Abstract
The main goal of this study is to elucidate the mechanisms of the signal transmission for radiation induced bystander response. The NF-κB-dependent gene expression of IL8, IL6, PTGS2/COX2, TNF and IL33 in directly irradiated human skin fibroblasts produced the cytokines and prostaglandin E2 (PGE2) with autocrine/paracrine functions, which further activated signaling pathways and induced NF-κB-dependent gene expression in bystander cells. As a result, bystander cells also started expression and production of interleukin-8, interleukin-6, COX-2-generated PGE2 and interleukin-33 (IL-33) followed by autocrine/paracrine stimulation of the NF-κB and MAPK pathways. A blockage of IL-33 transmitting functions with anti-IL-33 monoclonal antibody added into the culture media decreased NF-κB activation in directly irradiated and bystander cells. On the other hand, the IGF-1-Receptor kinase regulated the PI3K-AKT pathway in both directly irradiated and bystander fibroblasts. A pronounced and prolonged increase in AKT activity after irradiation was a characteristic feature of bystander cells. AKT positively regulated IL-33 protein expression levels. Suppression of the IGF-R1--AKT--IL-33 pathway substantially increased radiation-induced or TRAIL-induced apoptosis in fibroblasts. Taken together, our results demonstrated the early activation of NF-κB-dependent gene expression first in directly irradiated and then bystander fibroblasts, the further modulation of critical proteins, including IL-33, by AKT in bystander cells and late drastic changes in cell survival and in enhanced sensitivity to TRAIL-induced apoptosis after suppression of the IGF-1R--AKT--IL-33 signaling cascade in both directly irradiated and bystander cells.
Keywords: NF-κB, Interleukin-33, IGF-1-Receptor, AKT, Bystander response
1. Introduction
Ionizing radiation and chemotherapy are two principal therapeutic modalities used for cancer treatment. Ever since the discovery of the X-rays, it has always been accepted that the deleterious effects of ionizing radiation are due to direct damage of DNA. However, radiation-induced bystander response, which is defined as the induction of biological effects in cells that are not directly exposed to ionizing radiation, but in close proximity to cells that are irradiated, represents a paradigm shift in our understanding of radiobiological effects of ionizing radiation [1–3]. The most direct approach to bystander studies, precise α-irradiation of cells using a focused microbeam, has been successfully applied in numerous studies performed with the Columbia University charged particle microbeam [4–6], as well as with focused microbeams in other laboratories [7]. There is also is considerable evidence that media from irradiated culture (after transfer to non-irradiated cells) can induce biological effects in the latter, suggesting that irradiated cells secreted biologically active factors [8].
Although bystander effects have been described over the past decade, the precise mechanisms of these processes remain unclear. There are at least five aspects of the initiation and development of bystander response: i) radiation-induced stress reactions of exposed cells, which could be accompanied by release of pre-existing signal transmitters with nuclear localization, such as Interleukin-1α (IL-1α) and High-mobility group box-1 (HMGB1) protein, followed by initiation of autocrine and paracrine stimulation of cells by these ligands via the corresponding receptors [9]; ii) DNA-damage induced activation of ATM-p53 and ATM-NF-κB signaling pathways followed by the stimulation of NF-κB-dependent gene expression, including IL8, TNF, COX2, iNOS, and production of prostaglandin E2 (PGE2), reactive oxygen species (ROS) and nitric oxide (NO) [10–12]; iii) the activation and stimulation of bystander cells via a paracrine mechanism using cytokine or growth factor interactions with the correspondent receptors and an induction of cell signaling pathways and specific gene expression in bystander cells [6, 12]; iv) translocation of ROS and NO from directly irradiated to bystander cells through gap junction channels with the secondary damaging effects on the mitochondria and DNA in bystander cells [12]; v) induction of cell survival mechanisms or balancing between survival and apoptotic signaling at the late stages of bystander response [1].
The significance of ROS, NO, some cytokines, such as TGFβ and TNFα, and intercellular gap junctions for mediating bystander effects was confirmed in numerous studies, reviewed by Hei at al. [1] and Prise and O’Sullivan [13]. In our previous publications, we discovered a role for the transcription factor NF-κB, after direct exposure of human fibroblasts to α-particle irradiation, in the activation of COX2 and iNOS gene expression followed by production and translocation of ROS and NO into bystander cells [6, 12]. A recent investigation of global gene expression in directly irradiated and bystander cells further revealed transcription factor NF-κB as the dominant signaling hub in bystander response [14, 15], and introduced additional players, which have been investigated in the present study. These include NF-κB-dependent cytokines, IL-6 and IL-8 [16], and cytokines that induce the NF-κB signaling pathway via paracrine or autocrine mechanisms (IL-1β, TNFα and IL-33) [11, 17] in concert with receptor tyrosine kinase activities that control the PI3K-AKT and the MAPK pathways [18, 19]. The main aim of the present study was to elucidate IL33 (Interleukin-33) gene expression in human skin fibroblasts and the role of the IL-33--IL-33R/ST2--NF-κB and IGF-1R--PI3K--AKT--IL-33 signaling pathways in mediating bystander responses, including effects on cell survival and apoptosis. Our results indicated that IL33 expression played an important role in radiation-induced bystander effects via NF-κB-dependent regulation of expression of numerous genes, including several cytokines and COX2. However, in addition to damaging effects mediated by NF-κB-dependent COX-2--PGE2 and iNOS--NO expression, NF-κB-dependent expression of IL-33 controlled cell survival functions, balancing between cell life and death [20].
2. Materials and methods
2.1. Materials
Human Killer-TRAIL was purchased from Axxora (San Diego, CA, USA). PI3K inhibitor LY294002, IKK inhibitor BMS-345541, ATM inhibitor KU55933 and IGF-1 Receptor inhibitor picropodopyllin (PPP) were purchased from Calbiochem/EMD Chemicals (San Diego, CA, USA). Insulin-like Growth Factor Binding Protein-3 (IGFBP-3) was obtained from Sigma (St. Louis, MO, USA).
2.2. Cell culture
Human skin fibroblasts (HSF) immortalized by SV40 T antigen [12] were maintained in a 4.5 g/L glucose DMEM with 4 mM L-glutamine and 100 mg/L sodium pyruvate supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Suppression of p53 downstream activities, which were non-essential for bystander response [21], by SV40 T antigen allowed us to simplify the further analysis of gene expression that controls the development of bystander response in HSF. Normal human lung fibroblasts, IMR-90 (Coriell Cell Repository, Camden, NY, USA), were maintained in DMEM supplemented with 15% fetal bovine serum, antibiotics, vitamins and non-essential amino acids (Invitrogen, Carlsbad, CA, USA).
2.3. Irradiation procedure
The strip mylar dishes were used in the present studies as described [6]. Exponentially growing HSF were plated in the concentric strip mylar dishes two days before irradiation to ensure a confluent state. A 50 cGy dose of 4He ions (120 keV/μm) was delivered to the cells using the track segment irradiation facility of the 5.5-MV Singletron accelerator at the Radiological Research Accelerator Facility of Columbia University. After irradiation, at selected time points, the inner and outer mylar dishes were separated and the cells from each growth surface were trypsinized and individually pooled for endpoint analysis.
2.4. Survival of irradiated and non-irradiated cells
Non-irradiated control cells, α-irradiated and bystander cells were collected 24 h after irradiation. Cultures were trypsinized, counted with a Coulter counter, and aliquots of the cells were replated into 100-mm-diameter dishes for colony formation. Cultures for clonogenic survival assays were incubated for 12 days, at which time they were fixed with formaldehyde and stained with Giemsa. The number of colonies was counted to determine the surviving fraction as described [4].
