pH 3–10 nonlinear ReadyStrip IPG Strips (Bio-Rad). Cell viability and proliferation were determined by incubating cells with 1 mg/ml of 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) in DMEM for 1 h at 37 °C. The formazan product formed was solubilized with N-propyl alcohol and its absorbance measured at 560nm. Cell viability after different treatments was calculated as the percentage of the absorbance in untreated cells. For the filter retardation assays, samples were run through a 0.22 µm nitrocellulose membrane in a BioDot Blot apparatus (BioRad (Hercules, CA)), following resuspension of pellets in 100 µl of Tris Buffer and 100 µl of 4 % SDS-100mM DTT, and incubation at 100°C for 5 min. SYPRO Ruby Protein Stain (Bio-Rad) was used to highlight total proteins in bi-dimensional gels. Carbonyl groups in oxidized proteins were detected after derivatization with DNPH followed by immunoblot with an antibody against the DNP moiety using the OxyBlot™ Oxidized Protein Detection Kit from Chemicon International following the manufacturer’s recommendation. Images of immunoblots were prepared for publication using Adobe Photoshop CS3 Extended version 10.0.1. software. For those images in which brightness or contrast was modified to improve visibility of the bands the change was applied equally across the entire image and was applied equally to controls. Immunoblot cropping was limited to display at least six bands width above and below the band, but where large gel panels compromised clarity, the cropped blot is shown in the main figure and the full-length blots are presented in supplementary figures.
Supplementary Material
Acknowledgements
We thank Dr. Rajat Singh and Ms. Bindi Patel for technical assistance. This work was supported by a HDSA Grant (DS, AMC), NIH/NIA grants AG021904, AG031782, DK041918 (AMC) and a NINDS Udall Center of Excellence grant (DS), the Picower, Simons (DS) and Glenn (AMC) Foundations. E.W. is a fellow of the Hereditary Disease Foundation. E.A. is a Fulbright fellow. S.K. is supported by NIN/NIA T32AG023475.
Footnotes
Author’s contributions
MM–V, ZT, and EW, performed the experiments that constitute the main body of this work; RdV, HK, SK, EA and GT completed the rest of the experiments; SH analyzed the electron micrographs of lymphoblasts; AMC and DS designed the study and wrote the paper.
References
- 1.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinsztein DC. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar S, Rubinsztein DC. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–4270. doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- 4.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 5.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwata A, et al. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J Biol Chem. 2009;280:40282–40292. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 9.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 11.Kegel KB, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sapp E, et al. Huntingtin localization in brains of normal and Huntington's disease patients. Annals of Neurology. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 13.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 14.Nagata E, Sawa A, Ross CA, Snyder SH. Autophagosome-like vacuole formation in Huntington's disease lymphoblasts. Neuroreport. 2004;15:1325–1328. doi: 10.1097/01.wnr.0000127073.66692.8f. [DOI] [PubMed] [Google Scholar]
- 15.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. Journal of Biological Chemistry. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler V, et al. Length-dependent gametic CAG repat instability in the Huntington's disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky D, Cuervo A, Seglen P. Methods for Monitoring Autophagy from Yeast to Human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 20.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 21.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO Journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009 doi: 10.1038/nature07976. E-pub before print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington's disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen K, Fon E, Hastings T, Edwards R, Sulzer D. Methamphetamine-induced degeneration of dopaminergic neurons involves autophagy and upregulation of dopamine synthesis. J Neurosci. 2002;22:8951–8960. doi: 10.1523/JNEUROSCI.22-20-08951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinsztein DC, et al. Autophagy and its possible roles in nervous sytem diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 29.Chu CT, et al. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 32.Stefanis L, Larsen K, Rideout H, Sulzer D, Greene L. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu W, et al. Macroautophagy--a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulzer D, et al. Neuromelanin biosynthesis is driven by excess cytosolic catecholamines not accumulated by synaptic vesicles. Proc Natl Acad Sci U S A. 2000;97:11869–11874. doi: 10.1073/pnas.97.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae BI, et al. Mutant huntingtin: nuclear translocation and cytotoxicity mediated by GAPDH. Proc Natl Acad Sci U S A. 2006;103:3405–3409. doi: 10.1073/pnas.0511316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Browne SE. Mitochondria and Huntington's disease pathogenesis: insight from genetic and chemical models. Ann N Y Acad Sci. 2008;1147:358–382. doi: 10.1196/annals.1427.018. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Aparicio E, et al. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington's disease. J Neurosci. 2001;21:8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28:21. doi: 10.1002/0471142727.mb2801s70. Unit 28. [DOI] [PubMed] [Google Scholar]
- 41.Petersen A, et al. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum Mol Genet. 2001;10:1243–1254. doi: 10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 42.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohsumi Y, Ishikawa T, Kato K. A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J Biochem. 1983;93:547–556. [PubMed] [Google Scholar]
- 45.Storrie B, Madden E. Isolation of subcellular organelles. Meth Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.