Abstract
Many biological processes rely on the interaction of proteins with multiple DNA sites separated by thousands of base pairs. These long-range communication events can be driven by both the thermal motions of proteins and DNA, and directional protein motions that are rectified by ATP hydrolysis. The present review describes conflicting experiments that have sought to explain how the ATP-dependent Type III restriction–modification enzymes can cut DNA with two sites in an inverted repeat, but not DNA with two sites in direct repeat. We suggest that an ATPase activity may not automatically indicate a DNA translocase, but can alternatively indicate a molecular switch that triggers communication by thermally driven DNA sliding. The generality of this mechanism to other ATP-dependent communication processes such as mismatch repair is also discussed.
Keywords: ATPase, DNA sliding, helicase, mismatch repair, molecular switch, restriction–modification
Abbreviations: AFM, atomic force microscopy; MMR, mismatch repair; RM, restriction–modification
Introduction
The RM (restriction–modification) enzymes have an important biological role in protecting bacteria from infection by bacteriophages [1]. All RM systems include an endonuclease that cleaves DNA after binding a specific unmodified recognition sequence [2]. However, those systems that bind and cleave just one site in isolation may be in the minority: all of the Type I and Type III enzymes [3], as well as an ever growing list of Type II enzymes [4–6], require interaction with at least two DNA sequences before cleavage occurs. These multisite interactions are believed to have evolved to provide additional protection against accidental cleavage of the host DNA [7].
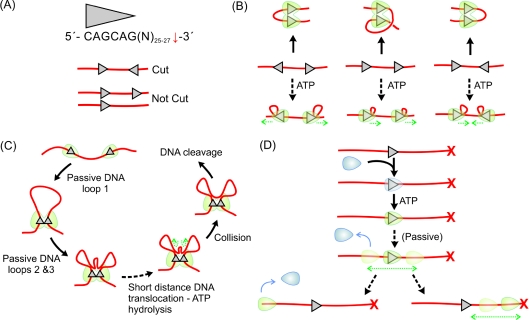
The Type III RM enzymes comprise two subunits, Res and Mod [2], that form a heterotetrameric Res2Mod2 complex [8,9]. They recognize short asymmetric DNA sequences with cleavage occurring downstream (Figure 1A) [2]. However, nuclease activity requires the presence of two sites on one DNA molecule [10]. The classical dogma states that cleavage only occurs when the two sites are in a specific inverted repeat, such that the sites are ‘head-to-head’ (Figure 1A) [3,11,12]. Only one of the two sites is then cut. DNA with sites in tail-to-tail or direct repeat, or with single sites, are reportedly not cut [11–13]. This suggested a directional communication mechanism [11,12]. Simple three-dimensional DNA looping cannot account for the site preference, as random motions can bring the sites together in the same way regardless of their relative orientation (Figure 1B) [14]. Since cleavage is independent of DNA topology, we can also rule out more complex DNA looping [10], as seen with supercoiling-dependent site-specific recombinases [15]. How then do the Type III enzymes not only communicate between two distant sites, but also recognize their relative orientation?
Figure 1. Models for long-range communications by Type III RM enzymes.
(A) Recognition sequence for the EcoP15I enzyme [2], showing the downstream non-specific cleavage site and outcome of an endonuclease reaction on different DNA substrates (red lines) [11,12]. (B) DNA looping and DNA-loop translocation models for Type III enzymes (in green) [3,12]. Only directional loop translocation can distinguish the head-to-head substrate. (C) Expanded Type III model proposed by Crampton et al. [29]. (D) DNA-sliding model proposed by Ramanathan et al. [25], and the role of DNA ends. The capped end (e.g. biotin plus streptavidin) is shown with a cross.
The potential role of the helicase domains in the Type III RM enzymes
Both the Type I and III enzymes contain domains with motifs and structures that can be classified as Superfamily 2 helicases [16–19]. Despite the nomenclature, neither type of RM enzyme unwinds DNA [20,21]. For the Type I enzymes, the mechanistic role of the helicase is now well established ([20] and references therein, [22–24]): the helicase domain uses ATP to translocate along intact double-stranded DNA away from the recognition sequence; the motor moves on the 3′→5′ strand, using the complementary strand to maintain processivity; approx. one ATP molecule is hydrolysed per base pair; throughout the reaction, the complex remains bound to the recognition site, so that supercoiled DNA loops are extruded. It was proposed that the Type III enzymes also use a similar loop translocation mechanism in which the helicase domains (in Res) translocates unidirectionally from the site while the DNA-recognition domain (in Mod) remains at the site [3,12]. Directional motion helps to explain the bias in cleavage: only when two enzymes converge will they collide in the correct orientation to form an active endonuclease domain (Figure 1B).