2.5. RNA isolation
Directly irradiated (outer dish) and bystander (inner dish) cells were separated at specified times after irradiation and RNA was isolated using Ribopure (Applied Biosytems, Foster City, CA). RNA concentration was measured using a NanoDrop-1000 spectrophotometer (Thermo Scientific, Waltham, MA) and RNA quality was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
2.6. Quantitative Real-Time PCR (qRT-PCR)
The High-Capacity cDNA Archive Kit (Applied Biosystems) was used to prepare cDNA from total RNA. A custom low-density TaqMan array was designed with validated assays and obtained from Applied Biosystems as previously described [15]. For gene validation studies, 100 ng cDNA was used as input for low-density arrays. qRT-PCR reactions were performed with the ABI 7900 Real Time PCR System using Universal PCR Master Mix (Applied Biosystems) with initial activation at 50°C for 120 seconds and 94.5°C for 10 minutes followed by 40 cycles of 97°C for 30 seconds and 59.7°C for 60 seconds. Individual TaqMan assays for IL33 (Hs00369211_m1), IGF1R (Hs00951562_m1) and IGFBP3 (Hs00426287_m1) were pre-made and validated assays from Applied Biosystems. Input cDNA was set at 10 ng for all samples and genes, except IL33, which required 100 ng input, and qRT-PCR reactions were performed with the ABI 7900 Real Time PCR System using Universal PCR Master Mix from Applied Biosystems. All single gene assays were run in duplicate reactions. Relative fold-inductions were calculated by the ΔΔCT method as previously used [22] and with SDS version 2.3 software (Applied Biosystems). We measured 7 housekeeping genes on the low-density arrays and applied Genorm [23] to determine the most appropriate genes for normalizing the results. Both the low-density array data and the individual gene assays were normalized to Ubiquitin-C (UBC).
2.7. FACS analysis of DR5 surface levels
Surface levels of DR5 were determined by staining with the PE-labeled mAb from eBioscience (San Diego, CA, USA). A FACS Calibur flow cytometer (Becton Dickinson, Mountain View, CA, USA) combined with the CellQuest program was used to perform flow cytometric analysis.
2.8. Apoptosis studies
Cells were exposed to soluble TRAIL (50 ng/ml) alone or in combination with cycloheximide (2 μg/ml). Different variants of combined treatment were used, including α-irradiation (0.5 Gy) followed by TRAIL treatment. Apoptosis was then assessed by quantifying the percentage of hypodiploid nuclei using FACS analysis.
2.9. Western blot analysis
Total cell lysates (50 μg protein) were resolved on SDS-PAGE, and processed according to standard protocols. The monoclonal antibodies used for Western blotting included: anti-β-Actin (Sigma, St. Louis, MO, USA); anti-FLIP (NF6) (Axxora); anti-ATM (D2E2) and anti-phospho-ATM (Ser1981); anti-phospho-p53 (Ser15); anti-Smac/Diablo; anti-basic FGF; anti-GSK-3β (Cell Signaling, Beverly, MA, USA); anti-XIAP (clone 48) BD Biosciences (San Diego, CA, USA); anti-IL33 (ProSci, Poway, CA, USA); anti-IL8 (R&D Systems, Minneapolis, MN, USA). The polyclonal antibodies used included: anti-DR5 (Axxora); anti-phospho-p53 (Ser20), and anti-total p53; anti-phospho-SAPK/JNK (Thr183/Tyr185) and anti-JNK; anti-phospho-p44/p42 MAP kinase (Thr202/Tyr204) and anti-p44/p42 MAP kinase; anti-phospho-AKT (Ser473) and anti-AKT; anti-phospho-IGF-1 Receptor (Tyr1135/1136) and anti-IGF-1 Receptor β anti-β-Catenin; anti-Phospho-GSK-3β; anti-FOXO3A (Cell Signaling). The secondary Abs were conjugated to horseradish peroxidase; signals were detected using the ECL system (Thermo Scientific, Rockford, IL, USA).
2.10. EMSA
Electrophoretic mobility shift assay (EMSA) was performed for the detection of NF-κB DNA-binding activity as previously described [24]. Ubiquitous NF-Y DNA-binding activity was used as an internal control. Furthermore, Western blot analysis followed by densitometric analysis of IκBα, an inhibitor of NF-κB, and β-actin levels was performed. Background corrected IκBα intensity measurements were normalized to β-actin protein levels.
2.11. Statistical analysis
Data of 3–4 independent experiments were calculated as means and standard deviations. Comparisons of results between treated and control groups were made by the Students’ t-tests. A P-value of 0.05 or less between groups was considered significant.
3. Results
3.1. Signaling pathways in directly α-irradiated and bystander fibroblasts
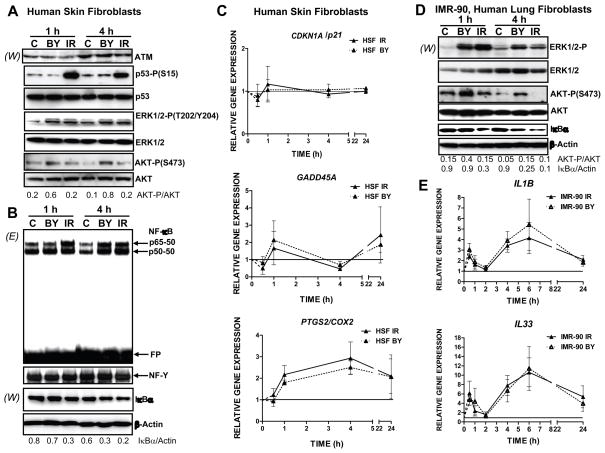
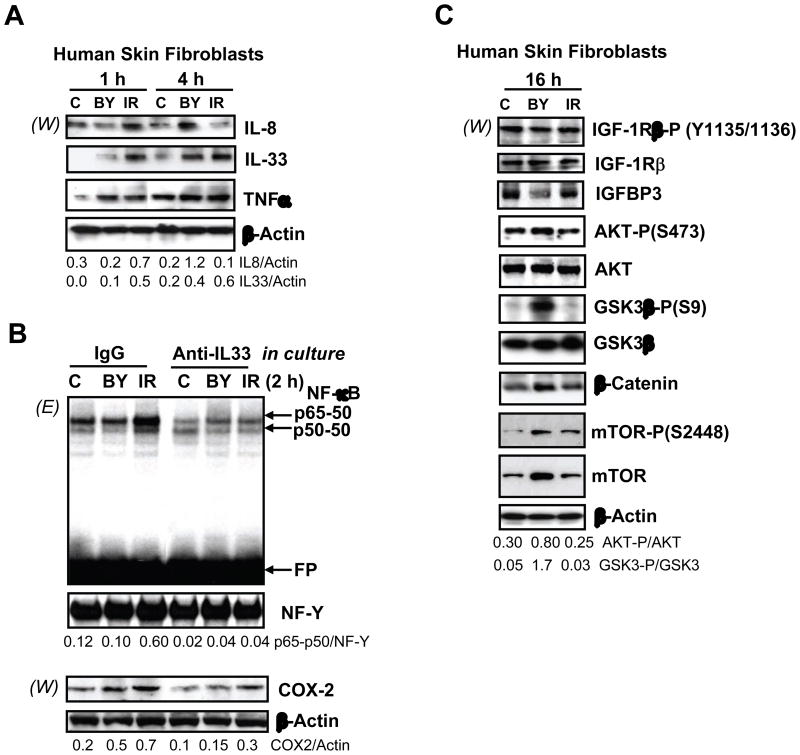
Ionizing radiation induces and accelerates numerous signaling pathways, some of which are initiated in the nucleus due to DNA damage, including two master signaling pathways, ATM-p53 and ATM-NF-κB [10, 25, 26]. Other signaling pathways are started at the cell surface by a ligand-receptor interaction, such as IGF-1/IGF-1R or FGF2/FGF-R, followed by activation of growth factor receptor tyrosine kinases [19, 27]. A direct modification of p53 via ATM-dependent phosphorylation of p53-Ser15, a characteristic feature of ATM activation, was observed one hour after α-particle irradiation of human skin fibroblasts (HSF), but no accompanying stabilization and accumulation of p53 protein (Fig. 1A), consistent with the presence of SV40 T-antigen used for immortalization of these cells [12]. Correspondingly, p53-dependent gene expression, such as CDKN1A/p21, and p53-dependent downstream pathways was mostly blocked in these fibroblasts (Fig. 1C and data not shown). GADD45A gene expression that could be p53-dependent or p53-independent [28] demonstrated modest activation in both directly irradiated and bystander cells (Fig. 1C). Since the p53 downstream pathway was not essential for induction of bystander response [15, 21], our cell model allowed us to focus on other crucial genes controlling the development of bystander response, especially, on gene expression regulated by transcription factor NF-κB. Up-regulation of nuclear NF-κB p65-p50 activity (determined by EMSA) that was accompanied by IκBα degradation (determined by Western) was detectable in irradiated fibroblasts one hour after treatment and then in both directly irradiated and bystander cells four hours after treatment, indicating an induction of bystander signaling (Fig. 1B). PTGS2/COX2 gene expression, a classical target of NF-κB, rapidly increased after irradiation in both directly exposed and bystander HSF (Fig. 1C).
Fig. 1.
The main signaling pathways induced in directly irradiated and bystander fibroblasts. (A, B) Expression levels of indicated proteins from the control, bystander and α-irradiated human skin fibroblasts (HSF) were determined 1 and 4 h after treatment by Western blot analysis (W). EMSA (E) was performed for determination of DNA-binding activity of nuclear NF-κB and NF-Y transcription factors. Two main NF-κB DNA-binding complexes are indicated. Ubiquitous NF-Y activity was used as an internal control of the nuclear protein loading. Position of the free labeled probe (FP) is indicated. (C) Time course of gene expression of CDKN1A/p21, GADD45A and PTGS2/COX2 in directly irradiated (IR) and bystander cells (BY) at 0.5, 1, 4, and 24 h after irradiation, was determined by quantitative real time PCR and normalized to Ubiquitin C (UBC). Values are relative to time matched controls (solid line at 1). Points are the mean and standard error of three independent experiments. (D) Western blot analysis of indicated proteins from the control, bystander and α-irradiated IMR-90, human lung fibroblasts. (E) Time course of gene expression of IL1B and IL33 in directly irradiated and bystander IMR-90 fibroblasts.
Furthermore, α-irradiation was linked with activation of the MAPK pathways and the PI3K-AKT pathway in exposed cells. The MEK-ERK1/2 pathway was substantially up-regulated in both directly irradiated and bystander HSF 1–4 h after treatment (Fig. 1A); while two other MAPK’s, JNK and p38, did not exhibit notable levels of phosphorylation in HSF (data not shown). Surprisingly, a significant increase in levels of active form phospho-AKT (S473) 1–4 h after irradiation was characteristic of bystander, but not directly irradiated HSF, which demonstrated more modest increase in AKT phosphorylation (Fig. 1A; see also Fig. 5A).
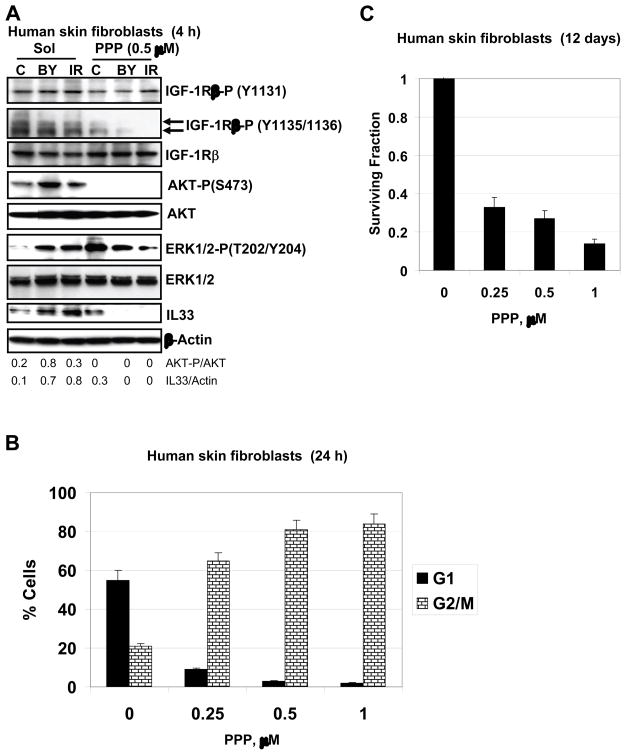
Fig. 5.
Effects of suppression of IGF-R1-mediated signaling pathway on AKT and ERK activation and apoptosis of HSF. (A) Western blot analysis of indicated proteins from HSF, which were irradiated in the presence and absence of PPP (0.5 μM). Actin was used as a protein loading control. (B) G2/M arrest of the cell cycle following PPP (0.25–1.0 μM) treatment of HSF. (C) Clonogenic survival assay of HSF 12 days after indicated treatment with PPP.
Strong up-regulation of ERK1/2 activities was also observed in directly irradiated and bystander human lung fibroblasts, IMR-90. Similarly with HSF, pronounced AKT phosphorylation was a characteristic feature of bystander IMR-90 fibroblasts (Fig. 1D). Up-regulation of NF-κB activity (determined by a notable decrease in IκBα levels) and NF-κB-dependent gene expression of PTGS2/COX2 [15], IL1B and IL33 was also observed in both directly irradiated and bystander IMR-90 cells (Fig. 1D and E), highlighting a role for NF-κB transcriptional targets in bystander response.
3.2. A role of ATM for regulation of bystander response
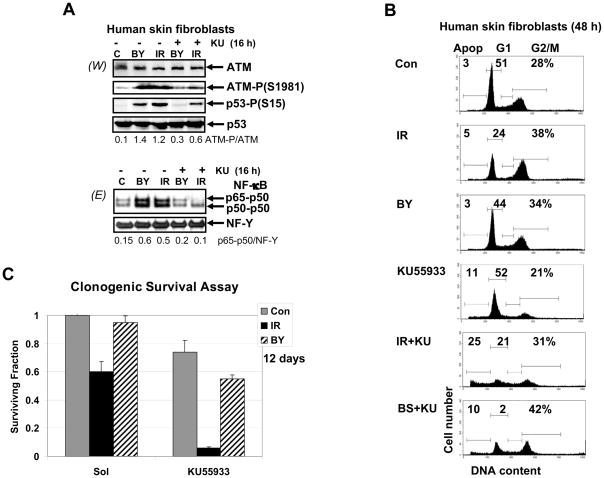
Sixteen hours after α-particle irradiation, ATM activation via autophosphorylation was observed in both directly irradiated and bystander fibroblasts (Fig. 2A). As we mentioned, two characteristic downstream targets of the ATM signaling pathway initiated by DNA damage are transcription factors p53 and NF-κB [10, 25, 29]. Using the specific pharmacological inhibitor of ATM autophosphorylation and activation, KU55933 (10 μM), we demonstrated effects of ATM inhibition on both p53- and NF-κB-signaling pathways: i) suppression or down-regulation of ATM-dependent Ser15 and Ser20 phosphorylation of p53 (Fig. 2A and data not shown) and ii) down-regulation of the nuclear NF-κB DNA binding activity in both directly irradiated and bystander HSF (Fig. 2A). These data established a dependence of NF-κB activation on ATM in HSF. Since ATM was involved in the regulation of the cell cycle and radiation-induced apoptosis, suppression of its activity by KU55933 drastically changed cell cycle profiles by up-regulating apoptotic levels in directly irradiated HSF and increasing levels of G2/M arrest for bystander cells (Fig. 2B). Furthermore, clonogenic survival assay revealed a dramatic decrease in general cell survival of directly irradiated HSF and notable decrease in survival of bystander fibroblasts in the presence of KU55933 (Fig. 2C). Taken together, results obtained confirmed a general regulatory function of ATM for both radioprotection and radiation-induced bystander response of HSF.