The evidence for DNA looping and translocation by Type III RM enzymes
A number of testable predictions can be made based on the loop translocation model (Figure 1B): (i) DNA cleavage will only occur when the sites are on the same DNA (in cis); (ii) one-dimensional DNA translocation must occur; (iii) DNA looping (active and possible passive) must occur; and (iv) translocation must be coupled to extensive ATPase activity. Whereas each of these predictions can be readily demonstrated for the Type I enzymes ([20] and references therein, [22–24]), the Type III enzymes have proven to be more enigmatic. Although it has been shown that the recognition sites must be in cis [10], a range of ensemble assays developed to measure DNA translocation and looping have failed to provide evidence for either [8,10,25]. ATP hydrolysis can be measured, but the activity is more than three orders of magnitude lower than that for the Type I enzymes [12,24–27].
To observe directly the Type III enzymes in action, a series of AFM (atomic force microscopy) studies measured the protein–DNA interactions on mica surfaces [28–30]. Both ATP-dependent and -independent DNA looping was observed. Moreover, it was possible to directly monitor in real time the formation of an apparent translocated DNA loop [30]. On the basis of these studies, a modified model was proposed in which passive DNA looping brings the motor domains into close proximity, followed by a final translocation stage that brings the motors into direct contact (Figure 1C) [29]. This accounts for the low ATPase activity as the final distance translocated is relatively short. However, closer examination of the experiments and model highlights a number of shortcomings. First, if the motor does not start immediately adjacent to its site, it will be just as likely for the passive looping events to deliver the motor on to an adjacent DNA molecule as on to the same DNA chain [14]. However, the reaction is constrained to sites in cis [10]. Moreover, when the intersite spacing is longer than the DNA persistence length, simple DNA looping cannot distinguish between different site orientations [14]. Secondly, the AFM studies did not measure both DNA communication and cleavage. Therefore it is not possible to state that the events observed are necessary steps before DNA cleavage. Thirdly, DNA attachment to a charged mica surface may have formed polynucleotide conformations not seen in free solution.
To address the Type III mechanism in a more holistic manner, an alternative multiplex magnetic tweezers assay was developed to follow both DNA cleavage and looping simultaneously as a function of tensional and torsional force [25]. DNA cleavage was shown to be force-independent and was never preceded by active DNA looping. Measured cleavage rates were the same as those in bulk solution, with DNA topology having no effect. In a modified experiment is was also possible to show that DNA cleavage was never preceded by passive DNA looping. This study also highlighted an important observation: that cleavage of linear DNA is very inefficient unless the ends are ‘capped’ with a bulky protein moiety, such as streptavidin. Circular DNA in solution was cleaved as efficiently as capped DNA [13]. It was shown subsequently that Type III enzymes could load on to linear DNA via the free ends [31]. The loop-translocation models (Figures 1B and 1C) are inconsistent will all these observations. An alternative model has also been suggested where translocation occurs without DNA looping [32], but this is not truly consistent with the ATP coupling of the Type III enzymes [12,25–27] or with the observed end effects [25,31].