Fig. 2.
The ATM signaling pathway induced NF-κB activation in directly irradiated and bystander human skin fibroblasts. (A) Expression levels of total and phosphorylated ATM and p53 from the control, bystander and α-irradiated human skin fibroblasts were determined 4 h after treatment by Western blot analysis. KU55933 (10 μM), a pharmacological inhibitor of ATM activity, was added 30 min before α-irradiation (0.5 Gy). EMSA was performed for determination of DNA-binding activity of nuclear NF-κB and NF-Y. (B) Cell cycle-apoptosis analysis of control non-irradiated, directly irradiated and bystander cells 48 h after treatment using PI staining of DNA and FACS analysis. Results of a typical experiment (one from four) are shown. KU55933 was added 30 min before α-irradiation. (C) Clonogenic survival assay of human skin fibroblasts (control, α-irradiated and bystander) 12 days after treatment in the presence or absence of KU55933. Error bars represent mean ± S.D. from four independent experiments.
3.3. Cytokine gene expression in directly irradiated and bystander HSF
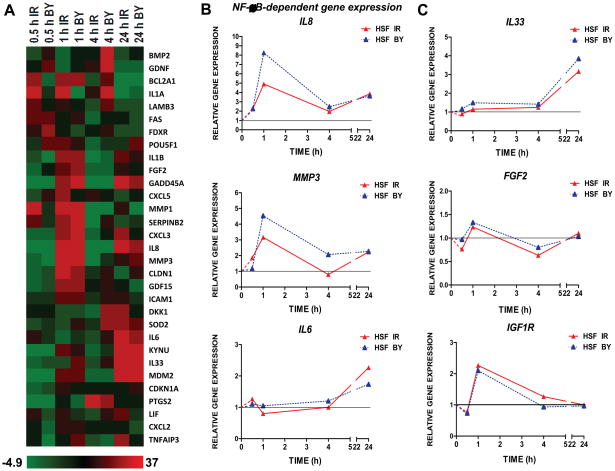
Within 0.5–1 h of irradiation, mRNA levels of the known NF-κB transcriptional targets PTGS2/COX2 and IL8, started to increase (Fig. 1D and 3A and 3B) in directly hit and bystander cells. A substantial up-regulation of IL-8 (Fig. 4A) and COX-2 [12] protein expression also occurred in bystander cells 4 h after treatment highlighting the generation of bystander response. The increase in IL8 mRNA levels preceded the corresponding change in protein levels in irradiated and bystander cells (Fig. 3B, and 4A).
Fig. 3.
Relative gene expression in α-irradiated and bystander HSF 0.5–24 h after treatment. (A) Heatmap of gene expression levels across the time course of 0.5, 1, 4 and 24 h in directly irradiated and bystander cells as measured by low density qPCR array. (B) Expression of IL8, MMP3 and IL6 in HSF in directly irradiated (red triangles) and bystander cells (blue triangles) at 0.5, 1, 4, and 24 h after irradiation, relative to time-matched controls and normalized to UBC. Gene expression was determined by quantitative real time PCR. (C) Relative gene expression changes in IL33, FGF2 and IGF-R1 were determined by quantitative real time PCR.
Fig. 4.
Protein expression levels of human skin fibroblasts following α-irradiation. (A) Western blot analysis of intracellular levels of indicated cytokines 1–4 h after α-irradiation. Actin was used as a protein loading control. Ratios of IL8/Actin and IL33/Actin are indicated. (B) EMSA for determination of nuclear NF-κB DNA-binding activity. Nonspecific IgG or anti-IL33 mAb (2 μg/ml) were introduced into the cell media 1 h before α-irradiation. Nuclear and cytoplasmic proteins were isolated from HSF 2 h after treatment. Nuclear protein was used for EMSA while cytoplasmic protein was used for detection of COX-2 and Actin by Western blot analysis. The p65-p50 NF-κB/NF-Y ratio and the COX-2/Actin ratio are indicated. (C) Western blot analysis of indicated proteins from human skin fibroblasts 16 h after treatment.
Additional NF-κB gene targets and some other genes identified in a prior global analysis of gene expression in directly irradiated and bystander IMR-90 fibroblasts [15] were screened for radiation and bystander response in HSF cells (Fig. 3A). Besides IL8, the NF-κB targets IL1A (interleukin-1A), IL1B and MMP3 (matrix metallopeptidase 3) also exhibited early increases in gene expression, while SOD2 (superoxide dismutase 2) was up-regulated 4 h and IL6-24 h after irradiation (Fig. 3A, B). Although IL33, a recently discovered member of the IL-1 cytokine family and a potential NF-κB target [20, 30], was transcribed at relatively low levels in HSF, its expression increased in both directly irradiated and bystander cells, modestly at 1–4 h after irradiation and pronouncedly by 24 h (Fig. 3A and 3C). Western blot analysis revealed increased protein levels of IL-33 (MW 31 kD) in bystander HSF 4 h after irradiation (Fig. 4A). Pronounced upregulation of IL33 and IL1B gene expression was also observed in directly irradiated and bystander IMR-90 cells (Fig. 1E).
On the other hand, endogenous TNFα protein (precursor form of 26 kD) was present at relatively high levels in both irradiated and bystander cells 1–4 h after treatment (Fig. 4A). A role for TNFα in the generation of ROS and regulation of bystander response was previously described [6, 12]. Skin fibroblasts are known to express receptors for IL-1, IL-6, IL-8 and IL-33, as well as TNF-R1 [31, 32]. Hence, our observations allowed us to consider cytokines IL-1α, IL-1β, IL-6, IL-8 and IL-33, in concert with TNFα, as potential intercellular signal transmitters following α-particle irradiation.
IL-33 is a known activator of the NF-κB signaling pathway upon its interaction with the specific IL-33-Receptor/ST2 [17]. To confirm IL-33-dependent NF-κB activation in HSF, we added the inhibitory anti-IL-33 mAb (2 μg/ml) to the cell culture media 1 h before irradiation. This notably decreased the nuclear basal and inducible NF-κB DNA-binding activities determined by EMSA in both directly exposed and bystander cells 2 h after treatment (Fig. 4B). Simultaneously, protein levels of COX-2, a classical NF-κB target, were also decreased in the cytoplasmic fraction of cells (Fig. 4B) indicating a dependence of COX-2 expression levels on the IL-33--NF-κB pathway in HSF. We previously observed regulation of COX-2 expression via the TNFα--NF-κB pathway [12]. Taken together, our data demonstrated that both cytokines IL-33 and TNFα are involved in activation of the NF-κB signaling pathway in HSF, which is also under general control of the nuclear ATM activity (see Fig. 2).
3.4. Growth factors and growth factor receptors expression in HSF
Another NF-κB target gene, FGF2, encoding Fibroblast Growth Factor-2, demonstrated relatively modest changes in kinetics of transcription after irradiation of fibroblasts (Fig. 3C). FGF2 protein levels were easily detectable in non-treated control cells and were additionally increased 1 h after irradiation. FGF-Receptor-1 protein expression, however, was not detectable in either control or treated HSF (data not shown). This means that FGF2 was probably not a player in radiation-induced autocrine/paracrine stimulation in human skin fibroblasts, although it was involved in paracrine stimulation of melanocyte and melanoblast proliferation in vivo [33].
Endogenous IGF-1 (Insulin-like Growth Factor-1) gene expression was detected at low levels and did not notably change following irradiation of HSF (data not shown). However, exogenous IGF-1 protein was a constitutive component of the HSF cell media. IGF-1R (Insulin-like Growth Factor-1 Receptor) mRNA and protein levels showed a modest and transient increase in expression 1 h after irradiation. At later time points, IGF-1-Receptor-β mRNA and total protein levels were relatively stable with high content of the active phosphorylated form, IGF-1Rβ-P (Y1135) (Fig. 3C and 4C). This suggests that IGF-1/IGF-1R interaction might be permanently involved in the regulation of cell signaling cascades in both control and exposed HSF.
3.5. IGF-R--AKT--β-catenin and AKT-mTOR axes in HSF
The IGF-1R-kinase activation was accompanied by upregulation of AKT activity that was still detected at higher levels in bystander cells 16 h after irradiation (Fig. 4C). A pronounced decrease in the endogenous Insulin-like Growth Factor Binding Protein-3 (IGFBP-3) levels, an upstream regulator of IGF-mediated signaling, might be a reason for upregulation of AKT activity in bystander cells (Fig. 4C). AKT inactivated GSK3β via Ser9-phosphorylation in bystander cells that was followed by notable up-regulation of β-catenin protein levels, due to β-catenin stabilization (Fig. 4C) [34]. Stabilization of β-catenin favors its nuclear translocation, leading to positive effects on transcriptional regulation (in complex with LEF1) of targets such as PTGS2/COX2 [12, 35].
Another established axis of the AKT signaling pathway, mTOR phosphorylation and activation, was substantially upregulated in bystander HSF 16 h after treatment (Fig. 4C). Taken together, these observations highlighted activation of the AKT signaling pathway and its pronounced effects on the classical targets in bystander fibroblasts.
3.6. Pro-survival role for the AKT pathway and IL-33 expression in bystander fibroblasts
To further confirm the major role of IGF1-R-mediated signaling in the activation of the downstream AKT signaling pathway in HSF, we used picropodophyllin (PPP), a specific inhibitor of IGF1-R-mediated signaling that blocks the critical Tyr1135 autophosphorylation [36] and LY294002, an inhibitor of the PI3K-like kinases and PI3K-AKT pathway. Treatment with PPP (0.5 μM) caused only a slight decrease in non-target phospho-(Y1131)-IGF-1R levels, but a strong decrease in phospho-(Y1135/Y1136)-IGF1R levels, which are crucial for kinase activity (Fig. 5A). A minor basal activity was only detected in control cells (Fig. 5A) disappearing at increased dose of PPP (1.0 μM) (data not shown). PPP at 0.5 μM completely blocked basal and inducible AKT phosphorylation in both bystander and irradiated cells (Fig. 5A). Furthermore, PPP decreased levels of ERK1/2 phosphorylation, but only in directly irradiated cells, while strongly increasing phosho-ERK1/2 levels in control cells (Fig. 5A). Mutual antagonism between ERK and AKT activation could be partially responsible for such regulation [37].
On the other hand, LY294002 (50 μM) modestly decreased the basal levels of AKT phosphorylation and abolished inducible phosphorylation in both bystander and irradiated cells (data not shown). This suggested that regulation of the basal and inducible AKT activity based on different mechanisms in HSF. Furthermore, there was no linear correlation between IGF-1R kinase activity and activation of AKT in distinct fibroblast types (control, irradiated and bystander). It suggested that additional modulation, for example, either by PTEN or IGFBP-3 binding activity might finally determine levels of AKT activation in bystander and irradiated fibroblasts. IL-33 expression was also substantially suppressed by LY294002 (50 μM) and completely blocked by PPP (0.5 μM) in bystander and irradiated cells, but incompletely in the control cells (Fig. 5A). Hence, IGF-1R-dependent AKT signaling was involved in regulation of IL-33 expression levels in both irradiated and bystander fibroblasts (Fig. 5A). This signaling pathway was necessary, but not sufficient for the full control of IL-33 expression, which demonstrated only partial correlation with levels of AKT activation (Fig. 5A).
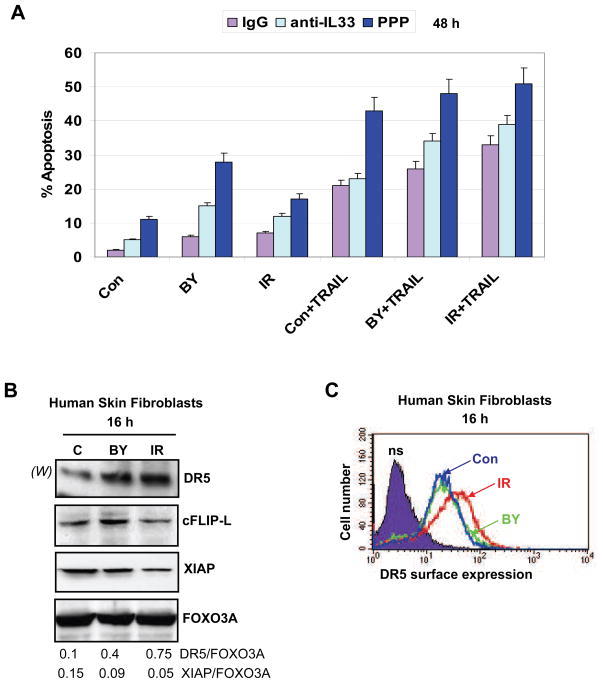
Treatment of HSF with PPP also resulted in a dose-dependent G2/M arrest (Fig. 5B), which was further accompanied by slow cell death, presumably by necrosis, based on the results of clonogenic survival analysis (Fig. 5C). However, combined treatment by PPP (0.5 μM) and α-irradiation (0.5 Gy) caused an up-regulation of apoptotic levels that were especially high in bystander cells (Fig. 6A), demonstrating a strong blockage of general survival functions in these cells. Pretreatment with the inhibitory anti-IL-33 mAb (2 μg/ml in the cell media), but not with non-specific IgG, also increased apoptotic levels in both directly irradiated and bystander cells, although this effect was less pronounced than that of PPP treatment (Fig. 6A).
Fig. 6.
Regulation of apoptosis in HSF. (A) Pretreatment with PPP or anti-IL-33 mAb increased radiation-induced or TRAIL-induced apoptosis levels in directly exposed or bystander cells. Apoptosis was induced by TRAIL (50 ng/ml) and cycloheximide (CHX, 2 μg/ml) in control, directly irradiated and bystander cells pretreated with nonspecific IgG, anti-IL-33 (2 μg/ml) mAb or PPP (0.5 μM). Apoptotic levels were determined using PI staining of DNA and FACS analysis. (B) Western blot analysis of indicated proteins from control, directly irradiated and bystander cells. FOXO3A levels were used as protein loading controls. (C) Surface expression of TRAIL-R2/DR5 was determined using immuno-staining and FACS analysis of control, α-irradiated and bystander HSF 16 h after treatment.