DNA sliding as an alternative mechanism
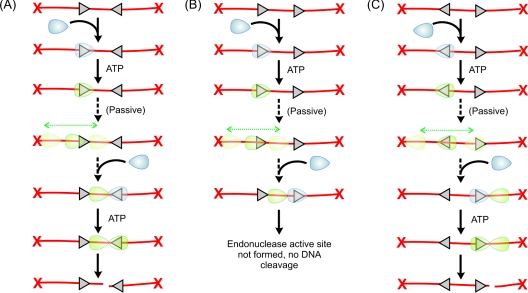
What alternative mechanism is possible given that looping and extensive translocation are unlikely to be on-pathway to DNA cleavage? A plausible alternative is ‘DNA sliding’ [33,34], in which a protein remains bound to DNA probably via weak electrostatic interactions and where the activation energy to move to an adjacent site is less than ~ 2·kBT (tighter binding would make sliding too slow) [35,36]. Thermal activation then leads to a one-dimensional random walk on DNA, in which both ‘backwards’ and ‘forwards’ steps occur with equal probability (assuming a uniform energy landscape) [33]. In this model (Figure 1D), the role of the helicase domain is to produce a conformational switch from a DNA-recognition mode to a DNA-sliding mode. ATP is not used for communication directly, so accounting for the low ATP consumption. Since the enzyme does not release the DNA, sliding retains the original binding orientation. Cleavage of a head-to-head DNA molecule can then occur if the enzyme eventually encounters an enzyme at the second target site (Figure 2A). On linear DNA, the directionally unbiased motion leads to the enzyme occasionally approaching a DNA end, where it can dissociate, making cleavage less efficient (Figure 1D). Where the end is capped by a bulky moiety, the sliding protein will be reflected, making cleavage as efficient as on circular DNA. The model in Figure 2(A) describes a dynamic binding, sliding and release scheme where not every sliding event would lead to cleavage. It also requires that the static motor can hydrolyse ATP, consistent with experimental observations [8].
Figure 2. The outcome of DNA sliding by Type III RM enzymes on different DNA substrates.
(A) Head-to-head DNA. (B) Head-to-tail DNA. (C) Tail-to-tail DNA.
An important mechanistic feature of the Type III enzymes is that cleavage only occurs at one of the two interacting sites, with the other site remaining intact [10,13]. The sliding model accounts for this as one protein moves away from its site and collides with a static protein bound at a distant site. Sliding therefore needs to be rapid compared with the site-binding/ATPase rates. Measurements of sliding by various DNA-binding proteins and enzymes indicate rates of 107–109 steps·min−1 (e.g. [36–38]). Simulations of kinetic models using this range of values and the Type III ATPase rates demonstrate that the sliding scheme can readily explain the observed cleavage [25]. Moreover, the relative distance of a pair of sites to an uncapped end also alters the relative proportion of cleavage at each site in a manner consistent with sliding and end-dependent dissociation (K. van Aelst, personal communication).
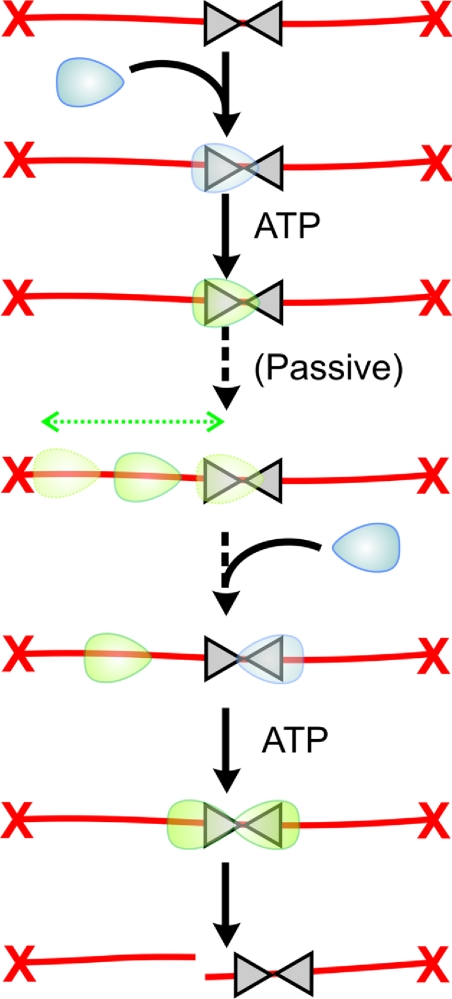
On DNA with two directly repeated sites, the enzymes load on to the DNA in the same direction (Figure 2B). Collisions between mobile and static enzymes cannot therefore produce DNA cleavage, as the correct interactions never occur. However, on DNA with sites in tail-to-tail repeat, the sliding model predicts that cleavage could occur if the first enzyme slides past the second site before the second enzyme binds (Figure 2C). This prediction has been confirmed using capped linear DNA substrates (K. van Aelst, personal communication). This observation was missed in earlier studies as they relied on uncapped linear DNA substrates where cleavage is very inefficient. The ability of an enzyme to distinguish both head-to-head and tail-to-tail substrates from head-to-tail substrates in a manner independent of DNA topology is unusual and cannot simply be explained by unidirectional translocation schemes. Sliding can also explain the previously mysterious observation that Type III enzymes can cut DNA even when the inverted sites are immediately adjacent, conditions where two enzymes cannot bind simultaneously (Figure 3) [39].