3.7. Suppression of IGF-1R--IL-33 increased TRAIL-mediated apoptosis
In spite of the blockade of the p53 downstream signaling (which positively regulates DR5 transcription), TRAIL-R2/DR5 expression was detected at average levels in human skin fibroblasts (Fig. 6B). Furthermore, total and surface expression of DR5 protein further increased upon irradiation (Fig. 6C). The total intracellular level of DR5 increased also in bystander cells; however, death receptor protein was not effectively translocated to the surface of bystander cells (Fig. 6C). Expression levels of the anti-apoptotic proteins cFLIP-L and XIAP were modestly higher in bystander cells than in directly irradiated cells (Fig. 6B). Exogenous TRAIL (50 ng/ml) induced apoptosis in skin fibroblasts, which was especially high in combination with ionizing radiation (0.5 Gy) (Fig. 6A). Pretreatment with inhibitory anti-IL-33 mAb that partially suppressed the nuclear NF-κB activity (Fig. 4B) additionally increased TRAIL-mediated apoptosis in the control, directly irradiated and bystander fibroblasts (Fig. 6A). As expected, pretreatment with PPP (0.5 μM) demonstrated more drastic effects on up-regulation of TRAIL-mediated apoptosis in the control, bystander and directly irradiated cells (Fig. 6A), due to its strong inhibition of the AKT survival pathway by PPP (Fig. 5A). These data further confirmed a general dependence of the resistance to TRAIL-mediated apoptosis on IGF-1R--AKT--IL-33-mediated protective functions in control, irradiated and bystander skin fibroblasts.
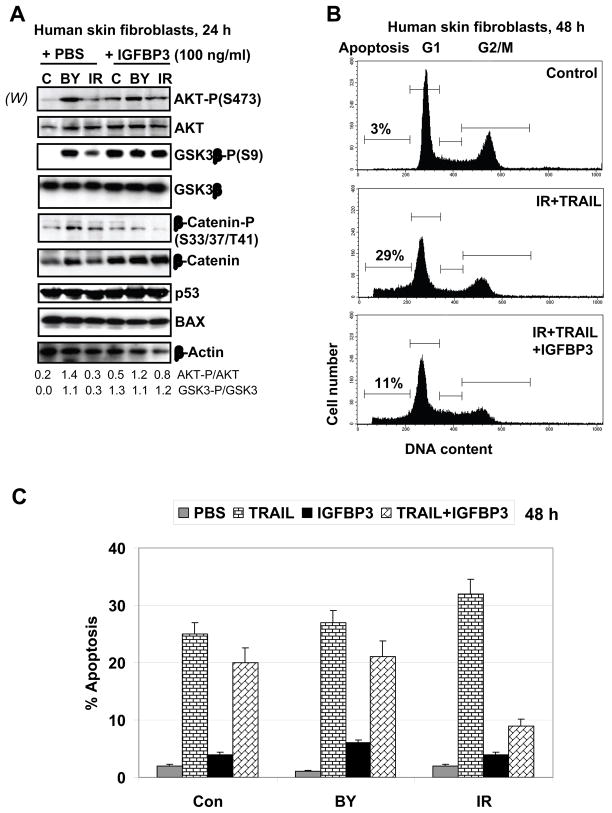
3.8. A role for exogenous IGFBP-3 in regulation of AKT activation and TRAIL-induced apoptosis in HSF
To further elucidate a role for natural physiological regulators, such as insulin-like growth factor binding protein-3, IGFBP-3 [38], in the control of IGF-1R-mediated signaling, AKT activation and TRAIL-mediated apoptosis in HSF, we used exogenous IGFBP-3 (100 ng/ml in the cell media) that reversibly binds IGF-1, presumably decreasing the current levels of activation of IGF-1R-mediated signaling, but extending the duration of its action (due to increased stability of IGF-1 in complex with IGFBP-3). Finally, after 24 h incubation in presence of IGFBP-3, we observed upregulation of the basal AKT activity, AKT-dependent phosphorylation and inactivation of GSK3β followed by decreased phosphorylation of β-catenin and the stabilization of β-catenin protein levels in the control and directly irradiated cells. No additional effects of IGFBP-3 were detected on already high levels of AKT activity in bystander fibroblasts 24 h after treatment (Fig. 7A). In general, these data suggested a preferential role of exogenous IGFBP-3 for functional extension of IGF-1R-mediated signaling in HSF. It was opposite to a negative role of the endogenous IGFBP-3 for down-regulation of AKT activity (see Fig. 4C). Levels of p53 and p53-dependent BAX were stable in this cell model before and after treatment, as expected (Fig. 7A). A physiological consequence of the exogenous IGFBP-3-induced upregulation of the AKT--β-catenin pathway was a decrease in levels of TRAIL-mediated apoptosis, especially in directly irradiated cells (Fig. 7B and 7C). It further suggested a critical role of AKT activity and, probably, AKT--β-catenin-dependent targets, in the attenuation of TRAIL-mediated apoptosis [39] in transformed human skin fibroblasts.
Fig. 7.
Effects of exogenous IGFBP-3 on cell signaling and TRAIL-mediated apoptosis in HSF. (A) Effects of IGFBP-3 (100 ng/ml in the cell media) on the AKT-dependent pathway, as well as on p53 and BAX in non-irradiated, irradiated and bystander cells were determined by Western blot analysis 24 h after treatment. (B, C) Effects of IGFBP-3 (100 ng/ml) on TRAIL-induced apoptosis in directly irradiated and bystander cells. Apoptotic levels were determined 48 h after treatment using PI staining of DNA and FACS analysis. Typical data are shown in B; Error bars represent mean ± S.D. from three independent experiments shown in C.
4. Discussion
The main cell model used in the present investigation is human skin fibroblasts (HSF) immortalized by SV40 T-antigen with suppressed p53-dependent functions. This cell model is characterized by features, such as blocked senescence and p53-mediated apoptosis that are common in both transformed cells and embryonic stem cells, as well as in induced pluripotent stem (iPS) cells [40]. Simultaneously, this cell model was suitable for studying bystander effects, due to independence of bystander response from p53 activation [1, 15, 21]. Our previous investigations, which were performed using these cells, demonstrated an important role of mitochondria in radiation-induced bystander effects through mitochondria-dependent regulation of the NF-κB--COX-2 and the NF-κB--iNOS pathways [12]. Suppression of NF-κB activation drastically decreased bystander response in HSF. Our next task was determining effects of the α-irradiation-induced gene expression (with special attention to NF-κB-mediated transcription) on the synthesis and secretion of signal transmitters that could initiate the secondary bystander signaling pathways in human fibroblasts.