Figure 3. How DNA sliding can explain the cleavage of DNA with two adjacent Type III recognition sites.
Model to explain the observation of Mücke et al. [39] that DNA cleavage can even occur when two head-to-head sites are immediately adjacent, where simultaneous binding of two enzymes is extremely unlikely.
A characteristic feature of a random walk is the quadratic dependence between distance moved and the number of steps taken [33]; e.g. doubling the intersite distance increases the communication time 4-fold. However, without a direct measure for protein motion, we can only measure DNA cleavage [13,25,27], and, under these conditions, there is no apparent relationship between rate and intersite distance. This is most likely because the cleavage rate is significantly lower than the communication rate. However, a relationship has been observed by varying the distance to an uncapped DNA end [25]: the nearer the uncapped end, the slower the cleavage. The next step is the development of fluorescent single-molecule assays [36–38], to allow direct observation of the communication and cleavage processes so that we can distinguish the bi-directional motion characteristic of a random walk [33].
The role of ATP in MMR (mismatch repair)
We suggest that the role of ATP-dependent DNA sliding may be more general than just the RM enzymes. For example, very similar arguments about long-range communications have taken place in the DNA MMR field, where translocation, looping and sliding models have all been discussed. Similarly, there are ATPase activities of two key MMR proteins, MutS and MutL, that are substantially slower than expected of bona fide motors [40]. MMR is a multistep pathway comprising mismatch recognition, strand discrimination, strand removal and DNA resynthesis [40]. It involves the communication between a mismatch recognized by MutS or related enzymes, and a strand-discrimination signal which can be pre-existing nick or, in the case of Escherichia coli, a hemimethylated GATC sequence that has to be nicked by the endonuclease MutH [40]. This nick can be 1000 bp or more either 5′ or 3′ to the mismatch, making the system bi-directional. Depending on this orientation, the MMR system executes strand removal towards the mismatch either in 5′→3′ or 3′→5′ direction.
Increasing experimental evidence supports the ATP-binding induced sliding clamp model involving a mobile MutS–MutL complex [41,42], at least for the communication of mismatch recognition and strand discrimination. First, as noted for the Type III RM enzymes, the efficiency of mismatch-provoked DNA incision depends on the relative distance of the GATC sites to the mismatch and a DNA end [40,43] and is increased when the DNA ends are capped (C. Jung, personal communication). Secondly, protein roadblocks between the mismatch and the GATC site block the mismatch-provoked activation of the MutH [44]. Furthermore, mismatch-provoked activation of MutH is possible even if the binding sites for MutS and MutH overlap, conditions that rule out simultaneous binding of MutS and MutH to their binding sites [45].
Although these data exclude a stationary MutS that remains bound at the mismatch during the reaction, they do not exclude a function for MutS-induced DNA loops that had been observed using electron microscopy [46]. DNA looping on top of sliding clamp formation may explain why roadblocks do inhibit the strand discrimination step, i.e. activation of MutH in the E. coli system, but not the activation of the excision step in the human system [47]. Similar experiments as described for the Type III RM enzymes, such as multiplex magnetic tweezers to follow DNA looping and unwinding/excision, are highly warranted to unravel the mechanistic details of this multifaceted system.
How have systems adapted to make DNA sliding efficient?
A critical assessment of sliding poses an important question: can a thermally activated random walk be an efficient process for motion on DNA? Because of the large number of steps required [33], sliding could be just too slow. However, the overall time to move, say, 1000 bp by sliding is similar to the rate for motor proteins which would consume thousands of extra ATP molecules. Even if sliding is rapid, the off-rate must be correspondingly low so that the enzyme remains bound long enough to move an appreciable distance [34]. The most straightforward way to achieve this is to encircle the DNA, as seen in many sliding clamp processivity factors [48]. But this solution is not universal, as some processivity factors are not toroidal [38]. An intriguing suggestion is that proteins that rely on sliding may have evolved a ‘catalytic’ DNA-binding surface that reduces the activation barrier for thermally driven movement to adjacent DNA sites, while retaining a low dissociation rate [36]. The question above can then be rephrased: what mechanisms do proteins use to make sliding more efficient? More structural data are required, since, with some notable exceptions (e.g. [49]), there are few structures of enzymes bound to non-specific double-stranded DNA.