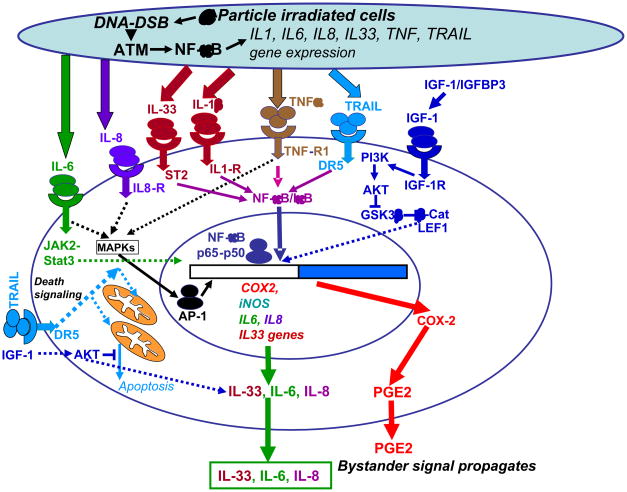
Fig. 8 summarizes our experimental data and demonstrates a unified current model of the bystander signaling pathways determined in our studies for factors transmitted through cell media. Technically, we could not detect gap junction translocation from directly irradiated to bystander cells in this type of experimental model. Since activation of the ATM-p53 and ATM-NF-κB pathways with the subsequent up-regulation of expression of the p53- and NF-κB-regulated genes is hallmark of the cell radiation response [10, 25], we determined expression levels and activation of signaling proteins from these cascades in directly α-irradiated and bystander human skin fibroblasts (HSF) at early and late time points after irradiation and monitored the development of bystander effects. We confirmed previously reported data on the general significance of ATM in bystander response [41] using our cell system. Since the downstream p53 signaling pathways were suppressed in the SV40 T-antigen immortalized HSF, the NF-κB-dependent pathway appears to be the main regulator of radiation-induced gene expression. An additional possibility for “immediate early” activation of this pathway might be linked with pre-existing IL-33 that together with IL-1α and HMGB1 play a role as “alarmins”, signaling proteins that are released by cells upon stress conditions or cell damage and may initiate both cell survival (using the IL-33-R/ST2–NF-κB loop) or cell death signaling [9, 30]. Hence, IL-33--IL-33R/ST2 via activation of the NF-κB pathway may control IL8, COX2, iNOS and, probably, its own expression, which is also dependent on AKT activation (see Fig. 5). Besides IL-33, several cytokines, such as TNFα and IL-1β, are known, to be inducers of the NF-κB signaling pathway [42]. A pronounced inhibition of NF-κB signaling and NF-κB-dependent proteins (such as COX-2) by the inhibitory anti-IL-33 mAb added to the cell media (Fig. 4B) demonstrated a role for IL-33 in the paracrine/autocrine NF-κB stimulation after irradiation of human skin fibroblasts. Upregulation of NF-κB-dependent IL1B and IL33 gene expression was also demonstrated in directly irradiated and bystander IMR-90, human lung fibroblasts (see Fig. 1E). Taken together, our results indicated that the primary ATM-NF-κB signaling pathway was tightly linked with gene expression and secretion of pro-inflammatory cytokines, IL-1β, IL-6 and IL-8, a characteristic feature of persistent DNA damage signaling [43], as well as with IL-33, a recently discovered member of the IL-1 family (see also Fig. 8 with general signaling pathways). These cytokines may initiate the secondary activation of cell signaling pathways, including the NF-κB pathway, via autocrine/paracrine mechanisms. To our knowledge, this is the first demonstration of the role of IL-33 in bystander response.
Fig. 8.
A simplified model of the signaling pathways regulating radiation-induced bystander effects. DNA damage induces ATM activation in α-particle irradiated cells. ATM further activates the NF-κB pathway, which targets gene expression of numerous cytokines and COX2. Cytokines initiate specific signaling pathways in directly irradiated and bystander cells resulting in the secondary activation of IKK-NF-κB, JAK2-STAT3 and the MAPK pathways with the subsequent induction of cytokine, COX2 and iNOS gene expression. COX-2-produced PGE2 is involved in regulation of ROS production, iNOS controls synthesis of NO. Effects of ROS and NO in the mitochondrial damage and bystander response were previously described [12]. On the other hand, NF-κB regulates expression of numerous genes controlling a general cell survival and anti-apoptotic activity. Exogenous IGF-1 via ligation of IGF-1R activates the PI3K-AKT pathway that using suppression of GSK3β stabilizes β-catenin protein levels; LEF1--β-catenin heterodimer goes to the nucleus and further controls gene expression. Furthermore, AKT controls numerous general metabolic, survival and anti-apoptotic functions in the cell. IGFBP-3 stabilizes the exogenous IGF-1 and extends its action time further activating the basal PI3K-AKT pathway.
Positive regulation of the IGF1-R--PI3K-AKT signaling pathway in bystander HSF and IMR-90 cells that was revealed in the present study appears to be one of the critical features of bystander response. An up-regulation of AKT activation in bystander cells might be directly linked with down-regulation of PTEN activity, the main negative regulator of the PI3K-AKT pathway. It is well known that PTEN is one of the main targets of reactive oxygen species (ROS), which could oxidize and inactivate this phosphatase [44] resulting in activation or up-regulation of the AKT pathway. An additional possibility for suppression of PTEN is dependent on NF-κB activation [45]. A typical downstream effect of the AKT signaling pathway, inactivation of GSK3β by AKT-dependent phosphorylation and the subsequent stabilization of β-catenin protein levels [34], was also observed in bystander cells and appeared to be important for the development of bystander response, for example via further activation of COX2 gene expression by β-catenin-LEF1 [35].
An additional level of regulation of the IGF-1R--PI3K--AKT pathway might be provided by IGFBP-3 [38]. The endogenous IGFBP-3 protein levels were decreased in bystander fibroblasts, and this was accompanied by a corresponding increase in AKT activation in these cells. Based on these observations, we further elucidated effects of exogenous IGFBP-3 (added to the culture media) on cell signaling and apoptosis. IGFBP-3 is the major member of the insulin-like growth factor binding protein family. The protein forms a ternary complex with either IGF-1 or IGF-2. Such relations, prolonging the half-life of IGFs and altering their interaction with cell surface receptors may have very different effects on acute and continuous responses induced by IGF in target cells [27, 38]. Surprisingly, exogenous IGFBP-3 (100 ng/ml) actually upregulated the AKT--GSK3β--β-catenin pathway in control and directly irradiated HSF after prolonged exposure (Fig. 7A). Subsequently, due to up-regulation of AKT activity, IGFBP-3 down-regulated TRAIL-mediated apoptosis levels in both irradiated and bystander HSF 24–48 h after treatment (Fig. 7B and C). In contrast to these observations, IGFBP-3 was previously described as an accelerator of TRAIL-mediated apoptosis in some human cancer cell lines [46]. If effects of IGFBP-3 are quite different in normal and cancer cells, it will allow us to use IGFBP-3 for attenuation of apoptotic signaling in normal cells during anti-cancer therapy. This hypothesis is under active investigation in our laboratory using human primary cell cultures and human cancer cell lines. The important task is to find differences in the IGFBP-3-dependent signaling pathways in normal and cancer cells.
Taken together, results of the present study demonstrated the early activation of NF-κB-dependent gene expression (such as IL8, MMP3 and PTGS2/COX2) in directly irradiated and bystander cells and the further modulation of critical protein levels, including IL-33, by the IGF1-R--PI3K--AKT pathway in bystander cells (see Fig. 8). IL-33 plays an important role for NF-κB activation in bystander cells, probably via autocrine/paracrine interaction with the IL-33R/ST2L, TRAF6 and IRAK complex [47]. Suppression of IL-33 expression, as well as suppression of its upstream regulator PI3K-AKT, was followed by late drastic changes in cell survival and an increase in TRAIL-mediated apoptosis in both directly irradiated and bystander human skin fibroblasts.
Acknowledgments
We would like to thank Mr. Stephen Marino and the Radiological Research Accelerator Facility (RARAF) staff for assistance with irradiations. Grant support: NIH grants: CA 49062, ES 012888-05S1; RARAF is an NIH supported Resource Center through NIBIB grant EB-002033.
Abbreviations
- ATM
Ataxia Telangiectasia Mutated protein kinase
- CHX
cycloheximide
- COX-2
cyclooxygenase-2
- DR4
death receptor-4
- DR5
death receptor-5
- EMSA
electrophoretic mobility shift assay
- ERK
extracellular signal-regulated kinase
- FACS
fluorescence-activated cell sorter
- GADD45α
Growth Arrest DNA Damage inducible protein 45
- GSK3β
Glycogen synthase kinase 3 beta
- IGF-1
insulin-like growth factor
- IGF-1R
insulin-like growth factor receptor
- IL-1β
interleukin-1β
- IL-6
interleukin-6
- IL-8
interleukin-8
- IL-33
Interleukin-33
- IL-33R
IL-33 receptor
- IκB
inhibitor of NF-κB
- IKK
inhibitor nuclear factor kappa B kinase
- JNK
Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- MFI
medium fluorescence intensity
- NF-κB
nuclear factor kappa B
- PI
propidium iodide
- PI3K
phosphoinositide 3-kinase
- PPP
picropodopyllin
- PTGS2
Prostaglandin-endoperoxide synthase 2
- ROS
reactive oxygen species
- TNFα
tumor necrosis factor alpha
- TRAIL
TNF-related apoptosis inducing ligand
- TRAIL-R
TRAIL-Receptor
- XIAP
X-linked inhibitor of apoptosis
Footnotes
Conflicts of interests
No potential conflicts of interest were disclosed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hei TK, Zhou H, Ivanov VN, Hong M, Lieberman HB, Brenner DJ, Amundson SA, Geard CR. J Pharm Pharmacol. 2008;60(8):943–950. doi: 10.1211/jpp.60.8.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagasawa H, Little JB. Cancer Res. 1992;52(22):6394–6396. [PubMed] [Google Scholar]
- 3.Little JB. Oncogene. 2003;22(45):6978–6987. doi: 10.1038/sj.onc.1206988. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Randers-Pehrson G, Waldren CA, Vannais D, Hall EJ, Hei TK. Proc Natl Acad Sci U S A. 2000;97(5):2099–2104. doi: 10.1073/pnas.030420797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H, Suzuki M, Randers-Pehrson G, Vannais D, Chen G, Trosko JE, Waldren CA, Hei TK. Proc Natl Acad Sci U S A. 2001;98(25):14410–14415. doi: 10.1073/pnas.251524798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Ivanov VN, Gillespie J, Geard CR, Amundson SA, Brenner DJ, Yu Z, Lieberman HB, Hei TK. Proc Natl Acad Sci U S A. 2005;102(41):14641–14646. doi: 10.1073/pnas.0505473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prise KM, Belyakov OV, Newman HC, Patel S, Schettino G, Folkard M, Michael BD. Radiat Prot Dosimetry. 2002;99(1–4):223–226. doi: 10.1093/oxfordjournals.rpd.a006768. [DOI] [PubMed] [Google Scholar]
- 8.Nagar S, Smith LE, Morgan WF. Cancer Res. 2003;63(2):324–328. [PubMed] [Google Scholar]
- 9.Luthi AU, Cullen SP, McNeela EA, Duriez PJ, Afonina IS, Sheridan C, Brumatti G, Taylor RC, Kersse K, Vandenabeele P, Lavelle EC, Martin SJ. Immunity. 2009;31(1):84–98. doi: 10.1016/j.immuni.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y. Trends Biochem Sci. 2006;31(7):402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Karin M, Lin A. Nat Immunol. 2002;3(3):221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Ivanov VN, Lien YC, Davidson M, Hei TK. Cancer Res. 2008;68(7):2233–2240. doi: 10.1158/0008-5472.CAN-07-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prise KM, O’Sullivan JM. Nat Rev Cancer. 2009;9(5):351–360. doi: 10.1038/nrc2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, Trent JM, Bittner ML, Meltzer PS, Fornace AJ., Jr Cancer Res. 2008;68(2):415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 15.Ghandhi SA, Yaghoubian B, Amundson SA. BMC Med Genomics. 2008;1:63. doi: 10.1186/1755-8794-1-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karin M. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Vivanco I, Sawyers CL. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 19.Sachdev D, Yee D. Mol Cancer Ther. 2007;6(1):1–12. doi: 10.1158/1535-7163.MCT-06-0080. [DOI] [PubMed] [Google Scholar]
- 20.Kakkar R, Lee RT. Nat Rev Drug Discov. 2008;7(10):827–840. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhou J, Held KD, Redmond RW, Prise KM, Liber HL. Radiat Res. 2008;169(2):197–206. doi: 10.1667/RR1189.1. [DOI] [PubMed] [Google Scholar]
- 22.Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ., Jr Cancer Res. 2004;64(18):6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov VN, Hei TK. J Biol Chem. 2004;279(21):22747–22758. doi: 10.1074/jbc.M314131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Science. 2006;311(5764):1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 26.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. Mol Cell. 2009;36(3):365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 27.Pollak M. Nat Rev Cancer. 2008;8(12):915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 28.Zhan Q, Fan S, Smith ML, Bae I, Yu K, Alamo I, Jr, O’Connor PM, Fornace AJ., Jr DNA Cell Biol. 1996;15(10):805–815. doi: 10.1089/dna.1996.15.805. [DOI] [PubMed] [Google Scholar]
- 29.Wu ZH, Miyamoto S. Embo J. 2008;27(14):1963–1973. doi: 10.1038/emboj.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. Proc Natl Acad Sci U S A. 2007;104(1):282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manetti M, Ibba-Manneschi L, Liakouli V, Guiducci S, Milia AF, Benelli G, Marrelli A, Conforti ML, Romano E, Giacomelli R, Matucci-Cerinic M, Cipriani P. Ann Rheum Dis. 2009 doi: 10.1136/ard.2009.119321. [DOI] [PubMed] [Google Scholar]
- 32.Mossner R, Schon MP, Reich K. Clin Dermatol. 2008;26(5):486–502. doi: 10.1016/j.clindermatol.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 33.Perlis C, Herlyn M. Oncologist. 2004;9(2):182–187. doi: 10.1634/theoncologist.9-2-182. [DOI] [PubMed] [Google Scholar]
- 34.Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Proc Natl Acad Sci U S A. 1998;95(19):11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Wen S, Bunnett NW, Leduc R, Hollenberg MD, MacNaughton WK. J Biol Chem. 2008;283(2):809–815. doi: 10.1074/jbc.M703021200. [DOI] [PubMed] [Google Scholar]
- 36.Girnita A, Girnita L, del Prete F, Bartolazzi A, Larsson O, Axelson M. Cancer Res. 2004;64(1):236–242. doi: 10.1158/0008-5472.can-03-2522. [DOI] [PubMed] [Google Scholar]
- 37.Krasilnikov M, Ivanov VN, Dong J, Ronai Z. Oncogene. 2003;22(26):4092–4101. doi: 10.1038/sj.onc.1206598. [DOI] [PubMed] [Google Scholar]
- 38.Hwa V, Oh Y, Rosenfeld RG. Endocr Rev. 1999;20(6):761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 39.De Toni EN, Thieme SE, Herbst A, Behrens A, Stieber P, Jung A, Blum H, Goke B, Kolligs FT. Clin Cancer Res. 2008;14(15):4713–4718. doi: 10.1158/1078-0432.CCR-07-5019. [DOI] [PubMed] [Google Scholar]
- 40.Krizhanovsky V, Lowe SW. Nature. 2009 doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burdak-Rothkamm S, Rothkamm K, Prise KM. Cancer Res. 2008;68(17):7059–7065. doi: 10.1158/0008-5472.CAN-08-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karin M, Yamamoto Y, Wang QM. Nat Rev Drug Discov. 2004;3(1):17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 43.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Nat Cell Biol. 2009;11(8):973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonks NK. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 45.Vasudevan KM, Gurumurthy S, Rangnekar VM. Mol Cell Biol. 2004;24(3):1007–1021. doi: 10.1128/MCB.24.3.1007-1021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams AC, Smartt H, AMHZ, Macfarlane M, Paraskeva C, Collard TJ. Cell Death Differ. 2007;14(1):137–145. doi: 10.1038/sj.cdd.4401919. [DOI] [PubMed] [Google Scholar]
- 47.Funakoshi-Tago M, Tago K, Hayakawa M, Tominaga S, Ohshio T, Sonoda Y, Kasahara T. Cell Signal. 2008;20(9):1679–1686. doi: 10.1016/j.cellsig.2008.05.013. [DOI] [PubMed] [Google Scholar]