The role of helicases as switches rather than translocases is emerging as an important theme, particularly among the RNA helicases [50]. Many enzymes deemed to be ‘DNA helicases’ on the basis of their motifs show extremely poor unwinding activity in vitro (often requiring a substantial molar excesses of enzyme over DNA substrate to elicit any activity). Alternative roles such as switches need to be considered. For the Type III RM enzymes, we await a crystal structure to fully understand how the helicase domain can produce movement on DNA in such an energy-efficient manner.
Acknowledgements
We thank Kara van Aelst and Caroline Jung for communicating unpublished data. We also thank members of our laboratories past and present for their contributions.
Funding
Our work is funded by The Wellcome Trust [grant numbers 067439 and 084086 (to M.D.S.)], Die Deutsche Forschungsgemeineschaft [grant numbers SE1646/1-1 and SE1646/2-1 (to R.S.), and FR1495/4-1 (to P.F.)] and the European Union (Marie Curie Research Training Network ‘DNA Enzymes’ to M.D.S. and P.F.).
References
- 1.King G., Murray N.E. Restriction enzymes in cells, not eppendorfs. Trends Microbiol. 1994;2:465–469. doi: 10.1016/0966-842x(94)90649-1. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE: enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dryden D.T., Murray N.E., Rao D.N. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bath A.J., Milsom S.E., Gormley N.A., Halford S.E. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 5.Gowers D.M., Bellamy S.R., Halford S.E. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan R.D., Dwinell E.A., Bhatia T.K., Lang E.M., Luyten Y.A. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009;37:5208–5221. doi: 10.1093/nar/gkp534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halford S.E., Gowers D.M., Sessions R.B. Two are better than one. Nat. Struct. Biol. 2000;7:705–707. doi: 10.1038/78911. [DOI] [PubMed] [Google Scholar]
- 8.Janscak P., Sandmeier U., Szczelkun M.D., Bickle T.A. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 9.Sears A., Szczelkun M.D. Subunit assembly modulates the activities of the Type III restriction-modification enzyme PstII in vitro. Nucleic Acids Res. 2005;33:4788–4796. doi: 10.1093/nar/gki788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peakman L.J., Szczelkun M.D. DNA communications by Type III restriction endonucleases: confirmation of 1D translocation over 3D looping. Nucleic Acids Res. 2004;32:4166–4174. doi: 10.1093/nar/gkh762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meisel A., Bickle T.A., Krüger D.H., Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 12.Meisel A., Mackeldanz P., Bickle T.A., Kruger D.H., Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peakman L.J., Antognozzi M., Bickle T.A., Janscak P., Szczelkun M.D. S-Adenosyl methionine prevents promiscuous DNA cleavage by the EcoP1I type III restriction enzyme. J. Mol. Biol. 2003;333:321–335. doi: 10.1016/j.jmb.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 14.Halford S.E, Welsh A.J., Szczelkun M.D. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 15.Stark W.M., Boocock M.R. Topological selectivity in site-specific recombination. In: Sherratt D.J., editor. Mobile Genetic Elements. Oxford: Oxford University Press; 1995. pp. 101–129. [Google Scholar]
- 16.Gorbalenya A.E., Koonin E.V. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991;291:277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- 17.McClelland S.E., Szczelkun M.D. Molecular motors that process DNA. In: Pingoud A., editor. Restriction Enzymes, Nucleic Acids and Molecular Biology, vol. 14. Berlin: Springer Verlag; 2004. pp. 111–135. [Google Scholar]
- 18.Uyen N.T., Park S.Y., Choi J.W., Lee H.J., Nishi K., Kim J.S. The fragment structure of a putative HsdR subunit of a type I restriction enzyme from Vibrio vulnificus YJ016: implications for DNA restriction and translocation activity. Nucleic Acids Res. 2009;37:6960–6969. doi: 10.1093/nar/gkp603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapkouski M., Panjikar S., Janscak P., Smatanova I.K., Carey J., Ettrich R., Csefalvay E. Structure of the motor subunit of type I restriction–modification complex EcoR124I. Nat. Struct. Mol. Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 20.Murray N.E. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha S., Rao D.N. Mutations in the Res subunit of the EcoPI restriction enzyme that affect ATP-dependent reactions. J. Mol. Biol. 1997;269:342–354. doi: 10.1006/jmbi.1997.1045. [DOI] [PubMed] [Google Scholar]
- 22.Seidel R., van Noort J., van der Scheer C., Bloom J.G., Dekker N.H., Dutta C.F., Blundell A., Robinson T., Firman K., Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 23.Stanley L.K., Seidel R., van der Scheer C., Dekker N.H., Szczelkun M.D., Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidel R., Bloom J.G., Dekker C., Szczelkun M.D. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanathan S.P., van Aelst K., Sears A., Peakman L.J., Diffin F.M., Szczelkun M.D., Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha S., Rao D.N. ATP hydrolysis is required for DNA cleavage by EcoPI restriction enzyme. J. Mol. Biol. 1995;247:559–567. doi: 10.1016/s0022-2836(05)80137-8. [DOI] [PubMed] [Google Scholar]
- 27.Sears A., Peakman L.J., Wilson G.G., Szczelkun M.D. Characterization of the Type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 2005;33:4775–4787. doi: 10.1093/nar/gki787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich S., Gössl I., Reuter M., Rabe J.P., Krüger D.H. Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I. J. Mol. Biol. 2004;341:337–343. doi: 10.1016/j.jmb.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 29.Crampton N., Roes S., Dryden D.T., Rao D.N., Edwardson J.M., Henderson R.M. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crampton N., Yokokawa M., Dryden D.T., Edwardson J.M., Rao D.N., Takeyasu K., Yoshimura S.H., Henderson R.M. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peakman L.J., Szczelkun M.D. S-adenosyl homocysteine and DNA ends stimulate promiscuous nuclease activities in the Type III restriction endonuclease EcoPI. Nucleic Acids Res. 2009;37:3934–3945. doi: 10.1093/nar/gkp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavendra N.K., Rao D.N. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berg H.C. Princeton: Princeton University Press; 1993. Random Walks in Biology. [Google Scholar]
- 34.Halford S.E. An end to 40 years of mistakes in DNA–protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 35.Slutsky M., Mirny L.A. Kinetics of protein–DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blainey P.C., van Oijen A.M., Banerjee A., Verdine G.L., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorman J., Chowdhury A., Surtees J.A., Shimada J., Reichman D.R., Alani E., Greene E.C. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2–Msh6. Mol. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komazin-Meredith G., Mirchev R., Golan D.E., van Oijen A.M., Coen D.M. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10721–10726. doi: 10.1073/pnas.0802676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mücke M., Reich S., Möncke-Buchner E., Reuter M., Krüger D.H. DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites. J. Mol. Biol. 2001;312:687–698. doi: 10.1006/jmbi.2001.4998. [DOI] [PubMed] [Google Scholar]
- 40.Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 41.Kolodner R.D., Mendillo M.L., Putnam C.D. Coupling distant sites in DNA during DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gradia S., Acharya S., Fishel R. The human mismatch recognition complex hMSH2–hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 43.Smith J., Modrich P. Mutation detection with MutH, MutL, and MutS mismatch repair proteins. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4374–4379. doi: 10.1073/pnas.93.9.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pluciennik A., Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinze R.J., Giron-Monzon L., Solovyova A., Elliot S.L., Geisler S., Cupples C.G., Connolly B.A., Friedhoff P. Physical and functional interactions between Escherichia coli MutL and the Vsr repair endonuclease. Nucleic Acids Res. 2009;37:4453–4463. doi: 10.1093/nar/gkp380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen D.J., Makhov A., Grilley M., Taylor J., Thresher R., Modrich P., Griffith J.D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Hays J.B. Signaling from DNA mispairs to mismatch-repair excision sites despite intervening blockades. EMBO J. 2004;23:2126–2133. doi: 10.1038/sj.emboj.7600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom L.B. Loading clamps for DNA replication and repair. DNA Repair. 2009;8:570–578. doi: 10.1016/j.dnarep.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viadiu H., Aggarwal A.K. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 50.Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]